Abstract
Background
A more accurate prognosis for non-small-cell lung cancer (NSCLC) patients could aid in the identification of patients at high risk for recurrence. Many NSCLC mRNA expression signatures claiming to be prognostic have been reported in the literature. The goal of this study was to identify the most promising mRNA prognostic signatures in NSCLC for further prospective clinical validation.
Experimental design
We carried out a systematic review and meta-analysis of published mRNA prognostic signatures for resected NSCLC. The prognostic performance of each signature was evaluated via a meta-analysis of 1927 early stage NSCLC patients collected from 15 studies using three evaluation metrics (hazard ratios, concordance scores, and time-dependent receiver-operating characteristic curves). The performance of each signature was then evaluated against 100 random signatures. The prognostic power independent of clinical risk factors was assessed by multivariate Cox models.
Results
Through a literature search, we identified 42 lung cancer prognostic signatures derived from genome-wide expression profiling analysis. Based on meta-analysis, 25 signatures were prognostic for survival after adjusting for clinical risk factors and 18 signatures carried out significantly better than random signatures. When analyzing histology types separately, 17 signatures and 8 signatures are prognostic for adenocarcinoma and squamous cell lung cancer, respectively. Despite little overlap among published gene signatures, the top-performing signatures are highly concordant in predicted patient outcomes.
Conclusions
Based on this large-scale meta-analysis, we identified a set of mRNA expression prognostic signatures appropriate for further validation in prospective clinical studies.
Keywords: non-small-cell lung cancer, prognostic gene signatures, meta-analysis
Introduction
Non-small-cell lung cancer (NSCLC) accounts for ∼85% of known lung cancer cases. Current guidelines for treating NSCLC are largely based on clinical and pathological staging systems, as well as additional factors such as smoking history and gender [1, 2]. However, clinical outcome varies widely—although roughly 60% of patients with stage I/II NSCLC will never recur after surgical resections, 40% will ultimately die of the disease. Identifying those patients at a higher risk for recurrence could help ‘tailor’ specific treatment plans for individual patients. In the last decade, large-scale genomic profiling technology has been used to obtain genome-wide mRNA expression levels in lung cancer, and many mRNA expression signatures have been developed for NSCLC prognosis (supplementary Table S1, available at Annals of Oncology online). However, there is a long-standing debate on the reproducibility, robustness and real clinical utility of these expression signatures [3, 4]; and prospective clinical studies are needed to validate these prognostic biomarkers/signatures before they are incorporated into clinical practice. Thus, determining which prognostic signatures are suitable for expensive and long-term prospective clinical validation becomes an important question. Consequently, the goal of our current study was to perform a systematic review and meta-analysis for published lung cancer prognostic signatures derived from the genome-wide studies, to identify the most promising ones for further prospective clinical validation. The main criteria of the evaluations were as follows: (i) the ability of the signature to predict patients’ survival outcomes across different studies; (ii) the ability of the signature to perform better than random signatures and (iii) the independent predictive power on top of clinical variables such as stage, age, and smoking history.
Although several studies have reviewed prognostic signatures in lung cancer, no systematic evaluations have yet been carried out by individual or meta-analysis. In this study, we carried out a comprehensive evaluation of 42 lung cancer prognostic signatures tested in 15 lung cancer datasets (summarized in supplementary Table S2, available at Annals of Oncology online) with 1927 NSCLC patients, based on a systematic literature search (see Methods). In this study, a gene signature refers to a gene list itself and does not include how the genes were combined numerically (using statistical models). The prediction performance of each gene signature was determined based on the gene list itself, the statistical methods used for prediction, and study populations. Different statistical models were used to combine mRNA expression levels of genes in the original signature studies to predict prognosis. Although the comparisons of effectiveness of those statistical models and analysis procedures are interesting, the focus of this study is to compare which gene lists are most promising for further prospective clinical validations. To ensure systematic and unbiased comparisons among different gene signatures, we evaluated all 42 signatures with the same statistical methods {principal component analysis (PCA) or supervised principal component analysis (SuperPCA) [5]} on the same patient data.
