Abstract
Background
Mutations in the androgen receptor (AR) ligand-binding domain (LBD), such as F877L and T878A, have been associated with resistance to next-generation AR-directed therapies. ARN-509-001 was a phase I/II study that evaluated apalutamide activity in castration-resistant prostate cancer (CRPC). Here, we evaluated the type and frequency of 11 relevant AR-LBD mutations in apalutamide-treated CRPC patients.
Patients and methods
Blood samples from men with nonmetastatic CRPC (nmCRPC) and metastatic CRPC (mCRPC) pre- or post-abiraterone acetate and prednisone (AAP) treatment (≥6 months’ exposure) were evaluated at baseline and disease progression in trial ARN-509-001. Mutations were detected in circulating tumor DNA using a digital polymerase chain reaction-based method known as BEAMing (beads, emulsification, amplification and magnetics) (Sysmex Inostics’ GmbH).
Results
Of the 97 total patients, 51 had nmCRPC, 25 had AAP-naïve mCRPC, and 21 had post-AAP mCRPC. Ninety-three were assessable for the mutation analysis at baseline and 82 of the 93 at progression. The overall frequency of detected AR mutations at baseline was 7/93 (7.5%) and at progression was 6/82 (7.3%). Three of the 82 (3.7%) mCRPC patients (2 AAP-naïve and 1 post-AAP) acquired AR F877L during apalutamide treatment. At baseline, 3 of the 93 (3.2%) post-AAP patients had detectable AR T878A, which was lost after apalutamide treatment in 1 patient who continued apalutamide treatment for 12 months.
Conclusions
The overall frequency of detected mutations at baseline (7.5%) and progression (7.3%) using the sensitive BEAMing assay was low, suggesting that, based on this assay, AR-LBD mutations such as F877L and T878A are not common contributors to de novo or acquired resistance to apalutamide.
ClinicalTrials.gov identifier
Keywords: apalutamide, ARN-509, castration-resistant prostate cancer, androgen receptor, mutations
Introduction
Castration-resistant prostate cancer (CRPC) is the lethal form of the disease that carries a poor prognosis [1, 2]. Molecular profiling studies have shown that androgen receptor (AR) overexpression is associated with resistance to conventional antiandrogens, and preclinical experiments confirm that AR overexpression contributes to CRPC progression [3]. This insight and the demonstration that androgen ligands persist in CRPC patient tumors despite medical castration led to the eventual clinical development of novel androgen-AR axis–signaling inhibitors, including most recently, apalutamide [4, 5].
Although the majority of patients respond to these next-generation AR-targeted agents, the durability of response is limited [6], and only a subset benefit from sequential AR-directed therapies [7–10]. Several potential mechanisms have been proposed to explain resistance to these agents, including DNA alterations in the AR gene, the production of AR mRNA splice variants such as AR-V7 [11, 12], increased mitogen-activated protein kinase signaling and alternative signaling pathways [3]. Point mutations in the AR ligand-binding domain (AR-LBD) have also been associated with resistance to AR-targeted therapy [13–20], including AR F877L and AR T878A (formerly AR F876L and AR T877A) [21], which have been associated with resistance to apalutamide, enzalutamide or the androgen biosynthesis inhibitor abiraterone acetate (hereafter abiraterone), respectively. Additionally, although all AR mutations alter the specificity of ligand binding, there are 2 types of AR mutations, those that convert AR antagonists to agonists (e.g. F877L, W742L/C) and those that result in broadened ligand specificity and a ‘promiscuous AR’ that can bind to other endogenous steroids [17].
To evaluate the relationship of AR-LBD mutations and resistance to next-generation antiandrogens, Balbas et al. [14] screened for human prostate cancer cell populations with persistent AR transcriptional activity, proliferative ability and tumorigenic potential in the presence of enzalutamide using an AR-regulated enhanced green fluorescent protein reporter and a randomly mutagenized AR library. These investigators identified a novel mutation, AR F877L, that spontaneously arose in cells with prolonged treatment with enzalutamide and apalutamide [14]. Joseph et al. [18] and Korpal et al. [19] confirmed these findings with AR F877L-expressing prostate cancer cell lines in castrated mice. Neither enzalutamide nor apalutamide inhibited tumor growth in the AR F877L-expressing tumors, but both drugs exhibited robust antitumor activity in wild-type AR-expressing tumors [18, 19]. Based on these preclinical data, Joseph et al. [18] used the BEAMing (beads, emulsification, amplification and magnetics) technique to evaluate serial circulating tumor DNA (ctDNA) samples from 29 patients with metastatic CRPC (mCRPC) treated on a phase I study of apalutamide. As expected, AR F877L was not found in pretreatment samples but the mutation was detected in 3 (10%) post-apalutamide patients with a rising prostate-specific antigen (PSA), suggesting a possible mechanism for acquired treatment resistance [18]. There is biochemical evidence based on engineered cell line models that enzalutamide is only a weak partial agonist of AR F877L, but a strong partial agonist of the double mutant AR F877L/T878A [22, 23].
