Abstract
Background
Deletion of the chromatin remodeler chromodomain helicase DNA-binding protein 1 (CHD1) is a common genomic alteration found in human prostate cancers (PCas). CHD1 loss represents a distinct PCa subtype characterized by SPOP mutation and higher genomic instability. However, the role of CHD1 in PCa development in vivo and its clinical utility remain unclear.
Patients and methods
To study the role of CHD1 in PCa development and its loss in clinical management, we generated a genetically engineered mouse model with prostate-specific deletion of murine Chd1 as well as isogenic CHD1 wild-type and homozygous deleted human benign and PCa lines. We also developed patient-derived organoid cultures and screened patients with metastatic PCa for CHD1 loss.
Results
We demonstrate that CHD1 loss sensitizes cells to DNA damage and causes a synthetic lethal response to DNA damaging therapy in vitro, in vivo, ex vivo, in patient-derived organoid cultures and in a patient with metastatic PCa. Mechanistically, CHD1 regulates 53BP1 stability and CHD1 loss leads to decreased error-free homologous recombination (HR) repair, which is compensated by increased error-prone non-homologous end joining (NHEJ) repair for DNA double-strand break (DSB) repair.
Conclusions
Our study provides the first in vivo and in patient evidence supporting the role of CHD1 in DSB repair and in response to DNA damaging therapy. We uncover mechanistic insights that CHD1 modulates the choice between HR and NHEJ DSB repair and suggest that CHD1 loss may contribute to the genomic instability seen in this subset of PCas.
Keywords: chromatin remodeler CHD1, PCa, DDR, homologous recombination, non-homologous end joining, synthetic lethality
Introduction
Genomic profiling of human localized and metastatic prostate cancers (PCas) identified chromodomain helicase DNA-binding protein 1 (CHD1) as a homozygously deleted putative tumor suppressor gene [1, 2, 4–6]. CHD1 deficient PCas comprise a genetic subtype that typically presents mutations in the SPOP gene but lacks TMPRSS-ERG translocations and PTEN deletions, the most common genomic alteration found in human PCas [7]. Furthermore, the CHD1-loss or SPOP-mutant/CHD1-loss subtype is characterized by increased genomic instability and high levels of chromosomal rearrangements, suggesting a potential defect in DNA damage repair [2, 8, 9]. Indeed, a recent in vitro study has linked CHD1 function with DNA double-strand break repair [10].
CHD1 is an evolutionarily highly conserved chromatin remodeler containing a chromodomain, a SNF2-related ATPase/helicase domain and a C-terminal DNA-binding domain [11]. CHD1 regulates chromatin assembly [11, 12] and active transcription by binding to H3K4me3 and elongation factors [13–15]. Loss of CHD1 leads to increased heterochromatin formation in murine embryonic stem cells (mESCs) [16] and embryonic lethality [17].
Recent studies indicate that DNA repair genes, including BRCA2, ATM, CDK12, FANCA and RAD51C, are frequently inactivated in primary and metastatic castration resistant PCas (mCRPC) [7, 18, 19]. Although some of these genetic defects sensitize mCRPC to Poly-ADP-ribose (PARP) inhibitor treatment, these genomic aberrations do not explain all responders [19], suggesting that other unidentified molecular and genetic events may control the responsiveness of some PCas to DNA damage related treatment.
So far, there are no genetically engineered in vivo models bearing prostate-specific deletion of CHD1. Furthermore, the existing in vitro models with stable CHD1 deletion were either not prostate-derived or did not reflect the genetic characteristics of the CHD1 loss subtype [1]. To study the role of CHD1 in PCa development, we generated a murine Chd1 prostate conditional knockout model, human prostate CHD1 knockout cell lines as well as human PCa-derived organoids with and without CHD1 deletion. Here we show that in preclinical models closely reflecting the genetic background of this disease subtype, CHD1 functions by orchestrating DSB repair independent of AR activity. Loss of CHD1 sensitizes prostatic epithelial cells to DNA damaging treatments, including irradiation and drugs such as carboplatin and PARP inhibitors. Mechanistically, loss of CHD1 stabilizes 53BP1, increases error-prone NHEJ activity and decreases error-free HR DSB repair. Therefore, CHD1 status may be used to stratify human PCa for effective treatments.
Methods
Mouse strains
All studies were carried out under the regulation of the division of Laboratory Animal Medicine at the University of California at Los Angeles (UCLA). Methods concerning in vivo experiments are provided in supplementary experimental procedures, available at Annals of Oncology online.
Vectors, celllines, antibodies, and in vitro assays
Details of cell lines, antibodies, and in vitro assays are provided in supplementary experimental procedures, available at Annals of Oncology online.
Human PCa metastasis biopsies and assays on patient-derived tissue
Details on human PCa metastasis biopsies and assays on patient-derived tissue are provided in supplementary experimental procedures, available at Annals of Oncology online.
Histology and IHC
Details of IHC on mouse prostates and 22Rv1 xenografts procedures are provided in supplementary experimental procedures, available at Annals of Oncology online.
HPRT-DRGFP assays
HR and NHEJ assays were carried out as described previously [27, 54]. Details for generation of HPRT-DRGFP clones and measuring repair proficiency are described in supplementary experimental procedures, available at Annals of Oncology online.
Statistical analysis
The results are represented as means of at least three independent experiments [standard error of the mean (SEM) are indicated by errors bars]. Details on statistical analysis are provided in supplementary experimental procedures, available at Annals of Oncology online.
Results
Pb- c re+;Chd1L/L mice do not develop PCa
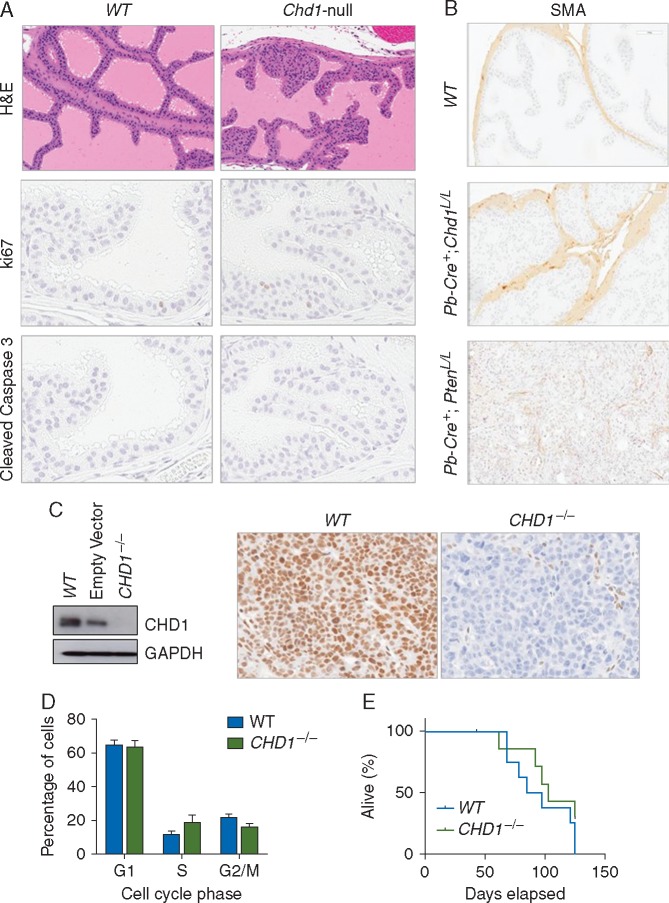
To determine the impact of CHD1 deletions on PCa development, we deleted Chd1 in murine prostate epithelial cells in vivo by crossing Chd1L/L conditional knockout females [17, 20] with probasin (Pb)-Cre transgenic males [21]. Pb-Cre+;Chd1L/+ and Pb-Cre+;Chd1L/L mice were born with normal Mendelian distributions (data not shown). Pb-Cre+;Chd1L/L prostates showed hyperplasia and prostatic intraepithelial neoplasia (PIN) lesions while Pb-Cre+;Chd1L/+ prostates were comparable to that of Pb-Cre− wild-type (WT) control mice (Figure 1A; supplementary Figure S1, available at Annals of Oncology online and data not shown). We therefore focused our analysis on the Pb-Cre+;Chd1L/L prostate (Chd1-null hereafter).
