Abstract
Background
Genomic profiling is increasingly incorporated into oncology research and the clinical care of cancer patients. We sought to determine physician perception and use of enterprise-scale clinical sequencing at our center, including whether testing changed management and the reasoning behind this decision-making.
Patients and methods
All physicians who consented patients to MSK-IMPACT, a next-generation hybridization capture assay, in tumor types where molecular profiling is not routinely performed were asked to complete a questionnaire for each patient. Physician determination of genomic ‘actionability’ was compared to an expertly curated knowledgebase of somatic variants. Reported management decisions were compared to chart review.
Results
Responses were received from 146 physicians pertaining to 1932 patients diagnosed with 1 of 49 cancer types. Physicians indicated that sequencing altered management in 21% (331/1593) of patients in need of a treatment change. Among those in whom treatment was not altered, physicians indicated the presence of an actionable alteration in 55% (805/1474), however, only 45% (362/805) of these cases had a genomic variant annotated as actionable by expert curators. Further evaluation of these patients revealed that 66% (291/443) had a variant in a gene associated with biologic but not clinical evidence of actionability or a variant of unknown significance in a gene with at least one known actionable alteration. Of the cases annotated as actionable by experts, physicians identified an actionable alteration in 81% (362/445). In total, 13% (245/1932) of patients were enrolled to a genomically matched trial.
Conclusion
Although physician and expert assessment differed, clinicians demonstrate substantial awareness of the genes associated with potential actionability and report using this knowledge to inform management in one in five patients.
Clinical Trial number
Keywords: precision medicine, targeted therapy, tumor sequencing, next-generation sequencing
Introduction
In recent years, several academic centers have embarked on precision oncology initiatives utilizing large-scale sequencing to study cancer genomics, facilitate accrual to genomically matched clinical trials, and ultimately personalize patient care [1–3]. Despite the growing adoption of next-generation sequencing (NGS) panels at the point of care, physicians’ perceptions and interpretations of these results and their influence on clinical decision-making are currently unknown. Beginning in 2014, Memorial Sloan Kettering Cancer Center (MSKCC) began offering enterprise-scale large gene panel NGS utilizing an internally developed and clinically validated hybridization capture assay, MSK-IMPACT [3]. MSK-IMPACT uses matched tissue and normal samples to detect all classes of genomic alterations including somatic mutations, copy number alterations, and select structural rearrangements in >300 key cancer-associated genes. The germline is masked and therefore inherited alterations are not routinely identified. As part of an institution-wide initiative, testing was offered through a research nonbillable protocol to patients, at the discretion of their treating physicians, including tumor types where genomic profiling is not routinely used to guide standard treatment decisions [4]. The scale of this program provided a unique opportunity to study physician use of genomic data within a large comprehensive cancer center, specifically whether it altered management and the reasoning behind this decision-making.
Patients and methods
Population
All patients consented to MSK-IMPACT testing under an IRB-approved biospecimen protocol. A key endpoint of this study was whether MSK-IMPACT altered patient management. To evaluate this, chart reviews were performed and a questionnaire sent to physicians. As the role of genomic profiling in tumor types for which it is routine is already established, questionnaires targeted physicians ordering MSK-IMPACT as an investigational test. This provided the opportunity to gauge the perception and use of genomic profiling in cancer subspecialties that may have less familiarity with interpreting genomic information. At MSKCC, cancers for which tumor sequencing was considered standard during the study period included lung adenocarcinoma, colorectal cancer, melanoma, thyroid cancer, and gastrointestinal stromal tumor. However, these tumor types were not completely excluded, as physicians did order MSK-IMPACT when retesting patients previously sequenced using single analyte or smaller gene panels.
Patients who consented for MSK-IMPACT could have any disease stage and sequenced samples could be a primary or metastatic lesion. Importantly, not all patients required active therapy or a treatment change. Participating physicians were from a spectrum of specialties.