Methods
Selection criteria of gene signatures
To identify the reported lung cancer prognosis signatures, we first searched the PubMed database using the Boolean phrase ‘prognostic gene expression signature AND lung’. We also checked recent reviews on this topic [3, 6]. In this study, we considered signatures that were derived from genome-wide mRNA expression profiling studies and were shown to be associated with patient survival outcomes by validation using independent datasets. Considering the limited feasibility of gene signatures containing too many genes, we included only signatures containing no more than 500 genes. Our final 42 signatures were collected from 34 studies published between 2001 and 2012 (supplementary Table S1, available at Annals of Oncology online).
Patient cohorts
To obtain patient cohorts, we did a comprehensive search for lung cancer studies with genome-wide mRNA expression data from human tumor tissues in literatures and public websites, such as Gene Expression Omnibus (GEO) and the Microarray Data Management System (caArray). Using these criteria, we identified 60 datasets, among which 15 provided patient survival information for more than 50 NSCLC patients. These 15 datasets were then used as testing sets to evaluate the signatures. Nine of the 15 datasets with relatively large numbers (>100) of patients were used as training sets for supervised models. Supplementary Figure S1, available at Annals of Oncology online shows the Survival curve, and supplementary Tables S2–S4, available at Annals of Oncology online provide details in for each of these datasets.
Statistical analysis
All mRNA expression data were preprocessed as described in supplementary methods, available at Annals of Oncology online. To evaluate signatures’ prognostic values, we applied both SuperPCA and unsupervised PCA. Three different survival association metrics were used to evaluate each prognostic prediction model: hazard ratios (HRs) estimated by the Cox proportional-hazards model [7], the time-dependent receiver-operating characteristics (ROC) curves [8], and the Concordance Index (C.index). Meta-analysis was then conducted on the multiple independent validation sets for each prognostic signature to summarize its overall performance.
To see which of the 42 published signatures predicted significantly better than random signatures, we generated 100 gene sets with 50 genes each as random signatures and calculated their survival associations using the same training and testing strategies. A linear mixed-effect model (see supplementary methods, available at Annals of Oncology online) was then used to compare the published signatures versus the 100 random ones.
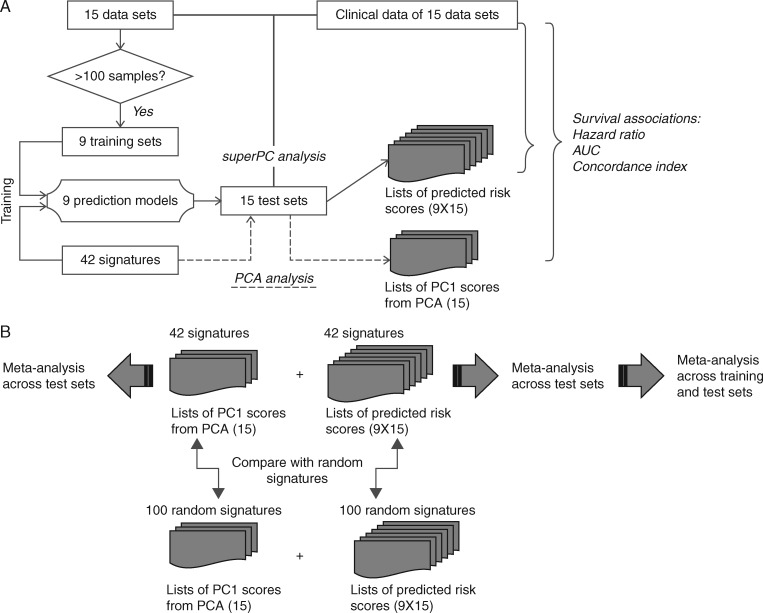
Figure 1 illustrates the workflow of the training and validation procedures used in this study.
Figure 1.
Flow chart summarizing the strategy used for the development (A) and validation (B) of the risk prediction models obtained from published gene signatures. Basically, 9 different superPCA predictors were trained for each of the 42 signatures using the nine training sets, respectively. The performance of a specific superPCA predictor was then evaluated based on a subset of 15 available data sets. The criteria to select the subset were as follows: (i), the data set was not the training set and (ii) the dataset was not used for developing the specific signature used in this superPCA predictor. Detailed descriptions of procedures and data analysis are provided in the supplementary material, available at Annals of Oncology online.