Figure 1.
Loss of CHD1 induces prostatic intraepithelial neoplasia in mice. (A) Knockout of Chd1 leads to PIN lesions. Representative IHC section images of hematoxylin and eosin (top), ki67 (middle), and cleaved caspase 3 (bottom) in the anterior lobe of Pb-Cre−;Chd1L/L and Pb-Cre+;Chd1L/L 8-week old mice (see also supplementary Figure S1, available at Annals of Oncology online). (B) Representative IHC sections of smooth muscle actin (SMA) in the anterior lobe of Pb-Cre−;Chd1L/L, Pb-Cre+;Chd1L/L, and Pb-Cre+;PtenL/L20-week old mice. (C) (Left) Western blot analyzing CHD1 levels in wild-type (WT), empty vector, and CHD1−/− isogenic 22Rv1 cells. (Right) Representative IHC section images of CHD1 staining in 22Rv1 xenograft tumors. (D) Cell cycle distribution of isogenic 22Rv1 cells with and without CHD1. Propidium iodide-based quantification of the percentage of WT and CHD1−/− isogenic 22Rv1 cells in G1, S, or G2/M phases of the cell cycle. Mean±SEM (n=3). (E) The growth of isogenic 22Rv1 xenograft tumors with (blue line) or without (green line) CHD1. Survival was determined when the xenograft tumor reached 1000 mm3, the maximal size allowance per institution guideline (n=3).
Previous in vitro studies have implied that CHD1 controls cell invasion [2, 5]. However, homozygous deletion of Chd1 in vivo showed no invasive adenocarcinoma in mice up to 1 year of age, as evidenced by well-maintained smooth muscle actin staining around acini (SMA; Figure 1B). In addition, Chd1-null mouse prostates showed no significant differences in cell proliferation and cell survival (Figure 1A, middle and lower panels).
In order to relate the data from our mouse model study with human PCa [7], we also generated isogenic CHD1 knockout clones in 22Rv1 and RWPE cells using the CRISPR/Cas9 approach. RWPE is a benign prostate epithelial line, while 22Rv1 is one of few human advanced PCa lines WT for PTEN with functional p53, which are known to regulate the DNA damage response (DDR) [22]. 22Rv1 cells are also TMPRSS2-ERG fusion negative and express AR, which best mimics the genetic background found in advanced human PCas with CHD1 loss [7].
CHD1 loss was confirmed by western blot and immunohistochemistry (IHC) analyses (CHD1−/− hereafter) (Figure 1C). Similar to the in vivo mouse model, CHD1 loss did not change cell cycle profile (Figure 1D) or in vivo xenograft tumor formation and growth (Figure 1E). These data suggest that CHD1 loss does not drive PCa in an otherwise unaltered genetic background.
Loss of CHD1 leads to increased sensitivity to ionizing radiation
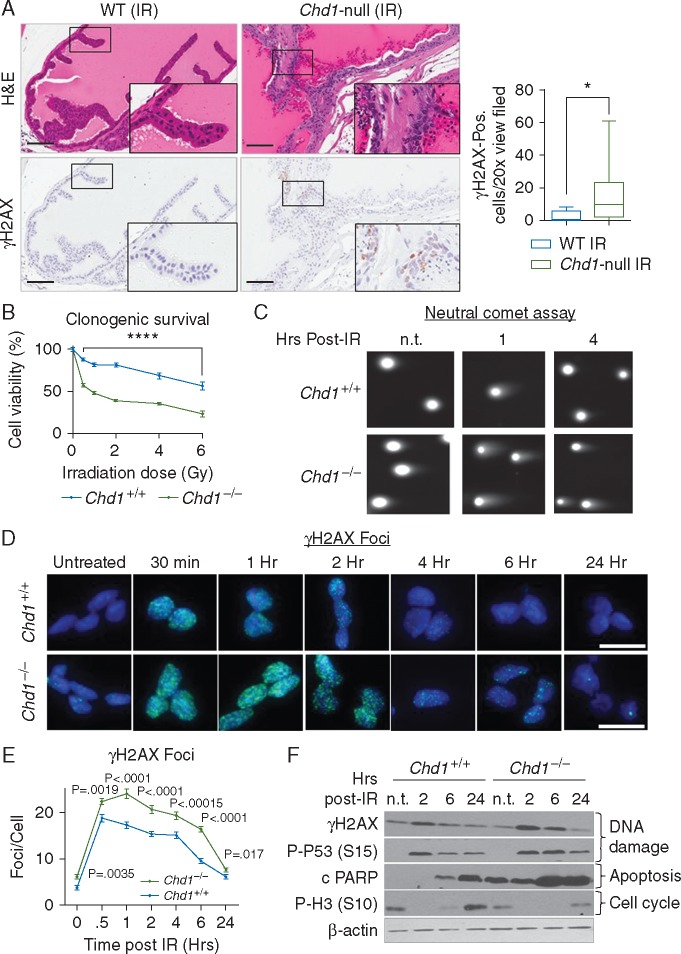
Since CHD1 loss is associated with genomic instability and a major increase in intrachromosomal rearrangements in human PCas [2, 8, 9], we tested the potential function of CHD1 in DDR in vivo. We treated WT and Chd1 null mice between 12 and 16 weeks of age with a single dose of 10 Gy of ionizing radiation (IR) and collected the prostates 24 h after IR [23]. Compared with WT prostates, Chd1-null prostates were more sensitive to IR as evidenced by increased γH2AX-positivity (Figure 2A, lower panel and quantified in graph on right).
Figure 2.
Loss of CHD1 leads to increased sensitivity to ionizing radiation. (A) (Left panel) Representative images of the anterior lobe of 16-week old mice that were treated with 10 Gy of whole body irradiation and collected 24 h later. Upper panels, H&E; lower, IHC for γH2AX; inserts, higher power images. Bar=100 mM. (Right panel) Quantification of γH2AX-positive cells per ×20 view field in the anterior lobes of WT and Chd1-null mice and 16 weeks of age. Mean±SEM (n=10). (B) Clonogenic survival of Chd1+/+ and Chd1−/− mESCs treated with the indicated doses of ionizing radiation. Mean±SEM (n=4). (C) Representative images from neutral comet assay of WT and Chd1-null mESCs, treated with 5 Gy of irradiation and collected at the indicated time points. Quantified in supplementary Figure S2B, available at Annals of Oncology online. (D) Representative images of γH2AX foci of Chd1+/+ and Chd1−/− mESCs treated with 5 Gy IR and collected at the indicated times. Bar=20 µm. (E) Quantification of γH2AX foci/nucleus in Chd1+/+ and Chd1−/− mESCs treated with 5 Gy of IR and analyzed at the indicated time points. More than 100 cells were analyzed per time point for each of 3 independent experiments. Mean±SEM (n=3). (F) Western blot shows the levels of γH2AX (Ser 139), P-P53 (Ser15), cleaved PARP and P-H3 (Ser10) in Chd1+/+ and Chd1−/− mESCs treated with 5 Gy of IR and analyzed at the indicated times.