Survey instrument and study procedures
During the study period, MSK-IMPACT reports included a list of genomic alterations without clinical or functional annotation. Specifically, reports provided the mRNA transcript identification, exon number, cDNA change, protein change, and coverage (minimum and mean). The absence of annotation offered an opportunity to evaluate how physicians perceived the actionability of results without prompting by curated sources. A survey queried physicians’ interpretations of each patient’s sequencing results and whether these results changed management (Table 1). Questionnaires were distributed approximately quarterly by email from August 2014 through August 2016 and included patients newly enrolled and those previously consented for whom there were no previous responses. Each questionnaire contained a summary of the sequencing results and IRB numbers for MSKCC clinical trials the patient had enrolled to since testing. The term ‘actionable’ was not specifically defined in the questionnaire to avoid influencing physicians’ use of this term. In addition to individual responses, demographic information about the patients and responding physicians was collected. Notably, the gene panel initially included coverage for the entire coding and select intronic regions of 341 cancer-related genes and was later expanded to 410 genes.
Table 1.
Questionnaire responses
| Questions | Responses, N (%) |
|---|---|
| DID alter treatment, as follows:a | |
| 1. Patient enrolled to a therapeutic protocol at MSKCC | 265 (14) |
| 2. Patient enrolled to a therapeutic protocol at another institution | 15 (1) |
| 3. Patient treated with off-label use of an FDA approved therapy | 43 (2) |
| DID NOT alter treatment, as follows: | |
| 4. Actionable mutation(s) identified, but no therapeutic protocol availableb | 175 (9) |
| 5. Actionable mutation(s) identified, but patient declined participation in, or was ineligible for, available therapeutic protocol | 115 (6) |
| 6. Actionable mutation(s) identified, but patient deteriorated, progressed, or died before results could be used | 176 (9) |
| 7. Actionable mutation(s) identified and therapeutic study available, but patient has not recurred/progressed since MSK-IMPACT result | 339 (18) |
| 8. No actionable mutation identified | 669 (35) |
| 9. Otherc | 135 (7) |
| TOTAL | 1932 |
Eight additional patients were noted to have been treated with therapy for an approved indication based on sequencing results.
This included patients for whom a protocol was identified but the patient was put on a wait list due to slot availability.
On manual review of cases, this includes patients placed on standard therapy, put on an alternate trial that was not genomically matched (i.e. immunotherapy or histology-based), lost to follow-up, and/or placed on targeted therapy for an approved indication.
Physician determination of genomic ‘actionability’ was compared to OncoKB (OncoKB.org), an openly accessible, expertly curated, knowledgebase of somatic variants developed at MSKCC that assigns each variant with a level of evidence corresponding to its actionability [5]. Levels range from 1 to 4. A level 1 alteration is an FDA-recognized biomarker that predicts response to an FDA-approved drug in the patient’s tumor type, a level 2 alteration is a biomarker routinely used to guide prescribing of an FDA-approved drug in the patient’s tumor type or another indication, a level 3 alteration has compelling clinical evidence to support use but neither the biomarker or drug is standard of care, and a level 4 alteration has compelling biologic evidence for use as a biomarker. OncoKB provides annotation at the level of the allele and cancer type such that the actionability varies by the allele mutated within a gene (i.e. BRAF V600E versus V600M) and the tumor type in which a mutation is observed. Variants ranked as levels 1–3 were considered actionable for this analysis. As level 4 alterations are those without clinical evidence supporting their use as predictive biomarkers, they were not considered actionable. Level 4 alterations include several commonly mutated genes such as KRAS. At the time of this analysis, 33 genes had at least one level 1–3 actionable variant (supplementary Table S1, available at Annals of Oncology online). For clinical trial matching, an expanded list of genes was used that included alterations with biologic or anecdotal clinical evidence to support their use as predictive biomarkers. Notably, the decision to screen a patient for a clinical trial was typically made by the individual provider or through referral to the Early Drug Development service. There was no formal molecular advisory board to assist with the interpretation of genomic data.