The AR T878A mutation has been associated with resistance to abiraterone in a xenograft model [15], which was subsequently detected in metastatic tumor biopsies from CRPC patients relapsing on the CYP17A1 inhibitors abiraterone or ketoconazole [16]. In a recent study, men harboring the AR T878A mutation in ctDNA showed inferior PSA response rates and shorter overall survival with abiraterone compared with men with a wild-type AR gene [24]. These studies and others underscore the need to further investigate predictive biomarkers for resistance to AR-targeted therapies.
The aim of the present study was to evaluate the frequency of F877L, T878A and other AR-LBD mutations at baseline and disease progression in nonmetastatic (nm) and mCRPC patients who were abiraterone plus prednisone naïve (AAP-naïve) or who had previously received abiraterone plus prednisone (post-AAP) [25, 26]. Eleven somatic AR-LBD mutations were evaluated at baseline and disease progression in ctDNA using BEAMing, a digital polymerase chain reaction (PCR)-based method (supplementary Table S1, available at Annals of Oncology online).
Methods
Patients with nmCRPC and mCRPC were enrolled in a phase II trial of apalutamide (ARN-509-001) [25, 26]. All patients had pathologically confirmed prostate cancer, had been medically or surgically castrated (serum testosterone of ≤50 ng/dl) and had an Eastern Cooperative Oncology Group performance status of 0–1. Patients were excluded if they had received prior enzalutamide, ketoconazole or chemotherapy for mCRPC or had distant metastases with nmCRPC. Patients in the mCRPC cohort had disease progression based on either PSA progression (≥2 ng/ml within 2 weeks of study enrollment) or radiographic progression (≥2 new bone lesions, Prostate Cancer Working Group 2 criteria) [27] and had no prior exposure to abiraterone plus prednisone (i.e. AAP-naïve cohort) or received ≥6 months of abiraterone plus prednisone treatment before disease progression (i.e. post-AAP cohort).
Plasma samples were sent to Sysmex Inostics’ GmbH (Hamburg, Germany) analytical facility on dry ice; samples were stored at –70 °C until they were analyzed. Samples were thawed at room temperature for 15–30 min before DNA preparation. BEAMing (Sysmex Inostics' GmbH), which combines emulsion PCR using magnetic beads coated with gene-specific primers to detect and quantify known mutations in ctDNA [28], was used to detect 11 possible somatic AR-LBD mutations in the patient samples (i.e. 11 of >30 known AR-LBD mutations available to assay via BEAMing at the time of the analysis) (supplementary Table S1, available at Annals of Oncology online). These 11 mutations affect 6 key amino acid residues (V716, W742, H875, F877, T878 and M896). Detection, quantification and validation are discussed in the supplementary methods, available at Annals of Oncology online).
Results
Baseline data (N = 97) were similar among cohorts, with the exception of percentage of black and Asian patients, baseline PSA and Gleason score (supplementary Table S2, available at Annals of Oncology online). Ninety-three of 97 (96%) patients in the phase II study were assessable for the AR mutation analysis at baseline (nmCRPC, n = 50; AAP-naïve mCRPC, n = 24; post-AAP mCRPC, n = 19); 82 of the 93 (88%) patients assessable at baseline were assessable for the mutation analysis at progression (nmCRPC, n = 47; AAP-naïve mCRPC, n = 20; post-AAP mCRPC, n = 15). The median (range) treatment duration was 26.9 (0.03–37.84) months for the nmCRPC cohort, 20.97 (2.63–37.54) months for the cohort with AAP-naïve mCRPC and 4.87 (1.28–23.2) months for those with post-AAP mCRPC. A low frequency of AR mutations was detected in the overall patient population (Table 1). AR F877L and AR T878A mutations were found in more than one patient, and these are the focus of this report.
Table 1.
Summary of overall androgen receptor mutation status
| AR point mutationb | Associated drug resistance | BaselineaN = 93 | Progression ‘acquired’ N = 82 | Total baseline and progression ‘acquired’ N = 93 |
|---|---|---|---|---|
| n (%) | n (%) | n (%) | ||
| F877Lc | Enzalutamide [14, 18, 19] | 2 (2.2) | 3 (3.7) | 5 (5.4) |
| Apalutamide [14, 18] | ||||
| T878Ad | Abiraterone [15, 16] | 3 (3.2) | 1 (1.2) | 4 (4.3) |
| W742Ce | Bicalutamide [17] | 1 (1.1) | 0 | 1 (1.1) |
| V716T | Flutamide [17] | 0 | 1 (1.2) | 1 (1.1) |
| H875Y | Flutamide [20] | 1 (1.1) | 1 (1.2) | 2 (2.2) |
| Abiraterone [13] |
Four nmCRPC patients were excluded from the efficacy analysis as they were later determined to have metastases on their screening scans.