To study the molecular mechanisms underlying the CHD1-regulated DDR, we needed experimental systems which minimize potentially confounding variables such as genomic alterations in DDR genes, which are frequently found in PCa cell lines [24]. Given CHD1’s ubiquitous expression pattern, isogenic mouse ES cell lines (mESCs) [16, 20] offer a clean genetic system for our mechanistic studies. Similar to our isogenic CHD1 WT and CHD1−/− 22Rv1 and RWPE human PCa cell lines, Chd1−/−mESCs have a cell cycle profile comparable to that of WT mESCs (supplementary Figure S2A, available at Annals of Oncology online), but are more sensitive to IR at all doses tested (0.5–6 Gy; Figure 2B), suggesting a conserved role for CHD1 in regulating DSB repair response. Neutral comet analysis showed that Chd1−/− mESCs have higher basal levels of DNA damage and increased comet tail length 1 and 4 h after irradiation (Figure 2C and supplementary Figure S2B, available at Annals of Oncology online). We further quantified γH2AX foci formation to monitor DSB formation and resolution after IR and found that Chd1−/− mESCs have higher γH2AX foci formation and slower resolution (Figure 2D and quantified in E), confirming the data from the comet assay. Western blot analysis showed that ATM-dependent phosphorylation of histone H2A and p53 at serine 139 (γH2AX) and serine 15 (p53), respectively, were also increased in Chd1−/− mESCs (Figure 2F) and failed to decrease 6 h post-irradiation, suggesting slower repair kinetics in Chd1−/− mESCs (Figure 2F). Similarly, Chd1−/− mESCs showed higher levels of the apoptotic response marker cleaved PARP with delayed reappearance of the mitosis marker phospho-H3 (Figure 2F). Together these results suggest that CHD1 plays an important role in DSB recognition and repair.
Loss of CHD1 leads to defect in error-free but increased error-prone DSB repair
Homologous recombination (HR) and non-homologous end joining (NHEJ) are the two major DSB repair mechanisms in response to DSB [25]. A recent published work by Kari et al. demonstrated in vitro that CHD1 loss specifically affects HR-mediated DNA repair but not NHEJ [10]. Since some of the major conclusions from this report were based on PC3 and VCaP cells, which are PTEN/p53 null and TMPRSS2-ERG fusion positive, respectively, we investigated the nature of CHD1-mediated DSB repair in our isogenic systems, which lack these alterations.
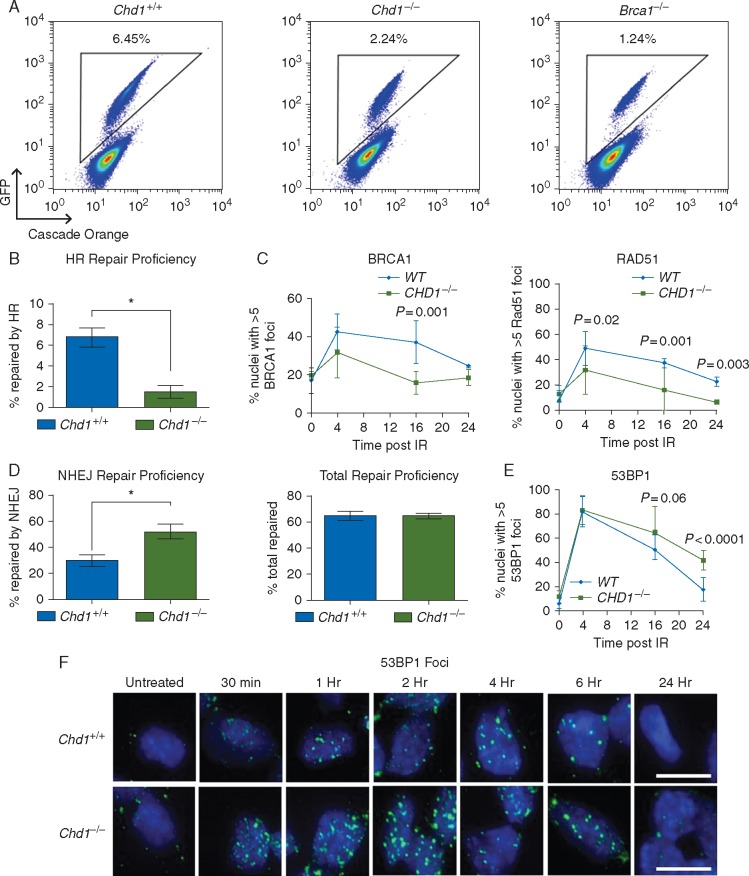
We first generated HDRGFP-Chd1+/+ and HDRGFP-Chd1−/− lines by knocking-in the previously described HDRGFP construct [26] into the endogenous Hprt locus in Chd1+/+ and Chd1−/− mESCs. This system enabled us to quantitatively compare the proficiency of these cells to repair DSBs by HR [27]. HR deficient HDRGFP-Brca1−/− mESCs were used as a control. HDRGFP-Chd1−/− cells showed an at least threefold reduction in HR competence compared with HDRGFP-Chd1+/+ WT cells (Figure 3A; an average of five independent clones were quantified in Figure 3B). Consistent with this finding, we also observed S-G2/M blockage after IR (supplementary Figure S3A, available at Annals of Oncology online). We examined RAD51 and BRCA1 foci, specific markers for HR repair [25] in CHD1-null 22Rv1 and RWPE cells and found decreased BRCA1 and RAD51 recruitment to DSBs after irradiation (Figure 3C; supplementary Figure S3B and C, available at Annals of Oncology online). Therefore our results, based on three independent isogenic cell lines further demonstrate the important role of CHD1 in modulating HR-mediated DSB repair.
Figure 3.
CHD1 regulates DSB repair pathway choices. (A) Representative FACS analysis of HDRGFP-Chd1+/+, HDRGFP-Chd1−/−, and HDRGFP-Brca1−/− mESCs 96 h after electroporation of I-SceI plasmid. Triangular gates represent GFP-positive cells that have repaired I-SceI mediated DSBs with HR. (B) HR repair proficiency in HDRGFP clones derived from Chd1+/+ and Chd1−/− mESCs. Mean±SEM (n=5). *P<0.05. (C) BRCA1 (left), and RAD51 (right) foci formation and resolution after irradiation-mediated DNA double-strand break induction (5 Gy) in isogenic 22Rv1 cells with or without CHD1. Mean±SEM (n=3) (see also supplementary Figure S3B, available at Annals of Oncology online). (D) NHEJ and total repair proficiency in HPRT-DRGFP clones derived from Chd1+/+ and Chd1−/− mESCs. Mean±SEM (n=5). *P<0.05. (E) 53BP1 foci formation and resolution after irradiation-mediated DNA double-strand break induction (5 Gy) in isogenic 22Rv1 cells with or without CHD1. Mean±SEM (n=3) (see also supplementary Figure S3B, available at Annals of Oncology online). (F) Representative images of P-53BP1 foci in Chd1+/+ and Chd1−/− mESCs treated with 5 Gy ionizing radiation and collected at the indicated times. Scale bar represents 20 µm (see also supplementary Figure S3D, available at Annals of Oncology online).
However, when quantifying total DDR as well as NHEJ repair proficiency using the well-established HDRGFP reporter assay [27], we found that NHEJ repair was significantly increased in HDRGFP-Chd1−/− cells compared with HDRGFP-Chd1+/+cells, whereas the total repair competence in all clones was similar (Figure 3D). Increased NHEJ in Chd1−/− mESCs could be further confirmed using 53BP1 foci quantification, a commonly used indicator for NHEJ repair (Figure 3F; supplementary Figure S3D, available at Annals of Oncology online). Consistently, CHD1 loss in 22Rv1 and RWPE cells also led to increased 53BP1 foci (Figure 3E; supplementary Figure S3B and E, available at Annals of Oncology online). These results demonstrate that loss of CHD1 impairs the proficiency of HR repair, which is compensated by error-prone NHEJ repair.