Results
Identification of 42 published gene signatures
We identified 42 published lung cancer prognostic signatures (supplementary Table S1, available at Annals of Oncology online) derived from genome-wide expression profiling analysis, containing ≤500 gene probes. These signatures were derived from a number of NSCLC histology subtypes, including adenocarcinomas (ADCs), squamous cell carcinomas (SCCs), mixed lung cancer types, and other (non-lung) cancer types, e.g. a 94-gene malignancy-risk gene signature [9] that was originally derived from breast cancer samples and later shown to be prognostic for NSCLC. Different microarray platforms, such as Affymetrix, Agillent and customized arrays, were used to measure gene mRNA expressions. Supplementary Table S1, available at Annals of Oncology online summarizes the 42 signatures in detail, including histology types, total number of probes used, microarray platforms, and literature citations.
Meta-analysis to evaluate the prediction performances of the signatures
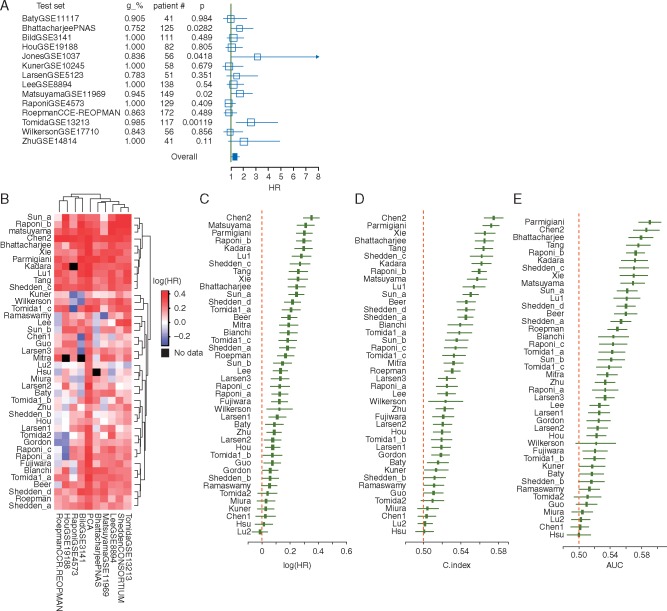
The performance of the same prediction model derived from the same signature can vary depend on the statistical algorithm used, the quality of the clinical outcome data provided, and the similarity between the training and testing patient datasets. For example, the prediction model derived from the same training dataset (the Director’s Consortium dataset) and gene list [10] showed different prognostic effects with the 14 datasets tested, with high- versus low-risk group HRs, respectively, ranging from 0.817 to 3.113 (Figure 2A). Thus, evaluating signatures based on any single testing set alone may introduce bias into the final conclusion. To overcome this problem, we used meta-analysis to summarize the prediction performance over all testing datasets (see Methods). The overall performance of the Shedden et al. signature [10] calculated from meta-analysis showed significant prediction for poor survival in high-risk patients (HR = 1.80; 95% CI 1.10–2.93). In Figure 2B, a heat map summarizes the meta-estimates of HRs for all signatures derived from different training sets. Sixteen signatures showed prognostic effects by using any of the nine training sets or by using unsupervised PCA analysis. Comparison of the signature models derived from different training data in Figure 2B demonstrated that some training sets (e.g. the Tomida dataset [11]) provided better training models for most signatures, while others (e.g. the RoepmanCCR dataset [12]) were less effective. One possible reason is that those training sets can map to a larger number of genes in the signatures. In fact, the Tomida dataset contained at least 80% genes for all signatures, but the RoepmanCCR dataset [12] did so for only 33 of 42 signatures. Similar pattern of model performances were observed for the summarized C.indexes and AUC (supplementary Figure S2, available at Annals of Oncology online). Finally, to get a single unbiased score for each gene signature, we summarized the prognostic performance of all the survival models that were derived from different training sets using a meta-analysis approach (supplementary methods, available at Annals of Oncology online) and found that 34 out of the 42 gene signatures could significantly separate high and low risk groups (with HR lower 95% CI >0, and P < 0.05, Figure 2C). The top-ranked signatures identified by HR, C.index and AUC (Figure 2C–E) are very similar. Therefore, to avoid ambiguity, we will present only our results based on HRs in the following sections.
Figure 2.