Results
Characteristics of consented patients
Surveys were returned on 1932 of 9147 (21%) patients who underwent MSK-IMPACT testing during the study period. A total of 49 cancer types were represented with the most common being breast cancer (20%), germ cell tumors (11%), nonsmall cell lung cancer (NSCLC) (6%), endometrial cancer (5%), and esophagogastric cancer (5%). By organ system, genitourinary cancers were the most frequent (21%), followed by breast (20%) and gynecologic cancers (11%) (Table 2). Sequenced samples included primary (52%) and metastatic tumors (48%). Approximately half of the patients were male (47%).
Table 2.
Patient and tumor characteristics
| Characteristics, N (%) | All patients | Therapeutic study type |
|||
|---|---|---|---|---|---|
| Genomically targeted | Immunotherapy | Other | No study | ||
| Sex | |||||
| Female | 1018 (53) | 179 (73) | 109 (52) | 221 (48) | 596 (51) |
| Male | 914 (47) | 66 (27) | 101 (48) | 235 (52) | 570 (49) |
| Sample typea | |||||
| Primary | 1003 (52) | 95 (39) | 89 (42) | 227 (50) | 645 (55) |
| Metastasis | 929 (48) | 150 (61) | 121 (58) | 229 (50) | 521 (45) |
| Sequencing platforma | |||||
| IMPACT 341 | 1297 (67) | 195 (80) | 146 (70) | 322 (71) | 745 (64) |
| IMPACT 410 | 635 (33) | 50 (21) | 64 (31) | 134 (29) | 421 (36) |
| Tumor type | |||||
| Genitourinary | 407 (21) | 15 (6) | 45 (21) | 117 (26) | 258 (22) |
| Breast | 393 (20) | 85 (35) | 23 (11) | 84 (18) | 254 (22) |
| Gynecologic | 221 (11) | 38 (15) | 34 (16) | 55 (12) | 117 (10) |
| Gastrointestinal | 143 (7) | 23 (9) | 13 (6) | 36 (8) | 89 (8) |
| Sarcoma | 134 (7) | 7 (3) | 12 (6) | 30 (7) | 82 (7) |
| Lung | 128 (7) | 21 (8) | 21 (10) | 14 (3) | 76 (7) |
| Head and neck | 101 (5) | 13 (5) | 29 (10) | 26 (6) | 48 (4) |
| Skin | 84 (4) | 14 (6) | 15 (7) | 5 (1) | 53 (5) |
| Thyroid | 83 (4) | 13 (5) | 0 (0) | 30 (7) | 41 (4) |
| Hepatobiliary | 82 (4) | 6 (3) | 1 (0) | 24 (5) | 52 (4) |
| Other | 156 (8) | 10 (4) | 31 (15) | 35 (8) | 97 (8) |
| Highest OncoKB levelb | |||||
| 1 | 115 (6) | 41 (17) | 15 (7) | 22 (5) | 52 (5) |
| 2 | 185 (10) | 53 (22) | 23 (11) | 44 (10) | 90 (8) |
| 3 | 410 (21) | 108 (44) | 38 (18) | 87 (19) | 231 (20) |
| Unranked | 1222 (63) | 43 (18) | 134 (64) | 303 (66) | 793 (68) |
| Total | 1932 | 245 | 210 | 456 | 1166 |
The sample type and sequencing platform data are specific to the sample inquired about in the survey.
If a patient had multiple samples sequenced, the highest OncoKB level is recorded here.
Characteristics of consenting physicians
Responses were received from 57% (146/258) of clinicians emailed, 55% of whom were male (80/146). Represented specialties included medical oncology (67%), pediatric oncology (8%), surgery (6%), radiation oncology (5%), neuro-oncology (5%), interventional radiology (5%), and gynecologic oncology (3%). Clinical experience, determined by years since completion of specialty training, varied with an average of 14 years in practice (range: 2–41 years). The average number of questionnaires completed per physician was 13 (median: 7, range: 1–202).