CHD1 regulates DDR independent of AR signaling pathway
Previous studies demonstrated that inhibition of AR signaling sensitizes PCa cells to IR [28–30]. Mechanistically, AR signaling regulates the expression of genes related to DDR and promotes classical NHEJ repair [28–30]. Since CHD1 loss could impair AR-dependent transcription in vitro [1, 31], it could also affect AR-regulated DNA repair gene expression, leading to altered NHEJ repair pathway. We therefore studied the impact of CHD1 loss on AR signaling in our preclinical models and publicly available PCa datasets [4].
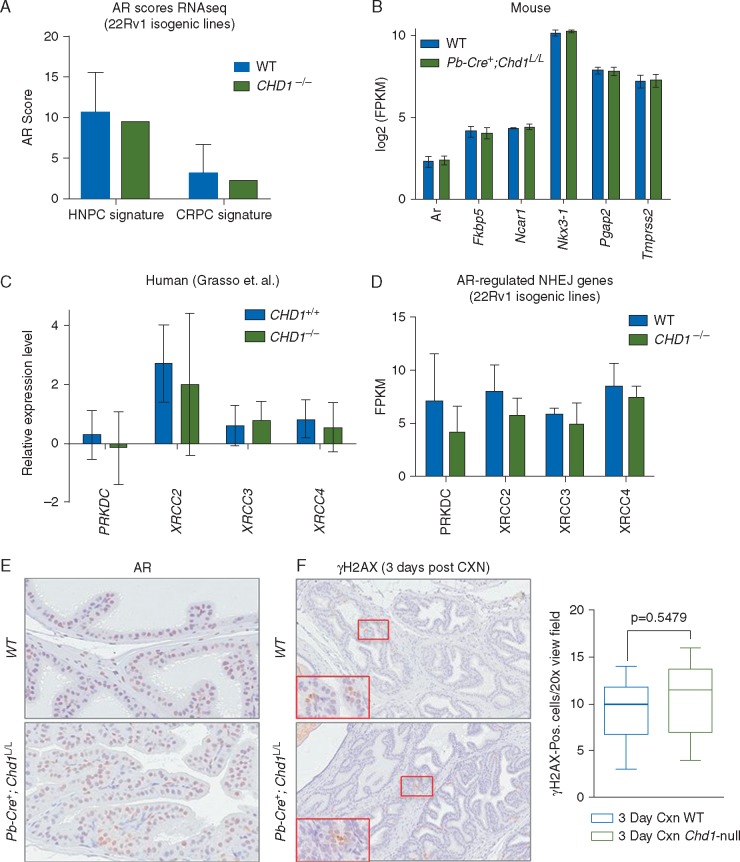
We first investigated whether AR transcriptional activity is altered by CHD1 loss in 22Rv1 cells by calculating their AR activity scores, which is based on the expression of established AR target genes [7, 32–34]. As shown in Figure 4A, there was no difference between WT and CHD1-deleted 22Rv1 cells. Similar results were also obtained when comparing the expression levels of these AR target genes in WT and Chd1-null prostates (Figure 4B; supplementary Figure S4A, available at Annals of Oncology online). We also analyzed the expression levels of AR-regulated DNA repair genes, such as PRKDC (encoding for DNAPKcs), XRCC2, XRCC3 and XRCC4 [28, 29] and found no significant differences between WT and CHD1-null human PCa patients [4], PCa cell lines, and our murine model (Figure 4C and D; supplementary Figure S4B, available at Annals of Oncology online).
Figure 4.
CHD1 regulates DDR independent of AR signaling pathway. (A) AR transcriptional activity is not altered by loss of CHD1 in 22Rv1 cells. The AR activity score was calculated based on AR target gene expression using a signature from hormone-sensitive PCa cells based on 27 AR-regulated genes defined in LnCaP cells after DHT stimulation (HNPC signature) [34] and castrate-resistant PCa patients based on 21 AR-regulated genes defined in 171 metastatic prostate tumors (CRPC signature) [35]. Mean±SEM (B) RNA expression levels of AR target genes in prostate derived from 10-week old WT (n=6) or Pb-Cre+;Chd1L/L (n=5) mice. Mean±SEM (C) No significant difference in the expression of DDR genes under AR control. Data from Grasso et al. [4]. Relative expression levels are presented as mean±SEM. (D) No significant difference in the expression of DDR genes under AR control. Data from 22Rv1 cells with or without knockout of CHD1 (n=3). Relative expression levels are quantified as FPKM and presented as mean±SEM (see also Figure S4B). (E) Representative IHC images of androgen receptor (AR) in the anterior lobe of age-matched, uncastrated WT (top) and Chd1-null (bottom) mice at 8 weeks of age. (F) Representative IHC images of γH2AX in the anterior lobe of age-matched WT (top) and Chd1-null (bottom) mice 3 days post-castration (CXN). (Right graph) Quantification of γH2AX-positive cells per x20 view-field in the anterior lobes of 3-day post-castrated WT and Chd1-null mice (n=6).
To further evaluate whether CHD1 regulates AR function, we castrated WT and Chd1-null animals at 6 weeks-of-age and evaluated prostate involution by H&E staining, Ki67 (for androgen-independent growth) and γH2AX (for castration-induced DDR) IHC analyses 3 days and 1, 2, 4 and 8 weeks post-castration (supplementary Figure 4C, available at Annals of Oncology online) [35–37]. We did not observe any significant differences in the kinetics/extent of prostatic involution when we compared WT and Chd1-null castrated prostates at these time points, nor in the number of Ki67 positive cells (supplementary Figure S4C, available at Annals of Oncology online and data not shown), indicating that CHD1 loss does not influence AR-dependent cell proliferation, which is very different from our previous studies of the Pten-null PCa model [36, 37]. Both WT and Chd1 null prostates have comparable AR levels before castration (Figure 4E) and responded similarly to castration-induced DNA damage as evidenced by the numbers of γH2AX positive cells (Figure 4F and data not shown). These in vivo results further support the notion that CHD1 does not play a major role in regulating AR pathway or AR-targeted DDR genes.