Evaluation of 42 gene expression signatures via random effect meta-analysis. (A) Prognostic performance of the Shedden et al. gene signature using the Director’s Consortium dataset as the training set (10). Hazard ratios (HRs) between the two predicted risk groups varied widely when using different data as the test sets. (B and C) Heatmap of the HR and Concordance index (C.index) estimates based on meta-analysis using different prediction models and gene signatures. SuperPCA models were trained by 1 of the 9 training sets, and tested on the remaining 14 data sets. The name of each training set is shown in the column title column. The PCA model was developed using the first principal component of the signature in the test set. (D) Meta-analysis results from all SuperPCA models using all pairs of training and test sets. In (B) and (D), the x-axis denotes the HR of survival differences between two predicted risk groups, with black squares denoting the summarized HR from different training and testing strategies, and the black solid line denoting the confidence interval. For each gene signature, datasets used in signature development were excluded from meta-analysis.
About half of the gene signatures showed significantly better prediction than random gene sets
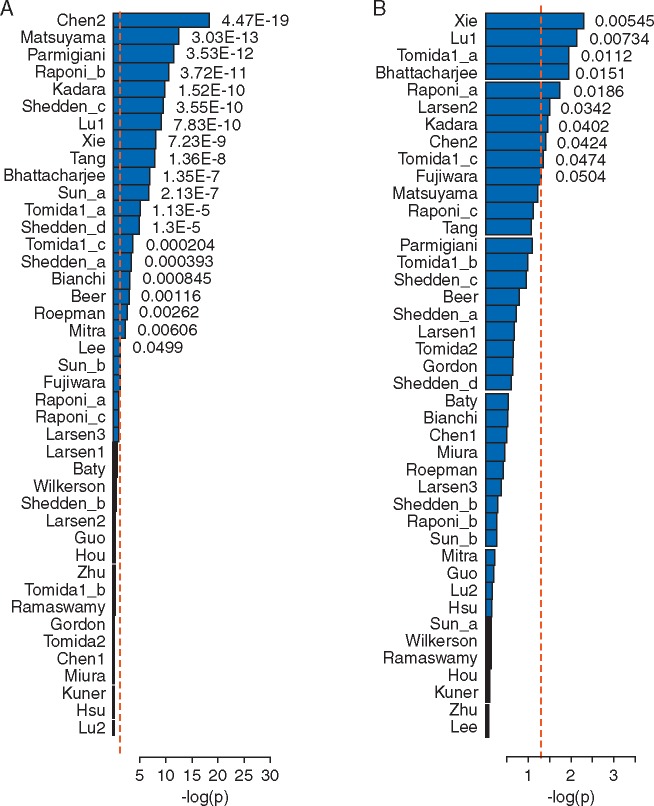
When compared with random signatures, 20 out of the 42 published signatures (48%) significantly outperformed (P <0.05) random signatures using the superPCA modeling approach (Figure 3A). It should also be noted that several of the signatures that significantly outperformed the random signatures actually contained a relatively small number of genes. In this study, we also generated random signatures with 100 genes and the results are similar.
Figure 3.
Over 50% of published signatures perform better than random signatures when using SuperPCA prediction models (A) and 20% perform better when using PCA prediction models (B). The x-axis denotes the P-values comparing gene signatures against 100 random signatures, based on linear mixed-effect models. Red and black bars indicate signatures that carried out better or worse than random signatures, respectively.
Our results showed that the unsupervised PCA models had better prognosis performance than supervised PCA models, but fewer unsupervised models were significant when compared with models from random signatures (Figure 2C versus Figure 3B). Figure 3B shows that only 9 out of the 42 signatures carried out significantly better than random signatures using the PCA-based method, in contrast to 20 out of the 42 signatures for superPCA. Therefore, unsupervised approaches are likely to overestimate the performance of random gene signatures, which diminished the differences between the real signatures and the random signatures. One reason for overestimation is that unsupervised methods can only divide patients into two groups. To assign high- versus low-risk groups, the traditional protocol compares the median survival times between the two groups and assigns the one with lower-median survival as the high-risk group. In this way, the same survival information was used during the modeling stage and again during the evaluation stage, leading to overrating of the performances of the unsupervised models. In contrast, the supervised approach largely reduces the risks of overestimation by using independent training and testing datasets. Therefore, the remaining analysis and evaluation in this study will be based on supervised approaches.