Genomic results
As classified by expert curators (i.e. OncoKB), 37% (710/1932) of all patients harbored at least one actionable alteration. The diseases with the highest frequency of actionable alterations were melanoma (76%, 28/37), glioma (66%, 21/32), breast cancer (62%, 244/393), endometrial cancer (56%, 55/98), and thyroid cancer (54%, 45/83). Tumor types accounting for the largest number of actionable alterations, unadjusted for frequency in the larger cohort, included breast cancer (34%, 244/710), endometrial cancer (8%, 55/710), NSCLC (7%, 50/710), and thyroid cancer (6%, 45/710).
Clinical use of sequencing results
Physicians indicated that MSK-IMPACT results altered management in 21% (331/1593) of patients in need of a treatment change by the time of survey response (i.e. excluding the 339 patients without disease recurrence or progression) (Table 1). Enrollment to a therapeutic protocol at MSKCC was cited as the most common way in which treatment was affected, followed by off-label treatment with a commercially available therapy. In eight additional patients, treatment was noted to be altered for another reason, including identification of an alteration for which there was approved therapy for that indication or reclassification of tumor type based on sequencing results.
At the time of data analysis, 13% of the entire cohort (245/1932) had been enrolled to at least one genomically matched trial at MSKCC. Of these, 69% (169/245) were enrolled after MSK-IMPACT profiling resulted, indicating that prior testing may have been used to guide enrollment in some patients. Of those patients in need of a treatment change and with alterations deemed actionable by experts, 43% (245/564) were treated on a genomically matched study. The median time from MSK-IMPACT report to trial matching was 5 months (range: 0–28), excluding patients who underwent sequencing after matching. Thirty-eight patients had more than one alteration qualifying as a match on the study to which they were enrolled. Eight patients had more than one alteration used to match them to two or more trials. Almost all the matched studies were early phase trials (98%, 284/291), with the majority evaluating targeted therapy alone (63%, 184/291). In contrast, patients without the presumptive biomarker that were enrolled to unmatched targeted therapy were more likely to receive combinations with another agent or radiotherapy (71%, 139/195 trials). Possible reasons include the presence of trials using targeted therapy to treat resistance mechanisms (for example, the addition of everolimus to exemestane to prevent endocrine resistance in hormone-receptor-positive breast cancer), and the added security of additional therapy in biomarker-negative patients felt less likely to respond to single-agent targeted therapy [6].
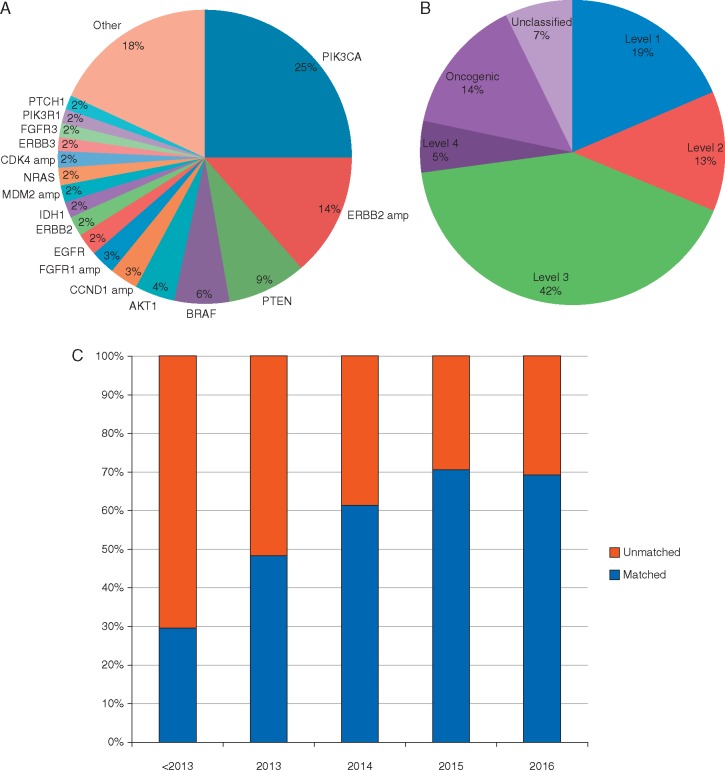
The most common genomic alterations used to enroll patients to matched therapy were PIK3CA mutations (25%), ERBB2 amplifications (14%), PTEN loss-of-function alterations (9%), BRAF mutations (6%), and AKT1 mutations (4%) (Figure 1A). The majority of matching alterations was classified as actionable by experts (74%), with level 3 being the most prevalent level of evidence (42%) (Figure 1B). Matching alterations included somatic mutations, copy number changes, and structural rearrangements. The rate of genomic matching to targeted therapy has increased over time, likely due to increasing use of panel testing and a rising number of trials testing biomarker-driven hypotheses (Figure 1C). Finally, the presence of an actionable alteration appeared to influence trial selection. Of the 337 patients with alterations expertly curated as actionable who went on any therapeutic trial, 60% (202/337) were treated on a genomically matched study.