CHD1 complexes with NHEJ components
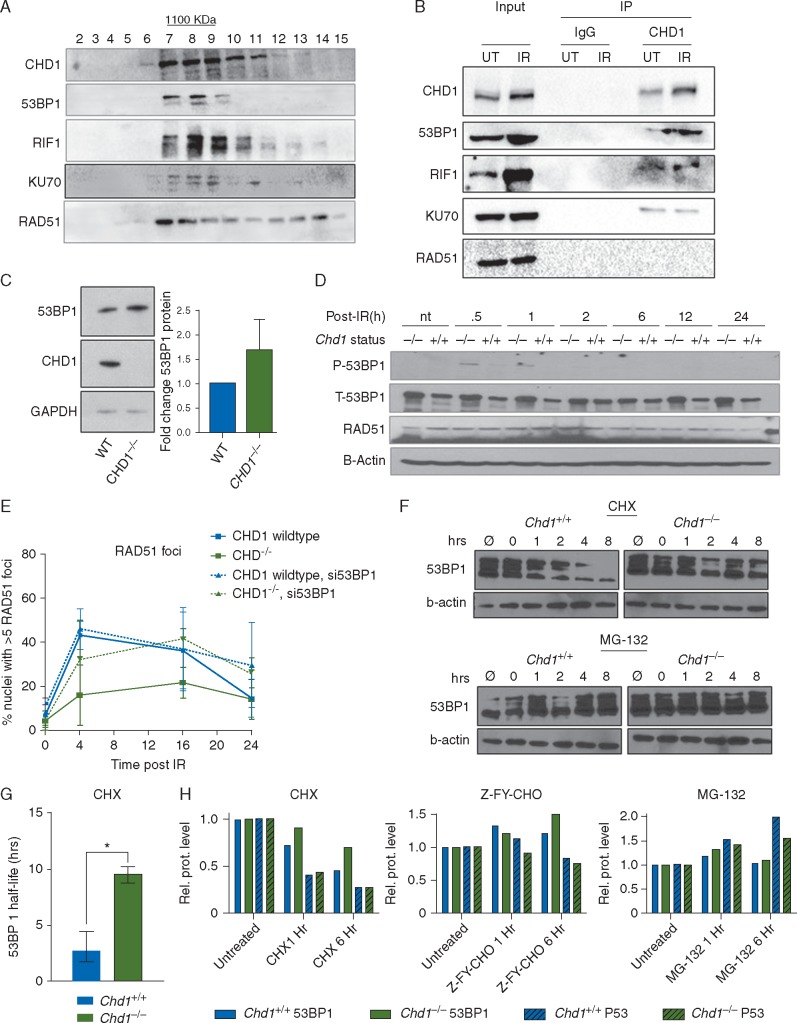
To understand how CHD1 regulates HR-mediated DSB repair, we investigated CHD1 interacting proteins. Gel filtration analysis indicated that endogenous CHD1 is in a high molecular weight complex (∼1100 kDa; Figure 5A), overlapping with several NHEJ components, such as 53BP1, RIF1 and Ku70 (Figure 5A). RAD51, on the other hand, was more evenly distributed (Figure 5A), indicating that it may not be in the same complex. To confirm the physical association of endogenous CHD1 with NHEJ components, we also conducted immunoprecipitation and western blot analyses on 22Rv1 human PCa cells and mESCs. As shown in Figure 5B, CHD1 physically associates with multiple endogenous NHEJ components, including 53BP1, RIF1 and KU70, but not RAD51 (Figure 5B; supplementary Figure S5A, available at Annals of Oncology online).
Figure 5.
CHD1 interacts with components of NHEJ and stabilizes 53BP1. (A) Protein lysates from Chd1+/+ mESCs were run through a gel filtration column. Fractions were collected and immunoblotted with anti-CHD1, 53BP1, RIF1, KU70 and RAD51 antibodies. Fractions 7–9 represent complexes of ∼1100 kDa. (B) Proteins from Chd1+/+ mESCs were immunoprecipitated with anti-CHD1 antibody and immunoblotted with anti-53BP1, KU70, and RAD51 antibodies (see also supplementary Figure S5A, available at Annals of Oncology online). (C) (Left) Western blot analysis of 53BP1 protein level in isogenic 22Rv1 cells with and without CHD1 deletion. (Right) Densitometry of western blots to quantify 53BP1 level; n=3. Mean±SEM (D) Western blot analysis of P-53BP1, T-53BP1, and RAD51 levels in WT and Chd1-null mESCs treated with 5Gy of irradiation and collected at the indicated time points (see also supplementary Figure S5C, available at Annals of Oncology online). (E) Quantification of HR activity in 22Rv1 cells with or without CHD1 based on RAD51 recruitment to DNA DSBs. Isogenic 22Rv1 cells were treated with control siRNA or siRNA targeting 53BP1; n=3. Mean±SEM (F) Western blot analysis of total 53BP1 and β-actin protein levels in Chd1+/+ and Chd1−/− mESCs treated with 50 µg/ml of cycloheximide (CHX, top panel) or 10 µM Mg-132 (bottom panel) for various amounts of time after 5 Gy irradiation and continuous drug treatment. (G) 53BP1 half-life was measured by western blot densitometry. WT and Chd1-null mESCs were treated with 5 Gy of irradiation 15 min after pre-treatment with 50 µg/ml cycloheximide (CHX). Mean±SEM (n=3, *P<0.05). (H) Quantification or total 53BP1 and P53 levels in WT and Chd1-null mESCs treated with 50 µg/ml cycloheximide (CHX), 10 µM ZY-F-CHO, or 10 µM MG-132 and analyzed 1 h and 6 h after continuous treatment. 53BP1 levels were normalized to vinculin and P53 levels were normalized to β-actin (see also supplementary Figure S5G, available at Annals of Oncology online).
To further investigate the impact of CHD1 loss on NHEJ components, we focused on 53BP1. 53BP1 is a major DDR protein and its levels are critical for DSB pathway choice, as higher levels of 53BP1 inhibit end resection of breaks, thus preventing HR [38]. Compared with WT 22Rv1 cells, CHD1−/− 22Rv1 cells have increased 53BP1 protein levels in vitro and in vivo (Figure 5C; supplementary Figure S5B, available at Annals of Oncology online); this was replicated in mESCs where Chd1−/− mESCs have significantly increased total 53BP1 (T-53BP1) with or without irradiation, but virtually no difference in RAD51 protein in the same setting (Figure 5D; supplementary Figure S5C, available at Annals of Oncology online). Total 53BP1 levels were also significantly increased in the in vivo Chd1-null mouse model (supplementary Figure S5D, available at Annals of Oncology online). On the other hand, 53BP1 mRNA levels were not changed in either our pre-clinical models or human PCa (supplementary Figure S5E, available at Annals of Oncology online), suggesting that CHD1 may modulate 53BP1 protein levels.
CHD1 regulates 53BP1 stability
To investigate whether the role of CHD1 in DSB pathway choice are mediated through 53BP1, we depleted 53BP1 in CHD1−/− 22Rv1 cells (supplementary Figure S5F, available at Annals of Oncology online). We show that 53BP1 knockdown can revert the HR deficiency seen in CHD1−/− 22Rv1 cells but have no obvious effect on HR-competent WT 22Rv1 cells (Figure 5E). This is consistent with previous studies showing that even a 50% reduction of 53BP1 expression could significantly rescue HR deficiency in BRCA1−/− cells [39].
53BP1 protein levels are regulated by post-translational mechanisms, i.e. degradation via either ubiquitin/proteasome- or cathepsin-l endosome/lysosome-mediated pathways [40–43]. To analyse whether 53BP1 protein stability is altered due to loss of CHD1, we treated mESCs with cycloheximide (CHX, 50 μg/ml, 15 min) before IR to block new protein synthesis (Figure 5F, top panel). We then calculated 53BP1 half-life based on western blot and densitometry. This analysis indicates that loss of CHD1 in mESCs leads to a fourfold increase of 53BP1 half-life (from 2 to 8 h) (Figure 5G). Furthermore, 53BP1 degradation was effectively blocked by either MG132 or Z-FY-CHO treatment, the proteasome and cathepsin-l inhibitors, respectively, similar to previous reports (Figure 5H; supplementary Figure S5G, available at Annals of Oncology online) [40–43], while the TP53 control was more strongly regulated by the ubiquitin/proteasome-mediated pathway (Figure 5H; supplementary Figure S5G, available at Annals of Oncology online). Taken together, our analyses demonstrate that CHD1 forms a complex with NHEJ components and negatively regulates 53BP1 stability and half-life, thereby modulating DSB repair choices.