AR M896T and AR M896V were not detected.
Three possible nucleotide changes (T → C, C → A and C → G).
Two possible amino acid changes (T → A and T → S).
Two possible amino acid changes (W → C and W → L).
AR, androgen receptor.
A R F877L
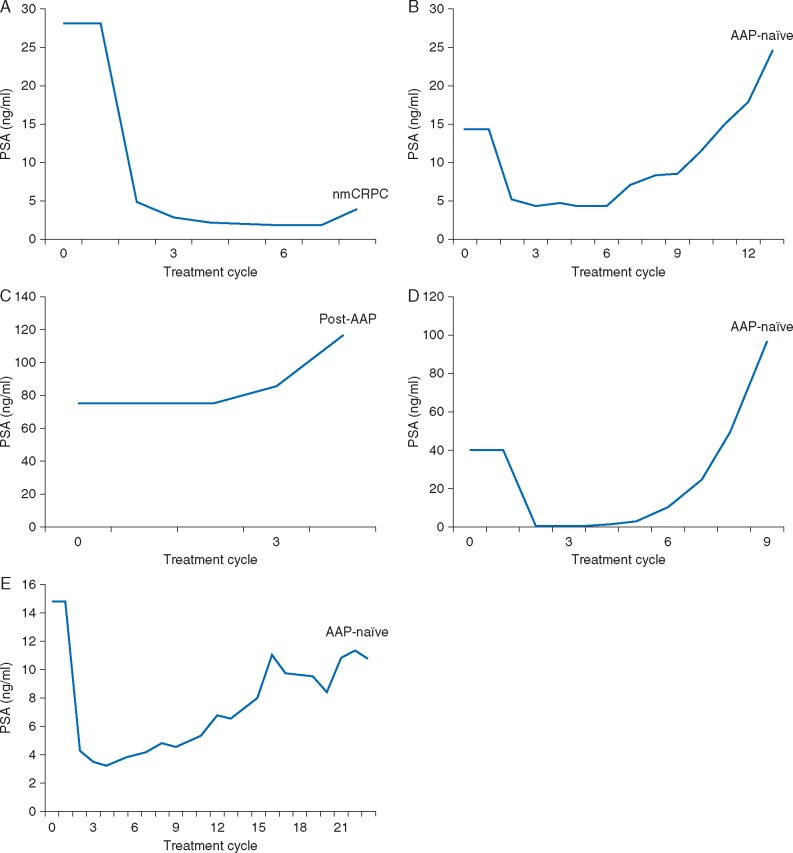
Two of the 93 (2.2%) patients harbored the AR F877L mutation at baseline at a mutation frequency of <0.05%, and both were subsequently found to have a PSA decline in response to apalutamide (Table 2; Figure 1A and B). One of these patients was in the nmCRPC cohort (12-week PSA change, –92.2%; treatment duration, 6.9 months) and the other was in the AAP-naïve cohort (12-week PSA change, –66.9%; treatment duration, 11.0 months). Both patients had detectable AR F877L and an increase in the mutation frequency at the time of progression.
Table 2.
Androgen receptor F877L and T878A mutation statusa in individual patients treated with apalutamide in the nmCRPC, AAP-naïve and post-AAP cohorts
| Cohort | Patient ID# | AR mutationb | Mutation fraction at baselinec | Cycle at which mutation fraction at progression detected | Mutation fraction at progressiond,e | 12-Week PSA changef | Treatment duration (months)g |
|---|---|---|---|---|---|---|---|
| nmCRPC | 1 | F877L | (0.02%) | 8 | (0.3%) | –92.2% | 6.9 |
| AAP-naïve | 2 | F877L | – | 22 | (0.721%) | –77.7% | 24.9 |
| 3 | F877L | (0.032%) | 11 | (0.41%) | –66.9% | 11.0 | |
| 4 | F877L | – | 9 | (0.18%) | –97.3% | 8.0 | |
| Post-AAP | 5 | F877L | – | 4 | (0.04%) | +55.9% | 3.4 |
| 6 | T878A | (0.84%) | 4 | (5.46%) | +112.7% | 2.8 | |
| 7 | T878A | (0.07%) | 14 | – | –62.7% | 12 | |
| 8 | T878A | (1.96%) | 6 | (0.4%) | –90.1% | 4.8 | |
| 9 | T878A | – | 10 | (0.02%) | –80.8% | 23.2 |
A plasma sample was deemed positive for a given mutation if the percentage of mutant beads was above the cutoff (0.02%).