Figure 1.
Genomically matched clinical trials. (A) Characterization of alterations used to genomically match patients to targeted therapy across tumor types. (B) Distribution of matching alterations by level of evidence. (C) Rate of genomic matching to clinical trials over time.
Knowledge of consenting physicians
Among those patients in whom treatment was not altered, physicians indicated the presence of an actionable alteration in 55% (805/1474). Of these patients, only 45% (362/805) had an alteration ranked as levels 1–3 to support this claim, suggesting that physicians may have a less stringent definition of actionability than expert curators (Figure 2). Further evaluation of patients for whom physicians reported an actionable alteration but expert curators did not revealed that 66% (291/443) had a variant with biologic but not clinical evidence (level 4) or a variant of unknown significance (VUS) in a gene with at least one level 1–3 alteration. By comparison, only 12% (83/669) of patients in whom physicians indicated there were no actionable alterations had a level 1–3 alteration (Figure 2). Of these 83 patients, 22% (18/83) had level 1 alterations including HER2 amplification (n = 17) or EGFR mutation (n = 1), suggesting that physicians chose ‘not actionable’ to indicate that MSK-IMPACT had not yielded additional actionable alterations beyond what had been previously identified through routine testing. Physicians correctly identified all actionable alterations considered standard of care (but not FDA-approved) biomarkers for approved drugs in the relevant indication. Of those patients with alterations ranked as having potential therapeutic importance (levels 1–3), 81% (362/445) were deemed actionable by physicians. When including the 18 alterations likely excluded due to parallel testing, this rate increases to 85%, suggesting that clinicians can identify actionable alterations in the majority of patients who harbor them.
Figure 2.
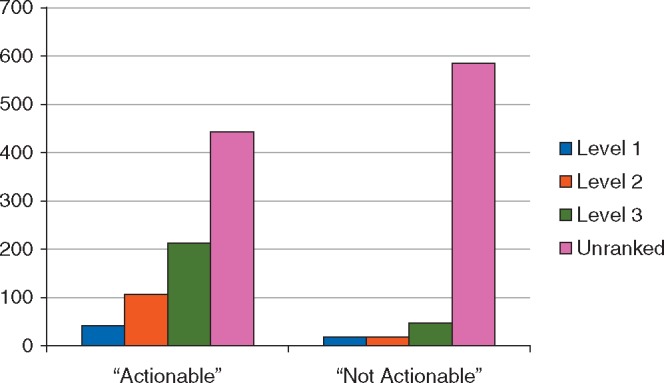
Physician determination of actionability stratified by level of evidence.
Discussion
A limited number of genomic biomarkers are currently used to successfully guide routine treatment in several cancers [7]. Efforts to broaden the benefits of this personalized approach have led to the adoption of large panel NGS within academic institutions and by commercial laboratories [1–3, 8]. Nevertheless, the utility of this approach and physicians’ ability to interpret the complex data these tests generate remains unknown. Clinical trials designed to measure the outcomes of patients treated in this manner have yielded mixed results [9, 10]. This report represents the first effort to prospectively elucidate physicians’ interpretations of sequencing results generated from active patients, compare their interpretations to expert consensus, and track the ultimate treatments undertaken. We found that although physician and expert assessment differed in meaningful ways, clinicians demonstrate substantial awareness of the genes and mutations associated with potential actionability and report using this knowledge to inform management in approximately one in five patients.