Loss of CHD1 leads to hypersensitivity to PARP inhibition and DNA damaging agents
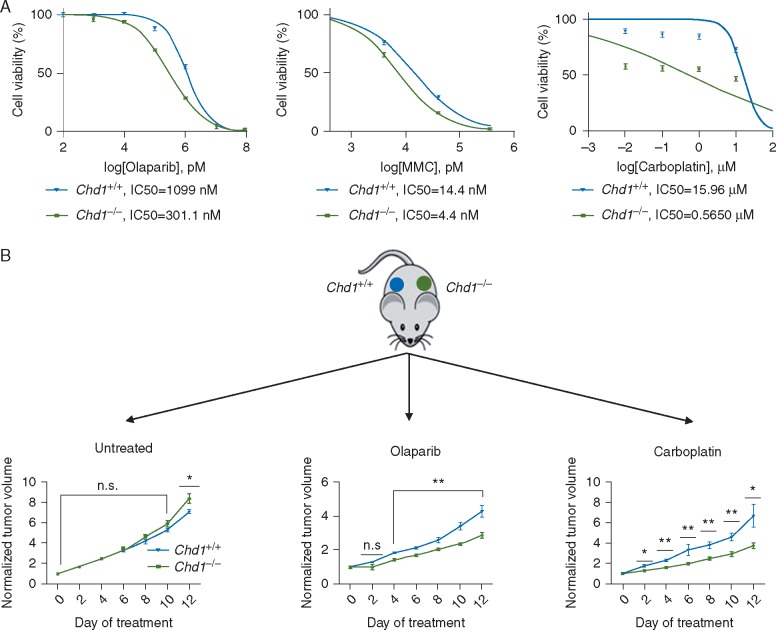
Tumors with defective HR repair, such as those with BRCA1 mutations, are sensitive to PARP inhibitors [44]. We treated WT and Chd1−/− mESCs with the PARP inhibitor olaparib, or the DNA cross-linking agents mitomycin C and carboplatin and found that Chd1−/− mESCs were more sensitive to all three drugs as single agents when compared with WT cells, although not as sensitive as Brca1-null mESCs (Figure 6A; supplementary Figure S6A, available at Annals of Oncology online).
Figure 6.
CHD1 loss leads to hypersensitivity to PARP inhibition and DNA damaging agents. (A) CHD1 loss leads to hypersensitivity to DNA damage agents in vitro. Cell viability, as determined by MTT assay, of Chd1+/+ and Chd1−/−mESCs after 48 h of continuous treatment of olaparib (left), mitomycin C (MMC) (middle), and carboplatin (right). IC50 was determined from the non-linear fit of normalized response curves (variable slope) (n=3) (see also supplementary Figure S6, available at Annals of Oncology online). (B) CHD1 loss leads to hypersensitivity to olaparib and carboplatin in vivo. Equal numbers of Chd1+/+ and Chd1−/− mESCs were implanted on to the bilateral flanks of nude mice. When tumors became palpable, mice were left untreated (Left) or treated with 100 mg/kg olaparib (middle) or 50 mg/kg carboplatin (right) twice daily for 2 weeks. Relative tumor volumes are presented with growth kinetics of untreated controls shown on the left (n=5; *P<0.05, **P<0.01).
To further confirm the hypersensitivity of Chd1 deficient cells to olaparib and carboplatin in vivo, we inoculated equal numbers of isogenic Chd1 WT and null mESCs into the bilateral flanks of the NSG mice (Figure 6B). When tumors became palpable, we treated the animals with olaparib (100 mg/kg) or carboplatin (50 mg/kg) for 2 weeks. While WT and Chd1−/− mESC-derived tumors grew at similar rates in the absence of any treatment, Chd1−/− mESC-derived tumors were more sensitive to both olaparib and carboplatin (Figure 6B). Taken together our study demonstrates that similar to BRCA1, CHD1 loss sensitizes cells to olaparib and carboplatin treatment in vitro and in vivo.
mCRPC with homozygous CHD1 loss is sensitive to olaparib and carboplatin ex vivo and in vivo
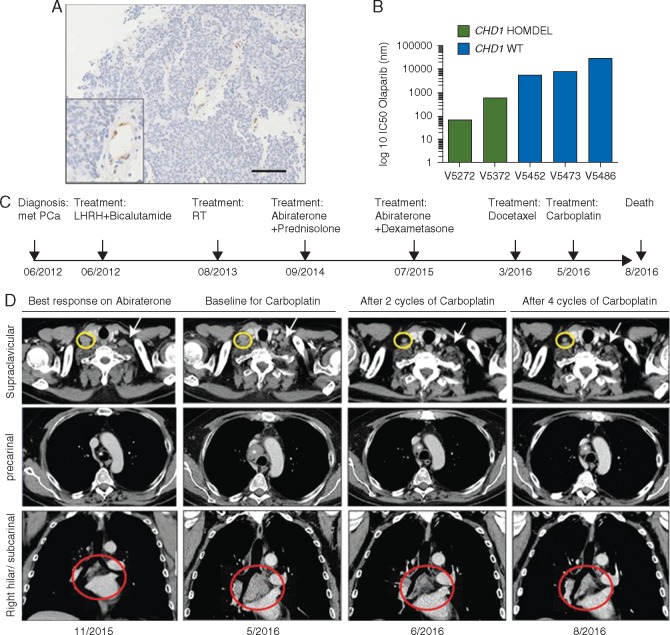
To investigate the relevance of our findings in the clinical setting, we generated organoids from mCRPC biopsies [6]. Patient-derived mCRPC organoids with homozygous deletion of CHD1, which was confirmed by digital droplet PCR, IHC (Figure 7A; supplementary Figure S7A, available at Annals of Oncology online) and FISH (supplementary Figure S7B, available at Annals of Oncology online), were more sensitive to olaparib compared with those organoids with normal CHD1 copy number (Figure 7B).
Figure 7.
Metastatic CRPCs with homozygous loss of CHD1 are sensitive to carboplatin and olaparib in vivo and ex vivo. (A) IHC analysis shows no CHD1 expression in tumors except blood vessels (boxed area) and stromal cells (see also supplementary Figure S7A and B, available at Annals of Oncology online). (B) mCRPC patient-derived organoids are sensitive to olaparib. Organoids from patients with homozygous loss of CHD1 (V5272 and V5372) show increased sensitivity to PARP inhibition. Copy number status was determined by digital droplet PCR. HOMDEL, homozygous deletion of CHD1; WT, wild-type. IC50, half maximal inhibitory concentration. (C) Clinical history of the patient represented as timeline. Arrows indicate clinical events. (D) Axial and coronal contrast enhanced CT images during disease progression (first and second columns) and after two and four cycles of carboplatin (third and fourth columns) showing significant reduction (37%) in the extent of the supraclavicular (yellow circles and white arrows), precarinal (*) and subcarinal (red circles) lymphadenopathy.
One of the patients, from whom CHD1 loss mCRPC organoids were derived (V5272), had rapidly progressing disease, fatigue, worsening performance status and liver function and rapidly rising LDH and ALP levels (supplementary Figure S7C, available at Annals of Oncology online). CT scanning revealed extensive liver and thoracic lymph-nodes metastasis (Figure 7D, left and middle panels). He also had a rapidly increasing circulating tumor cell count (CTC, supplementary Figure S7D, available at Annals of Oncology online) and PSA (supplementary Figure S7E, available at Annals of Oncology online) when his biopsy was taken for organoid culture. The patient had previously received castration, radical prostate radiotherapy (74Gy), abiraterone and docetaxel (Figure 7C). Since he was too unwell to be treated on a PARPi clinical trial, he received i.v. carboplatin treatment (700 mg, AUC 6, 3 weekly). After two carboplatin doses, his symptoms had fully resolved, his CT scan indicated a major response, his CTC count had decreased from 1157 to 30 cells in 7.5 ml of blood, and his PSA had dropped from 1300 to 806 µg/L (Figure 7D, right panels; supplementary Figure S7C, D, and E, available at Annals of Oncology online). His liver enzymes, ALP and LDH also normalized (supplementary Figure S7C, available at Annals of Oncology online). After 4 cycles of carboplatin, the patient continued to respond in lymph nodes and had a mixed response in the liver metastasis. However, the patient was discontinued due to progressing of the bone disease and clinical deterioration. These clinical data support our preclinical evidence and suggest that CHD1 status may be a predictive biomarker for DNA damaging agents such as carboplatin or PARPi for mCRPC.