No F877L/T878A double mutants were detected.
Number of mutation positive patients at baseline (F877L, n = 2/93; T878A, n = 3/93).
Number of mutation positive patients at progression (F877L, n = 5/82; T878A, n = 3/82).
Disease progression on apalutamide was defined as evidence of both PSA progression (≥25% and >2 ng/ml above PSA nadir confirmed ≥3 weeks later or >2 ng/ml above baseline PSA after 12 weeks) and radiographic progression (soft tissue metastases by modified Response Evaluation Criteria In Solid Tumors 1.0) seen on computed tomography/magnetic resonance imaging scans and/or bone metastases by 99mTc-methylene diphosphate bone scans by Prostate Cancer Working Group 2 criteria, and clinically by the occurrence of a skeletal-related event, pain progression, or worsening of disease-related symptoms requiring new systemic anti-prostate cancer therapy.
Median 12-week PSA change in F877L mutation negative patients (n = 86) was –79.8% (range, –99.9 to +175). Median 12-week PSA change in T878A mutation negative patients (n = 87) was –81.2% (range, –99.9 to +175).
Median treatment duration in F877L mutation-negative patients (n = 92) was 19.6 months (range, 0.03–37.8). Median treatment duration in T878A mutation-negative patients (n = 93) was 18.4 months (range, 0.03–37.8).
–, undetected. AAP, abiraterone acetate plus prednisone; AR, androgen receptor; mCRPC, metastatic castration-resistant prostate cancer; PSA, prostate-specific antigen.
Figure 1.
PSA changes in patients with androgen receptor F877L mutations detected at baseline [(A) Pt ID#1, (B) Pt ID#3] and at progression on apalutamide [(C) Pt ID#5, (D) Pt ID#4, (E) Pt ID#2]. AA, abiraterone acetate; PSA, prostate-specific antigen.
Three additional patients [3/82 (3.7%)] were found to have the mutation at progression that had not been detected at baseline (Table 2); the PSA trajectory is shown in Figure 1C–E. The single patient in the post-AAP cohort who acquired AR F877L demonstrated no PSA decline (12-week PSA change, +55.9%; treatment duration, 3.4 months) and had a relatively low mutation frequency of 0.04% (Table 2; Figure 1C). The other 2 patients with acquired AR F877L were both in the AAP-naïve cohort with 12-week PSA changes of –97.3% and –77.7%, treatment durations of 8.0 and 24.9 months, respectively, and mutation frequencies of 0.18% and 0.72%, respectively (Table 2; Figure 1D and E, respectively).
A R T878A
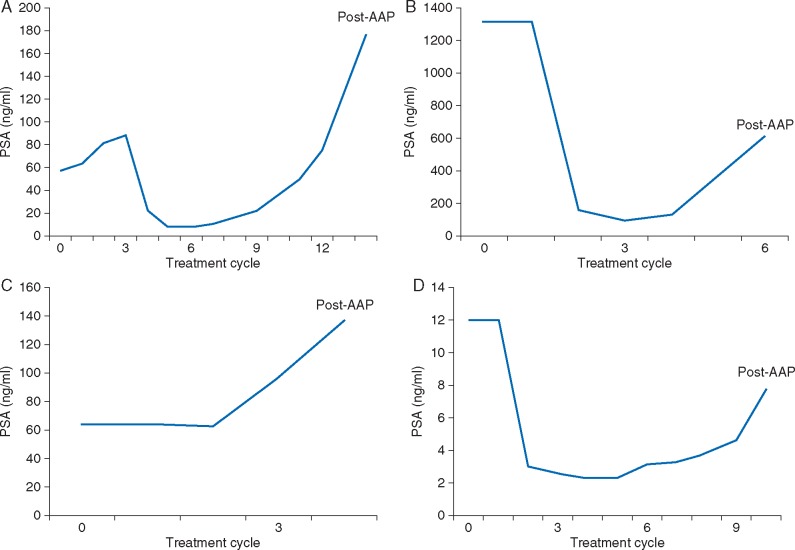
Three of 93 (3.2%) patients had the AR T878A mutation at baseline (Table 2); all had previously received at least 6 months of abiraterone and demonstrated similar baseline characteristics. Two had a PSA decline while on treatment with apalutamide, including 1 who had lost the mutation by the time of progression on apalutamide (Figure 2A; Table 3) (12-week PSA change, −62.7%; treatment duration, 12.0 months), and a second who had a decreased mutation fraction from 1.96% at baseline to 0.4% at progression (Figure 2B; Table 3) (12-week PSA change, −90.1%; treatment duration, 4.8 months). The third patient had an increased mutation fraction from 0.84% at baseline to 5.46% at progression and had no PSA decline (12-week PSA change, +112.7%; treatment duration, 2.8 months) (Figure 2C; Table 3). The PSA kinetics increased for these patients after AR T878A detection at progression (Figure 2A–D). One post-AAP patient acquired the AR T878 mutation at progression at a relatively low frequency of 0.02%. This patient had a PSA decline in response to apalutamide (12-week PSA change, –80.8%; treatment duration, 23.2 months) (Figure 2D; Table 3).