Prior work by Gray et al. [11] provided insight into the self-confidence of physicians regarding their knowledge of genomics and its clinical application. They found that the average clinician was ‘somewhat’ confident in his or her genomic knowledge but a sizable minority (22%) was not comfortable using genomic information. Importantly, this study was performed prior to the introduction of large panel NGS at the center and we do not know how closely results align to the ultimate adoption of sequencing data. Moreover, as the field of cancer genomics has matured over the past decade and this testing is increasingly utilized, clinicians’ understanding of genomics has likely grown.
Here we show that when compared to an expert consensus, physicians accurately identify actionable alterations in >80% of patients. This number likely underestimates the true rate as some physicians did not acknowledge standard-of-care alterations (HER2 amplification, EGFR mutation) previously identified through routine testing. Furthermore, while OncoKB annotates alterations at the allele and tumor level, it does not consider co-mutations that may alter the actionability. For example, one clinician noted that presence of an NRAS hotspot mutation made a concurrent BRAF mutation nonactionable. Moreover, actionability is a dynamic concept that changes as new data is generated. Although AKT1 E17K was not considered actionable by experts when this sequencing initiative began, it later became the basis of a basket study (ClinicalTrials.gov, NCT01226316) and based on preliminary results was reclassified as actionable [12]. In general, physicians were more liberal with their definition of actionability compared to experts. Approximately half of patients deemed actionable by clinicians were not categorized as such by experts. Most discrepant interpretations occurred in patients whose tumors had a variant with biologic significance but no clinical data, or a VUS in a gene with other known actionable variants. In some circumstances, biologic evidence may be sufficient for enrollment on a trial and therefore identification of these alterations can alter management. This reinforces the importance of using expert databases that aid in the interpretation of variants, understanding that some circumstances may make it appropriate to act on alterations with less evidence, especially in the context of a clinical trial that will ultimately feed back into these knowledgebases. As our understanding of the effect and clinical consequence of genomic alterations grows, these resources will become especially important for clinicians.
Using our survey, physicians reported that sequencing altered treatment in approximately one in five patients. This was most often through enrollment to a clinical trial at our institution. Auditing of these records revealed that in addition to those patients enrolled to a genotype-matched trial, several were treated on unmatched trials including immunotherapy studies. This indicates that physicians utilized knowledge of hypermutation or the absence of actionable genomic targets to select immunotherapy studies over unmatched targeted therapy. Physicians also reported that sequencing altered management through identification of a resistance mutation, recategorization of tumor type, and treatment with standard targeted therapy.
Ultimately we found that 13% of patients in our cohort composed primarily of tumor types for which tumor sequencing is not routine were treated on a genomically matched clinical trial, somewhat higher than the 4–5% rate previously reported with similar efforts [3, 13, 14]. Several factors may explain this difference. First, the two largest efforts utilized small gene panels, do not provide full exon coverage of each gene, and do not detect copy number alterations or structural rearrangements. Evolution of clinical trial design including adoption of basket studies and expansion cohorts has also improved the ability to offer patients genomically matched therapy. Our center has also invested in automated alert systems that notify treating physicians of study availability. The result of these efforts is that approximately half (43%) of patients with actionable alterations in need of a treatment change were enrolled to genomically matched studies. Since this survey was completed, we incorporated expert annotations from OncoKB into the static and dynamic web-based molecular pathology reports.