Discussion
By generating a murine Chd1 prostate conditional knockout model, isogenic human prostate and PCa CHD1 WT and deleted lines, as well as human PCa-derived organoids with and without CHD1 deletion, we evaluated the function of CHD1 in vitro and in vivo in: (i) DNA damage repair in a genetic background reflecting this subtype of PCa, and (ii) during PCa development. Our results indicate that: (i) homozygous deletion of Chd1 causes PIN lesions without a significant impact on cell proliferation and survival; (ii) CHD1 is involved in modulating the stability of 53BP1; (iii) CHD1 loss results in increased NHEJ activity and decreased HR which can be reversed by 53BP1 knockdown; (iv) CHD1 deficient cells are hypersensitive to DNA damage by radiation, carboplatin and mitomycin C, as well as PARPi treatment. We also report for the first time clinical evidence that advanced PCa with CHD1 loss is sensitive to carboplatin therapy, further validating these data. Overall, our study provides a rationale to evaluate whether CHD1 status predicts treatment outcome in prospective clinical trials of DNA damaging agents such as carboplatin or PARPi.
In contrast to our previous studies of the Pten prostate conditional deletion model, which produces highly invasive adenocarcinomas [36], homozygous deletion of Chd1 causes only low-grade PIN lesions (Figure 1A and B; supplementary Figure S1B, available at Annals of Oncology online). Similarly, we did not observe significant growth differences in vitro and in vivo in isogenic CHD1-WT and CHD1−/− 22Rv1 human PCa cell lines (Figure 1D and E), similar to a recent report [45]. Our data suggest that loss of CHD1 alone is not sufficient to cause aggressive adenocarcinoma. Therefore CHD1 is a new addition to the list of tumor suppressor genes associated with human PCas, such p53, Rb, Brca2 and NKX3.1, whose loss alone in mice does not cause an aggressive phenotype but only hyperplasia and PIN lesions [46–50]. Similar to the role of CHD1 in DNA damage repair, which we are describing here (Figure 2), many of these tumor suppressors also fulfill a critical role in the DDR.
Our results provide a mechanistic explanation for the increased genomic instability observed in PCas with homozygous deletion of CHD1 [2, 3, 8]. We demonstrate that CHD1 loss suppresses error-free HR DSB repair while promoting error-prone NHEJ (Figure 3). Thus CHD1 may function as a molecular switch between HR and NHEJ. Loss of CHD1 sensitizes cells to olaparib, carboplatin and mitomycin C (Figure 6), very similar to BRCA1 deleted cells. However, CHD1 may regulate HR upstream of BRCA1 by (i) recruiting CtIP to sites of DSB as demonstrated in the previous report [10] and/or (ii) modulating the stability of 53BP1 and enhancing NHEJ as shown in our study (Figure 5).
Genetic studies in mice have indicated the importance of NHEJ components in maintaining genomic stability [51]. However, next generation sequencing studies of human PCas have so far not identified alterations in components of canonical NHEJ [4, 7]. On the other hand, several genes that are frequently altered in PCa are required for maintaining genomic stability, particularly via deregulation of DSB repair. For example, we have shown that SPOP mutations are associated with increased genomic instability in PCa by inhibiting HR and promoting NHEJ [8], similar to what we have reported here. Although a molecular link between SPOP mutation and CHD1 loss in NHEJ has not yet been established, it is possible that in patients with combined CHD1 homozygous deletion and SPOP mutations, an additive effect of these alterations may further sensitize cells to carboplatin or olaparib. Further studies are now needed to answer this question functionally and clinically.
Previous studies indicated that CHD1 regulates AR transcriptional signaling by mediating the binding of AR to its target promoters [1, 31], while other studies found that antagonizing AR signaling by ADT sensitizes cells to IR by inhibiting NHEJ without impacting HR DSB repair [28–30, 52]. This would suggest that loss of CHD1 sensitizes cancer cells to IR by reducing AR signaling, consequently decreasing NHEJ repair. However, in our models, loss of CHD1 does not affect AR transcriptional output, although our studies could not rule out paracrine AR signaling from stromal prostate cells (Figure 4) [53]. The fact that loss of CHD1 promotes NHEJ and suppresses HR without affecting AR function or AR target expression suggests that AR blockage might further sensitize this PCa subtype to radiotherapy.
The protein stability of DNA-repair proteins is tightly controlled to ensure timely and spatially restricted activity. 53BP1 protein half-life ranges from 0.5 to 2 h in the context of DNA damage induction [42]. We show here that CHD1 contributes to the regulation of 53BP1 stability (Figure 5) although the exact mechanism remains to be resolved. Based on our analyses and those published, both ubiquitin/proteasome and endosome/lysosome pathways are likely to play roles.
In summary, we report that the loss of CHD1 leads to changes in DDR. Using functional genetic approaches, we report that CHD1 loss decreases HR-mediated DSB repair and increases error-prone NHEJ activity. Importantly, CHD1 loss is associated with an increased sensitivity to PARP inhibition and anti-cancer drugs that induce DNA intercross-strand links including carboplatin. These observations may provide a mechanistic explanation for the high number of chromosomal rearrangements observed in PCas with CHD1 homozygous deletion. Our data provide a rationale for treating patients bearing tumors with CHD1 deletion in prospective clinical trials of PARP inhibitors or DNA damaging agents such as carboplatin to evaluate their potential clinical benefit in this subclass of PCa.
Supplementary Material
Acknowledgements
We thank Dr Maria Jasin for providing BRCA1 null mESC line, HDRGFP construct and helpful discussions; Wenkai Yi and Bohan Chen for technical assistance; members of the Wu and De Bono laboratories for comments and suggestions. We thank Dr. Christopher E. Barbieri and Dr. Michael A. Augello for many helpful discussions and the Rubin laboratory for technical assistance with the organoid culture.
Funding
This work was supported in part by awards from the Peking-Tsinghua Center for Life sciences (to HW); grants from the NIH (P50 CA092131 and U01 CA164188 to HW and R01GM113014 to MR-S); MYW is supported by the Presidential Fellowship of Peking University and WLG is supported by a General Financial Grant from the China Postdoctoral Science Foundation (2015M570010). Prostate Cancer UK project grant (PG13-036 to JDB), a Cancer Research UK Centre grant (C347/A18077 to JDB), a RMH/ ICR NIHR Biomedical Research Centre Flagship Grant (A34 to JDB), a Marie Sklodowska-Curie Fellowship of the European Union's FP7-PEOPLE programme (625792 to GB), and a Prostate Cancer Foundation Challenge Award (to JDB).
Disclosure
The authors have declared no conflicts of interest.
Key Message
Homozygous deletion of CHD1 represents a distinct molecular subtype of PCa with genomic instability. CHD1 loss causes DDR defects and sensitizes cells to drugs such as olaparib (PARPi) and carboplatin (Pt) in preclinical models, patient-derived organoids and a metastatic PCa patient. Prospective clinical trials are now needed to evaluate CHD1 deletion as a predictive biomarker for Pt and PARPi treatment.