Figure 2.
PSA changes in patients with androgen receptor T878A mutations detected at baseline [(A) Pt ID#7, (B) Pt ID#8, (C) Pt ID#6] and at progression on apalutamide [(D) Pt ID#9]. AA, abiraterone acetate; PSA, prostate-specific antigen. Baseline characteristics for these patients are shown in Table 3.
Table 3.
Baseline demographics and disease characteristics of post-AAP patients with T878A mutations at baseline corresponding to patients shown in Figure 2 (per Figure 2A–C) and progression on apalutamide (per Figure 2D)
| Patient | A (Pt ID#7) | B (Pt ID#8) | C (Pt ID#6) | D (Pt ID#9) |
|---|---|---|---|---|
| Age | 83 | 64 | 74 | 58 |
| Race | White | White | White | White |
| Baseline PSA (ng/ml) | 58.4 | 1315.2 | 64.1 | 12.0 |
| ECOG PS | 1 | 1 | 1 | 0 |
| Gleason score | 4+5 | 4+3 | 4+3 | N/A |
AAP, abiraterone acetate plus prednisone; ECOG PS, Eastern Cooperative Oncology Group performance status; PSA, prostate-specific antigen.
Discussion
The survival benefits seen with agents that target the AR-signaling pathway have transformed the management of mCRPC. Nevertheless, one-third of patients do not respond to second-generation AR-targeted therapies, and the majority of those who initially respond, will acquire resistance to these agents. The optimal treatment of these patients, and how best to sequence available life-prolonging therapies, have not been established due to the inability to identify patients most likely to respond (or not respond) to specific AR-targeted drugs. This demonstrates the need for predictive molecular biomarkers to better inform treatment selection [11, 12, 24, 29]. Here, we report results of ctDNA sequencing using the BEAMing assay on samples from a phase II study of apalutamide in 3 distinct cohorts (nm, metastatic AAP-naïve, and metastatic post-AAP). The assay was selected because of its increased sensitivity versus an AR exon 8 sequencing approach used by others [13].
Overall, we tested 5 mutations derived from 11 possible amino acid alterations in 5 codons (supplementary Table S1, available at Annals of Oncology online) for which the assay was designed, including: F877L (n = 5), T878A (n = 4), W742C (n = 1), V716M (n = 1) and H875Y (n = 2). The most common (occurring in more than one subject) were AR F877L and AR T878A, LBD mutations associated in laboratory models and in the clinic with resistance to enzalutamide and apalutamide (AR F877L) [14, 18, 19] and abiraterone (AR T878A) [15, 16].
The frequency of AR F877L mutations (i.e. copies of mutant AR per genomic equivalent) increased in the mCRPC cohort after exposure to apalutamide, suggesting the possibility of preexisting clones that underwent positive selection with treatment. The 2 patients with the AR F877L mutations at baseline had a 12-week PSA decline of >50% after treatment with apalutamide. Notably, both had a relatively low frequency of the mutation at baseline (<0.05%) that increased at the time of progression. Another AAP-naïve mCRPC patient remained on study for 24.9 months and acquired the AR F877L mutation at progression (mutation frequency, 0.72%), suggesting a possible mechanism for secondary resistance. AR F877L was not detected in any post-AAP mCRPC patients at baseline. One post-AAP patient acquired the AR F877L mutation at progression on apalutamide; however, this patient had a low frequency of the mutation (0.04%) and was only on study for 3.4 months with a rising PSA, potentially suggesting a method of resistance other than development of AR F877L in the setting of prior AAP exposure.