Multivariate analysis adjusting for clinical variables
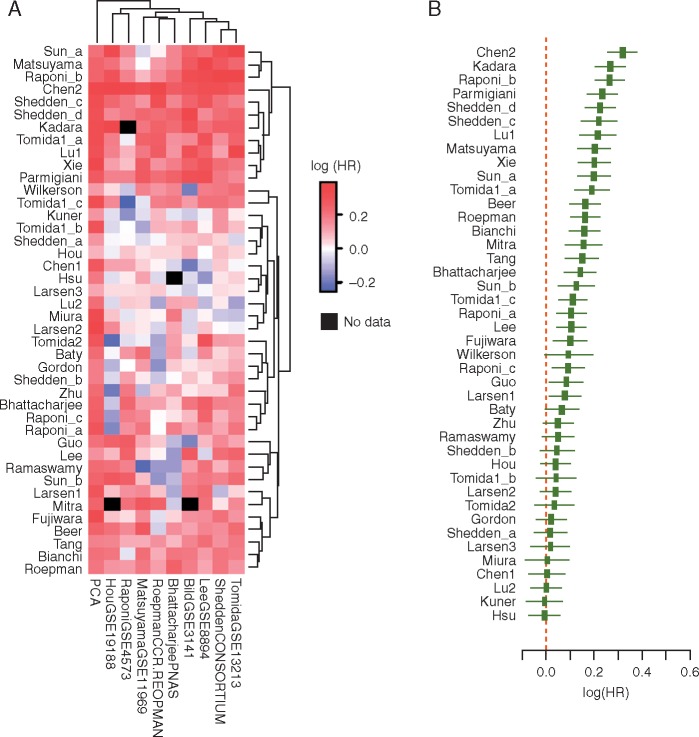
To develop prognostic signatures having potential clinical applicability, it was important to test whether each signature had predictive power in addition to various clinical prognostic factors. Therefore, we carried out multivariate analysis, adjusting for several clinical factors, including tumor histology, tumor stage, patient age/gender, and smoking history. As observed in previous review papers, most public datasets did not provide complete clinical information and the missing clinical variables were excluded from the multivariate analysis. Supplementary Table S3, available at Annals of Oncology online lists the available clinical factors provided and used in this study. Applying the same meta-analysis approach as above, we showed that most prognostic signature models remained significant after adjusting for clinical factors (Figure 4A). Indeed, the summarized HR showed that 29 out of the 35 signatures that were prognostic in the univariate analysis remained significant in multivariate analysis (Figure 4B).
Figure 4.
Meta-analysis results for clinical risk factor-adjusted prognostic power. (A) Heatmap of the meta-analysis results based on different prediction models and gene signatures. SuperPCA models were trained by one of the nine training sets and tested on the remaining 14 datasets. The names of the training sets are shown in the column titles. The PCA model was developed using the first principal component of the signature in the test set. (B). Meta-analysis results from all SuperPCA models using any one of the training sets and any one of the test sets. The x-axis denotes the HR of survival differences between the two predicted risk groups. Black squares denote the summarized HR from different training and testing strategies, while the black broken solid line denotes the confidence interval. For each gene signature, datasets used in signature development were excluded from meta-analysis.
Prognosis for different histology types
ADC and SCC are two major NSCLC histology subtypes having dissimilar molecular foundations in tumor pathogenesis and progression [13]. Several research groups have suggested using subtype-specific molecular biomarkers to investigate their biological differences and develop more accurate predictors. For over 23 of the 42 signatures collected in our studies, there were clear designations of the applicable cancer subtypes, or the signature was derived from one particular histology type (supplementary Table S1, available at Annals of Oncology online). We thus carried out meta-analysis again on ADC and SCC patient subgroups (supplementary Figure S3A–D, available at Annals of Oncology online), revealing more prognostic gene signatures in ADC than SCC NSCLC patients. In total, there were 17 (45%) signatures that carried out significantly better (P value <0.05) than random signatures for the ADC patients (supplementary Figure S3B, available at Annals of Oncology online), while 8 (19%) useful prognostic signatures were found for the SCC patients (supplementary Figure S3D, available at Annals of Oncology online). Interestingly, some signatures derived from ADC-only datasets also provided good predictions for SCC patient prognosis (Shedden_c, HR = 1.72, 95% CI: 1.56–1.89, q = 5.5E−14; Tang, HR = 1.39, 95% CI: 1.27–1.53, q = 2.6E−6; and Kadara, HR = 1.67, 95% CI: 1.53–1.81, q = 2.7E−13). Table 1 summarizes all the signatures significantly predictive for either ADC or SCC patients. The detailed gene information for these signatures was listed in (supplementary Table S5, available at Annals of Oncology online).