References
- 1. Burkhardt L, Fuchs S, Krohn A. et al. CHD1 is a 5q21 tumor suppressor required for ERG rearrangement in prostate cancer. Cancer Res 2013; 73: 2795–2805. [DOI] [PubMed] [Google Scholar]
- 2. Liu W, Lindberg J, Sui G. et al. Identification of novel CHD1-associated collaborative alterations of genomic structure and functional assessment of CHD1 in prostate cancer. Oncogene 2012; 31: 3939–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baca SC, Prandi D, Lawrence MS. et al. Punctuated evolution of prostate cancer genomes. Cell 2013; 153: 666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grasso CS, Wu YM, Robinson DR. et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012; 487: 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang S, Gulzar ZG, Salari K. et al. Recurrent deletion of CHD1 in prostate cancer with relevance to cell invasiveness. Oncogene 2012; 31: 4164–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gao D, Vela I, Sboner A. et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014; 159: 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Abeshouse A, Ahn J, Akbani R. et al. The molecular taxonomy of primary prostate cancer. Cell 2015; 163: 1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boysen G, Barbieri CE, Prandi D. et al. SPOP mutation leads to genomic instability in prostate cancer. Elife 2015; 4: e09207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baca SC, Prandi D, Lawrence MS. et al. Punctuated evolution of prostate cancer genomes. Cell 2013; 153: 666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kari V, Mansour WY, Raul SK. et al. Loss of CHD1 causes DNA repair defects and enhances prostate cancer therapeutic responsiveness. EMBO Rep 2016; 17: 1609–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lusser A, Urwin DL, Kadonaga JT.. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat Struct Mol Biol 2005; 12: 160–166. [DOI] [PubMed] [Google Scholar]
- 12. Konev AY, Tribus M, Park SY. et al. CHD1 motor protein is required for deposition of histone variant h3.3 into chromatin in vivo. Science 2007; 317: 1087–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Simic R, Lindstrom DL, Tran HG. et al. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J 2003; 22: 1846–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sims RJ, Millhouse S, Chen CF. et al. Recognition of trimethylated histone h3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell 2007; 28: 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin JJ, Lehmann LW, Bonora G. et al. Mediator coordinates PIC assembly with recruitment of CHD1. Genes Dev 2011; 25: 2198–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gaspar-Maia A, Alajem A, Polesso F. et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 2009; 460: 863–U897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guzman-Ayala M, Sachs M, Koh FM. et al. Chd1 is essential for the high transcriptional output and rapid growth of the mouse epiblast. Development 2015; 142: 118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robinson D, Van Allen EM, Wu YM. et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015; 161: 1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mateo J, Carreira S, Sandhu S. et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med 2015; 373: 1697–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koh FM, Lizama CO, Wong P. et al. Emergence of hematopoietic stem and progenitor cells involves a Chd1-dependent increase in total nascent transcription. Proc Natl Acad Sci USA 2015; 112: E1734–E1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu X, Wu J, Huang J. et al. Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mech Dev 2001; 101: 61–69. [DOI] [PubMed] [Google Scholar]
- 22. Pearl LH, Schierz AC, Ward SE. et al. Therapeutic opportunities within the DNA damage response. Nat Rev Cancer 2015; 15: 166–180. [DOI] [PubMed] [Google Scholar]
- 23. Brown CE, Warren S.. Carcinoma of the prostate in irradiated parabiotic rats. Cancer Res 1978; 38: 159–162. [PubMed] [Google Scholar]
- 24. Taylor BS, Schultz N, Hieronymus H. et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010; 18: 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chapman JR, Taylor MR, Boulton SJ.. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 2012; 47: 497–510. [DOI] [PubMed] [Google Scholar]
- 26. Pierce AJ, Johnson RD, Thompson LH, Jasin M.. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 1999; 13: 2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Weinstock DM, Nakanishi K, Helgadottir HR, Jasin M.. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Meth Enzymol 2006; 409: 524–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Polkinghorn WR, Parker JS, Lee MX. et al. Androgen receptor signaling regulates DNA repair in prostate cancers. Cancer Discov 2013; 3: 1245–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Goodwin JF, Schiewer MJ, Dean JL. et al. A hormone-DNA repair circuit governs the response to genotoxic insult. Cancer Discov 2013; 3: 1254–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tarish FL, Schultz N, Tanoglidi A. et al. Castration radiosensitizes prostate cancer tissue by impairing DNA double-strand break repair. Sci Transl Med 2015; 7: 312re311.. [DOI] [PubMed] [Google Scholar]
- 31. Metzger E, Willmann D, McMillan J. et al. Assembly of methylated KDM1A and CHD1 drives androgen receptor-dependent transcription and translocation. Nat Struct Mol Biol 2016; 23: 132–139. [DOI] [PubMed] [Google Scholar]
- 32. Nelson PS, Clegg N, Arnold H. et al. The program of androgen-responsive genes in neoplastic prostate epithelium. Proc Natl Acad Sci USA 2002; 99: 11890–11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hieronymus H, Lamb J, Ross KN. et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell 2006; 10: 321–330. [DOI] [PubMed] [Google Scholar]
- 34. Kumar A, Coleman I, Morrissey C. et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med 2016; 22: 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kyprianou N, Isaacs JT.. Activation of programmed cell death in the rat ventral prostate after castration. Endocrinology 1988; 122: 552–562. [DOI] [PubMed] [Google Scholar]
- 36. Wang S, Gao J, Lei Q. et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell 2003; 4: 209–221. [DOI] [PubMed] [Google Scholar]
- 37. Mulholland DJ, Tran LM, Li Y. et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell 2011; 19: 792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bunting SF, Callen E, Wong N. et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 2010; 141: 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Johnson N, Johnson SF, Yao W. et al. Stabilization of mutant BRCA1 protein confers PARP inhibitor and platinum resistance. Proc Natl Acad Sci USA 2013; 110: 17041–17046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mallette FA, Richard S.. K48-linked ubiquitination and protein degradation regulate 53BP1 recruitment at DNA damage sites. Cell Res 2012; 22: 1221–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gonzalez-Suarez I, Redwood AB, Grotsky DA. et al. A new pathway that regulates 53BP1 stability implicates cathepsin L and vitamin D in DNA repair. EMBO J 2011; 30: 3383–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hu Y, Wang C, Huang K. et al. Regulation of 53BP1 protein stability by RNF8 and RNF168 is important for efficient DNA double-strand break repair. PLoS ONE 2014; 9: e110522.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Han X, Zhang L, Chung J. et al. UbcH7 regulates 53BP1 stability and DSB repair. Proc Natl Acad Sci USA 2014; 111: 17456–17461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bryant HE, Schultz N, Thomas HD. et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005; 434: 913–917. [DOI] [PubMed] [Google Scholar]
- 45. Zhao D, Lu X, Wang G. et al. Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer. Nature 2017542: 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Francis JC, McCarthy A, Thomsen MK. et al. Brca2 and Trp53 deficiency cooperate in the progression of mouse prostate tumourigenesis. PLoS Genet 2010; 6: e1000995.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maddison LA, Sutherland BW, Barrios RJ, Greenberg NM.. Conditional deletion of Rb causes early stage prostate cancer. Cancer Res 2004; 64: 6018–6025. [DOI] [PubMed] [Google Scholar]
- 48. Zhou Z, Flesken-Nikitin A, Corney DC. et al. Synergy of p53 and Rb deficiency in a conditional mouse model for metastatic prostate cancer. Cancer Res 2006; 66: 7889–7898. [DOI] [PubMed] [Google Scholar]
- 49. Chen Z, Trotman LC, Shaffer D. et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 2005; 436: 725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bhatia-Gaur R, Donjacour AA, Sciavolino PJ. et al. Roles for Nkx3.1 in prostate development and cancer. Genes Dev 1999; 13: 966–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bunting SF, Nussenzweig A.. End-joining, translocations and cancer. Nat Rev Cancer 2013; 13: 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Al-Ubaidi FL, Schultz N, Loseva O. et al. Castration therapy results in decreased Ku70 levels in prostate cancer. Clin Cancer Res 2013; 19: 1547–1556. [DOI] [PubMed] [Google Scholar]
- 53. Cunha GR. The role of androgens in the epithelio-mesenchymal interactions involved in prostatic morphogenesis in embryonic mice. Anat Rec 1973; 175: 87–96. [DOI] [PubMed] [Google Scholar]
- 54. Pierce AJ, Jasin M.. Measuring recombination proficiency in mouse embryonic stem cells. Methods Mol Biol 2005; 291: 373–384. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.