All patients in our study who harbored the AR T878A mutation were in the mCRPC post-AAP cohort, consistent with the results of a recent analysis showing that AR T878A was associated with resistance to abiraterone [13] and consistent with prevalence reported in prior studies [13, 16], whereas AR T878A was never detected in patients with nmCRPC or in those in the AAP-naïve mCRPC cohort. One of the patients who lost the AR T878A mutation at progression initially had a PSA elevation but subsequently experienced a robust PSA decline and was on treatment for 12 months until treatment discontinuation due to PSA, radiographic and clinical progression. The decrease or loss of the AR T878A mutation observed in 2 of the 3 post-AAP patients who received treatment with apalutamide suggests 3 possibilities: apalutamide may have selectively inhibited the clone with this mutation and restored sensitivity to AR-directed treatment; discontinuation of abiraterone may have removed the evolutionary selection pressure that encouraged this AR mutation to emerge during abiraterone treatment; or discontinuation may have removed selective advantage of progesterones with the availability of endogenous steroids.
Blood samples were collected from 93 patients at baseline and from 82 patients at progression using a BEAMing assay designed to detect 11 selected AR-LBD mutations. These mutations were found at a relatively low incidence and frequency. Potential limitations of the analysis include the use of only one assay (limited to one assay per study sample availability) predesigned to detect 11 AR-LBD mutations already known to be associated with resistance to AR signaling-directed therapies. There may be other as of yet not well defined AR-LBD mutations that contribute to resistance. For example, the clinical significance of emergence of AR L702H in patients treated with exogenous glucocorticoids was not known when this study was designed [30]. Larger, prospective studies using assays that can detect mutations as well as other alterations in the receptor such as the AR splice variants [12, 31] to more completely address the question of the role of AR-LBD mutations in both de novo and acquired resistance would require a different type of blood sample. Given the high sensitivity of the BEAMing assay, it is likely that the AR F877L and AR T878A mutations are not major contributors to de novo or acquired resistance with apalutamide. It is also possible that the presence of AR F877L and AR T878A mutations in apalutamide-treated patients is an epiphenomenon associated with clonal selection pressures rather than being a driver of apalutamide resistance. Notably, however, preclinical data strongly suggest that these AR mutations confer resistance to AR-targeting agents. Ultimately, an integrated analysis of tumor-specific mRNA and DNA would be required to study the full complement of AR aberrations in men receiving novel hormonal therapies.
Conclusions
Although AR F877L has previously been associated with resistance to apalutamide and enzalutamide, patients with CRPC who were treated with apalutamide in our study had a low rate of de novo acquisition of the AR F877L mutation [3 of 82 patients (4%)] even using the sensitive BEAMing method. Not surprisingly, in patients without prior exposure to second-generation AR antagonists, AR F877L was detected at a low frequency at baseline [2 of 93 (2%)], and the presence of these mutations did not preclude PSA declines with apalutamide. The increased frequency of the mutation at the time of progression does suggest that AR F877L mutation may contribute to apalutamide resistance, although the frequency of these mutations in patients progressing on apalutamide in this study was low.
Second-line therapy with apalutamide in 2 post-AAP patients resulted in either a decrease or a loss of the preexisting AR T878A mutation while on therapy. Given the low frequency of the AR F877L and AR T878A mutations, they are unlikely to play a dominant role in the mechanism of primary or acquired resistance to apalutamide in CRPC patients.
Supplementary Material
Acknowledgements
Writing assistance was provided by Lashon Pringle, PhD, and Ira Mills, PhD, of PAREXEL, and was funded by Janssen Global Services, LLC.
Funding
This study was funded by Aragon Pharmaceuticals, Inc. Janssen Research & Development, LLC, is performing work on behalf of Aragon. No grant number is applicable. Dr. Rathkopf and Dr. Scher received support through the NIH/NCI Cancer Center Support Grant P30 CA008748.