Table 1.
Top signatures for ADC and SCC patients
| No. of genes | HR (95% CI) | Q | P value | |
|---|---|---|---|---|
| ADC patients | ||||
| Chen2 | 92 | 1.69 (1.56–1.83) | 63.29 | 5.54E−16 |
| Shedden_c | 452 | 1.72 (1.56–1.89) | 51.73 | 5.55E−14 |
| Kadara | 5 | 1.67 (1.53–1.81) | 59.15 | 2.70E−13 |
| Tomida1_a | 23 | 1.41 (1.30–1.52) | 62.49 | 4.63E−08 |
| Lu1 | 62 | 1.47 (1.33–1.63) | 81.93 | 8.68E−08 |
| Xie | 59 | 1.54 (1.42–1.66) | 67.27 | 1.15E−07 |
| Bianchi | 10 | 1.46 (1.32–1.62) | 81.69 | 3.41E−07 |
| Tang | 18 | 1.39 (1.27–1.53) | 45.50 | 2.58E−06 |
| Matsuyama | 169 | 1.42 (1.30–1.54) | 65.50 | 8.94E−06 |
| Sun_a | 46 | 1.34 (1.24–1.45) | 53.51 | 2.69E−05 |
| Bhattacharjee | 151 | 1.39 (1.27–1.51) | 47.96 | 0.000113 |
| Parmigiani | 14 | 1.33 (1.22–1.45) | 54.03 | 0.001416 |
| Raponi_b | 45 | 1.30 (1.21–1.41) | 49.89 | 0.001657 |
| Beer | 95 | 1.28 (1.18–1.38) | 62.77 | 0.002085 |
| Tomida1_c | 11 | 1.25 (1.15–1.35) | 75.89 | 0.00560 |
| Fujiwara | 70 | 1.25 (1.13–1.38) | 104.52 | 0.0110 |
| Sun_b | 43 | 1.25 (1.15–1.36) | 82.50 | 0.0143 |
| SCC patients | ||||
| Shedden_d | 332 | 1.41 (1.25–1.59) | 84.41 | 2.19E−11 |
| Raponi_b | 45 | 1.29 (1.15–1.44) | 50.99 | 1.54E−05 |
| Kadara | 5 | 1.21 (1.10–1.34) | 37.02 | 2.96E−05 |
| Roepman | 66 | 1.27 (1.13–1.44) | 65.38 | 3.40E−05 |
| Parmigiani | 14 | 1.23 (1.12–1.36) | 48.70 | 6.86E−05 |
| Chen2 | 92 | 1.18 (1.07–1.30) | 57.93 | 0.000130 |
| Tang | 18 | 1.18 (1.07–1.29) | 24.98 | 0.000444 |
| Mitra | 4 | 1.20 (1.05–1.37) | 47.30 | 0.00581 |
Information included are number of genes in the signatures, meta-estimated hazard ratios (HR), and 95% confidence intervals for the predicted high risk groups, Q-statistics for heterogeneity from meta-analysis, and P-values for comparison to random signatures.
Discussion
In this work, we provided a comprehensive systematic review and meta-analysis for a wide range of published lung cancer prognostic signatures. Although several previous reviews [3, 4, 6] summarized some microarray-based gene signatures, none of them systematically compared the prognostic performances of published signatures. Compared with previous studies, this study completed the following: (i) carried out a systematic literature search for both signatures and datasets, thereby yielding a comprehensive collection of lung cancer prognostic signatures; (ii) evaluated the signatures in a large collection of training and testing datasets, using meta-analysis for an objective and unbiased comparison; (iii) compared the published signatures with sets of random ones and (iv) studied the usefulness of the signatures in concert with known clinical factors. We compared 42 signatures that were validated by at least one independent patient data set in the original paper. This is the largest number of lung cancer prognostic signatures ever evaluated, tested on 1927 patient samples from 15 lung cancer genome-wide mRNA expression studies.