Even the 13% match rate likely underestimates the true rate of genomic matching in this cohort. Nearly one in five patients did not require a treatment change and therefore were not eligible to enroll to a matched trial. As time elapses and patients progress through standard therapy, sequencing results may provide additional therapeutic options. Physicians also reported using information beyond individual genomic variants to make therapeutic decisions. For example, MSK-IMPACT has been clinically validated to report microsatellite instability and can be used to infer mutation rate. However, for this analysis, we did not consider patients with these patterns of genomic alteration enrolled to immunotherapy as matched. Additionally, identification of germline alterations on sequencing can inform treatment. Eight patients were matched to a trial based on germline BRCA1/2 alterations detected by MSK-IMPACT. Here we only considered matches based on somatic alterations because pathologic germline variants are only returned to patients who specifically consent to this secondary analysis. Finally, our cohort was highly enriched for patients with tumor types in which genomic sequencing is not standard and therefore is expected to have a lower rate of actionability than the overall cancer population.
As clinical tumor sequencing becomes more prevalent, physicians are increasingly charged with interpreting these results and applying this knowledge to maximize the treatment options for individual patients. This study reinforces that physicians at our center have become skilled at identifying actionable alterations, including both standard of care biomarkers and those under investigation. Importantly, this study was conducted at a comprehensive cancer center where testing has become increasingly utilized. A multifaceted approach including the use of expertly curated databases will be important in disseminating this knowledge to community oncologists.
Funding
MSK Cancer Center Support Grant (P30 CA008748); National Institutes of Health awards T32-CA009207; Cycle for Survival; and Marie-Josée and Henry R. Kravis Foundation.
Disclosure
DMH: Atara Biotherapeutics (consulting), CytomX (consulting), Chugai (consulting), Puma Biotechnology (research funding), AstraZeneca (research funding) and LOXO (research funding). All remaining authors have declared no conflicts of interest.
Supplementary Material
References
- 1. Sholl LM, Do K, Shivdasani P. et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight 2016; 1: e87062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stockley TL, Oza AM, Berman HK. et al. Molecular profiling of advanced solid tumors and patient outcomes with genotype-matched clinical trials: the Princess Margaret IMPACT/COMPACT trial. Genome Med 2016; 8: 109.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cheng DT, Mitchell TN, Zehir A. et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor. Mol Oncol J Mol Diagn 2015; 17: 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hyman DM, Solit DB, Arcila ME. et al. Precision medicine at Memorial Sloan Kettering Cancer Center: clinical next-generation sequencing enabling next-generation targeted therapy trials. Drug Discov Today 2015; 20: 1422–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chakravarty D, Gao J, Phillips S. et al. OncoKB: a precision oncology knowledge base. J Clin Oncol 2017; 1: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baselga J, Campone M, Piccart M. et al. Everolimus in postmenopausal hormone-receptor–positive advanced breast cancer. N Engl J Med 2012; 366: 520–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hyman DM, Taylor BS, Baselga J.. Implementing genome-driven oncology. Cell 2017; 168: 584–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frampton GM, Fichtenholtz A, Otto GA. et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013; 31: 1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tourneau CL, Delord J-P, Gonçalves A. et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol 2015; 16: 1324–1334. [DOI] [PubMed] [Google Scholar]
- 10. Massard C, Michiels S, Ferté C. et al. High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: results of the MOSCATO 01 trial. Cancer Discov 2017; April 1 [Epub ahead of print] doi:10.1158/2159-8290.CD-16-1396 [DOI] [PubMed] [Google Scholar]
- 11. Gray SW, Hicks-Courant K, Cronin A. et al. Physicians’ attitudes about multiplex tumor genomic testing. J Clin Oncol 2014; 32: 1317–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hyman DM, Smyth LM, Donoghue MTA. et al. AKT inhibition in solid tumors with AKT1 mutations. J Clin Oncol 2017; May 10 [Epub ahead of print] doi:10.1200/JCO.2017.73.0143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Beltran H, Eng K, Mosquera JM. et al. Whole-exome sequencing of metastatic cancer and biomarkers of treatment response. JAMA Oncol 2015; 1: 466–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Meric-Bernstam F, Brusco L, Shaw K. et al. Feasibility of large-scale genomic testing to facilitate enrollment onto genomically matched clinical trials. J Clin Oncol 2015; 33: 2753–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.