Disclosure
DER reports a consultant or advisory role at Janssen Oncology and research funding from Janssen Oncology, Medivation, Celgene, Takeda, Millennium, Ferring and Novartis; MRS reports a consultant role at Janssen Research & Development; CJR reports a consultant or advisory role at Bayer and Millennium; honoraria from Janssen Oncology and Astellas Pharma; and research funding from BIND Biosciences, Karyopharm Therapeutics and Novartis; WRB reports research grants from AHRQ; NDS reports a consultant or advisory role at Astellas Pharma, Bayer, Janssen Scientific Affairs, Dendreon, Sanofi, Takeda, Tolmar and Ferring; GL and MS have declared no conflicts of interest; CSH reports consulting fees or honoraria from AbbVie, Algeta, Astellas, Bayer, Dendreon, Genentech, Johnson & Johnson, Medivation, Novartis, Pfizer and Veridex; grants or research support from Amgen, Aragon Pharmaceuticals, AstraZeneca, Bayer, Dendreon, Exelixis, Genentech, Johnson & Johnson, Medivation, Millennium, Novartis, OncoGenex, Sanofi-Aventis US, Taxynergy and Teva; and other financial benefit from Cell Therapeutics; JJA reports institutional research funding from Aragon Pharmaceuticals, Janssen Oncology, Astellas Pharma, Millenium, Novartis and Zenith Epigenetics; income for consulting with Astellas Pharma and for educational sessions with Bayer HealthCare Pharmaceuticals; RJH reports stock ownership in Aethlon; honoraria from Best Doctors, Inc.; research funding from US Oncology, Bavarian Nordic, Bristol-Myers Squibb, Merck and Amgen; remuneration for an ABIM Subspecialty Board; and has a patent pending on an immunotherapeutic agent; RFT reports a consultant or advisory role at Medivation/Astellas; stock ownership in Nymox and Sophiris Bio Inc.; honoraria from Nymox; research funding from Nymox, Medivation/Astellas, Janssen Oncology, Sophiris Bio Inc., Bayer, American Medical Systems, Boston Scientific, Advaxis, MDxHealth and Genomic Health; and has served on speakers’ bureaus for Medivation/Astellas and Dendreon; ECM reports employment at Aragon Pharmaceuticals (past); ST, DSR, MKY, AT, TK and RB report employment at Janssen Research & Development and stock ownership in Johnson & Johnson; CJdB reports employment at Janssen Biologics and stock ownership in Johnson & Johnson; HIS reports institutional research funding from Exelixis, Innocrin, Medivation, Janssen, Aragon Pharmaceuticals, and Illumina; a consultant or advisory role at Astellas, Sanofi, Millenium, and WCG Oncology (compensated), and at Medivation, Janssen, and Aragon Pharmaceuticals (uncompensated); and receipt of travel, accommodations, and expenses from Astellas, Janssen, Millenium, and Sanofi; ESA reports a consultant or advisory role at Sanofi, Dendreon, Medivation, Janssen Biotech, ESSA and Astellas Pharma; honoraria from Sanofi, Dendreon, Medivation, Janssen Biotech, ESSA and Astellas Pharma; research funding from Janssen Biotech, Johnson & Johnson, Sanofi, Dendreon, Aragon Pharmaceuticals, Exelixis, Millennium, Genentech, Novartis, Astellas Pharma and Tokai Pharmaceuticals; and receipt of travel, accommodations and expenses from Sanofi, Dendreon and Medivation.
References
- 1. Chi KN, Bjartell A, Dearnaley D. et al. Castration-resistant prostate cancer: from new pathophysiology to new treatment targets. Eur Urol 2009; 56: 594–605. [DOI] [PubMed] [Google Scholar]
- 2. Antonarakis ES, Eisenberger MA.. Expanding treatment options for metastatic prostate cancer. N Engl J Med 2011; 364: 2055–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen CD, Welsbie DS, Tran C. et al. Molecular determinants of resistance to antiandrogen therapy. Nat Med 2004; 10: 33–39. [DOI] [PubMed] [Google Scholar]
- 4. Clegg NJ, Wongvipat J, Joseph JD. et al. ARN-509: a novel antiandrogen for prostate cancer treatment. Cancer Res 2012; 72: 1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rathkopf DE, Morris MJ, Fox JJ. et al. Phase I study of ARN-509, a novel antiandrogen, in the treatment of castration-resistant prostate cancer. J Clin Oncol 2013; 31: 3525–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rathkopf D, Scher HI.. Androgen receptor antagonists in castration-resistant prostate cancer. Cance J 2013; 19: 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Azad AA, Eigl BJ, Murray RN. et al. Efficacy of enzalutamide following abiraterone acetate in chemotherapy-naive metastatic castration-resistant prostate cancer patients. Eur Urol 2015; 67: 23–29. [DOI] [PubMed] [Google Scholar]
- 8. Brasso K, Thomsen FB, Schrader AJ. et al. Enzalutamide antitumour activity against metastatic castration-resistant prostate cancer previously treated with docetaxel and abiraterone: a multicentre analysis. Eur Urol 2015; 68: 317–324. [DOI] [PubMed] [Google Scholar]
- 9. Loriot Y, Bianchini D, Ileana E. et al. Antitumour activity of abiraterone acetate against metastatic castration-resistant prostate cancer progressing after docetaxel and enzalutamide (MDV3100). Ann Oncol 2013; 24: 1807–1812. [DOI] [PubMed] [Google Scholar]
- 10. Noonan KL, North S, Bitting RL. et al. Clinical activity of abiraterone acetate in patients with metastatic castration-resistant prostate cancer progressing after enzalutamide. Ann Oncol 2013; 24: 1802–1807. [DOI] [PubMed] [Google Scholar]
- 11. Antonarakis ES, Lu C, Wang H. et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 2014; 371: 1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Scher HI, Lu D, Schreiber NA. et al. Association of AR-V7 on circulating tumor cells as a treatment-specific biomarker with outcomes and survival in castration-resistant prostate cancer. JAMA Oncol 2016; 2: 1441–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Azad AA, Volik SV, Wyatt AW. et al. Androgen receptor gene aberrations in circulating cell-free DNA: biomarkers of therapeutic resistance in castration-resistant prostate cancer. Clin Cancer Res 2015; 21: 2315–2324. [DOI] [PubMed] [Google Scholar]
- 14. Balbas MD, Evans MJ, Hosfield DJ. et al. Overcoming mutation-based resistance to antiandrogens with rational drug design. Elife 2013; 2: e00499.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cai C, Chen S, Ng P. et al. Intratumoral de novo steroid synthesis activates androgen receptor in castration-resistant prostate cancer and is upregulated by treatment with CYP17A1 inhibitors. Cancer Res 2011; 71: 6503–6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen EJ, Sowalsky AG, Gao S. et al. Abiraterone treatment in castration-resistant prostate cancer selects for progesterone responsive mutant androgen receptors. Clin Cancer Res 2015; 21: 1273–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hara T, Miyazaki J, Araki H. et al. Novel mutations of androgen receptor: a possible mechanism of bicalutamide withdrawal syndrome. Cancer Res 2003; 63: 149–153. [PubMed] [Google Scholar]
- 18. Joseph JD, Lu N, Qian J. et al. A clinically relevant androgen receptor mutation confers resistance to second-generation antiandrogens enzalutamide and ARN-509. Cancer Discov 2013; 3: 1020–1029. [DOI] [PubMed] [Google Scholar]
- 19. Korpal M, Korn JM, Gao X. et al. An F876L mutation in androgen receptor confers genetic and phenotypic resistance to MDV3100 (enzalutamide). Cancer Discov 2013; 3: 1030–1043. [DOI] [PubMed] [Google Scholar]
- 20. Taplin ME, Bubley GJ, Ko YJ. et al. Selection for androgen receptor mutations in prostate cancers treated with androgen antagonist. Cancer Res 1999; 59: 2511–2515. [PubMed] [Google Scholar]
- 21. Gottleib B, Beitel LK, Nadarajah A. et al. The androgen receptor gene mutations database (ARDB): 2012 update. Hum Mutat 2012; 33: 887–894. [DOI] [PubMed] [Google Scholar]
- 22. Lallous N, Volik SV, Awrey S. et al. Functional analysis of androgen receptor mutations that confer anti-androgen resistance identified in circulating cell-free DNA from prostate cancer patients. Genome Biol 2016; 17: 10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Prekovic S, van Royen ME, Voet AR. et al. The effect of F877L and T878A mutations on androgen receptor response to enzalutamide. Mol Cancer Ther 2016; 15: 1702–1712. [DOI] [PubMed] [Google Scholar]
- 24. Romanel A, Gasi TD, Conteduca V. et al. Plasma AR and abiraterone-resistant prostate cancer. Sci Transl Med 2015; 7: 312re10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smith MR, Antonarakis ES, Ryan CJ. et al. Phase 2 study of the safety and antitumor activity of apalutamide (ARN-509), a potent androgen receptor antagonist, in the high-risk nonmetastatic castration-resistant prostate cancer cohort. Eur Urol 2016; 70: 963–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rathkopf DE, Antonarakis ES, Shore ND. et al. Safety and antitumor activity of apalutamide (ARN-509) in metastatic castration-resistant prostate cancer with and without prior abiraterone acetate and prednisone. Clin Cancer Res 2017 February 17. [epub ahead of print], doi:10.1158/1078-0432.CCR-16-2509. [DOI] [PMC free article] [PubMed]
- 27. Scher HI, Halabi S, Tannock I. et al. Design and end points of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: recommendations of the Prostate Cancer Clinical Trials Working Group. J Clin Oncol 2008; 26: 1148–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Richardson AL, Iglehart JD.. BEAMing up personalized medicine: mutation detection in blood. Clin Cancer Res 2012; 18: 3209–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wyatt AW, Azad AA, Volik SV. et al. Genomic alterations in cell-free DNA and enzalutamide resistance in castration-resistant prostate cancer. JAMA Oncol 2016; 2: 1598–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carreira S, Romanel A, Goodall J. et al. Tumor clone dynamics in lethal prostate cancer. Sci Transl Med 2014; 6: 254ra125.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Antonarakis ES, Lu C, Luber B. et al. Androgen receptor splice variant 7 and efficacy of taxane chemotherapy in patients with metastatic castration-resistant prostate cancer. JAMA Oncol 2015; 1: 582–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.