As show in supplementary Figure S1 and Tables S3 and S4, available at Annals of Oncology online, these 15 lung cancer datasets are heterogeneous in terms of patient survival outcomes, histology, and tumor stage. In addition, the diversity of datasets collected from different study cohorts, countries, and microarray platforms allowed us a comprehensive evaluation of the lung cancer prognostic signatures. The heterogeneity of the datasets is a potential issue for meta-analysis, so we used the random-effect model for meta-analysis, which assumes the prognostic effects of a specific signature in different datasets are different. Finally, for each gene signature, the performances were also evaluated in either ADC-only or SCC-only patient cohorts, as some signatures were developed for a specific histology type of either ADC or SCC. We also carried out multivariate analysis to adjust for other clinical variables, including stage.
In this study, both P-value (that compares the gene signature with a set of random signatures) and the HR were used to evaluate the performance. In addition, concordance scores and time-dependent ROC curves were also used to evaluate the performance.
The purpose of this study is to select the most promising prognostic gene lists for further expensive prospective clinical validations, but not to reproduce the results of the original publications. The prognostic power of a gene signature depends on two factors: the set of genes and how the measurements from the gene set are combined. Ideally, the best evaluation is to implement the algorithm associated with each gene signature, and then evaluate the performance. However, it is very hard and sometime impossible to reproduce all the algorithms in published gene signatures. Furthermore, based on our experiences, how the measurements from the gene set are combined is usually less important when the number of genes in the signature is very large. So, we decided to use one method (supervised PCA) to represent the supervised approaches and one method (PCA) to represent unsupervised approaches. This standard procedure can lead to systematic and unbiased comparisons among different gene signatures for the selection of the most promising gene lists for clinical testing. However, it is possible that a set of genes has no prognostic power when combined using PCA or supervise PCA but does have it under another technique. Thus the conclusion that there is (or is not) prognostic power in this study might be limited. The focus on this study is to evaluate published signature lists using the same statistical methods and same datasets. Further studies that focus on the evaluation of different statistical methods and/or patient cohorts will be important for the field.
A recent review by Venet et al. [14] on prognostic signatures in breast cancer claimed that most signatures are no more significantly associated with survival than randomly generated ones, and even those associated with survival actually owe their high correlations to increased cell proliferation. Venet’s study triggered extensive discussion on the practicability of prognostic gene signatures. We wanted to evaluate whether similar conclusions hold for lung cancer and published lung cancer prognostic signatures. Our study showed that 20 out of the 42 signatures significantly outperformed random signatures using the SuperPCA method, clearly demonstrating the usefulness of gene expression signatures in predicting survival outcomes. In addition to the difference in the studied cancer types, we think there are several factors contributing to the different observations between Venet et al.’s and this study: (i) the conclusions in Venet et al. were based on separate evaluations for three datasets, while this study was based on the meta-analysis results from 15 datasets with a total of 1927 patient samples. The much larger sample size significantly increased the statistical power to detect even slight differences in performance between real versus random signatures; (ii) Venet et al. compared signatures based on HRs only, while our study used HRs, time-dependent ROC, and C.index. Thus, by design, this study provided a more general and unbiased evaluation; and (iii) Venet et al. used only an unsupervised approach, while our evaluations were based on the supervised approaches. We found that unsupervised approaches use the survival outcome to determine patients’ risk groups, which can overestimate the performance of random gene signatures. On the other hand, supervised models with independent training and testing datasets could provide an unbiased estimate of the prognostic performance.
Recent advances in lung cancer targeted therapy (e.g. for EGFR and EML4-ALK fusion mutations) have changed the therapeutic strategy for those patients. The mutation profiles of NSCLC patients play important roles in select the targeted therapies. For example, EGFR mutation is an important marker for gefitinib response [15]. This indicates that using a mutation profile to select therapies is important. However, only a small proportion of NSCLC patients have mutations that could be targeted by a limited number of targeted drugs. Furthermore, for molecular targeting drugs, although the mutation status of the ‘gene target’ is the natural biomarker for the drug, it cannot provide a comprehensive prediction for drug response. For example, although EGFR is the target mutation for tyrosine kinase inhibitors (TKIs), there are a subset of EGFR wild-type patients that have shown benefit from EGFR TKIs in several clinical trials [16–18]. Recently, an mRNA expression-based gene signature has been developed to predict the erlotinib (a type of TKIs) sensitivity in both EGFR-mutant and wild-type patients [19]. Therefore, mRNA expression profiles can provide additional information for targeted therapy response prediction. In addition, for early stage NSCLC patients undergoing potentially curative surgical resection, it is important to be able to predict the patient prognosis, so that patients with higher risk can be managed with more aggressive treatments and those with lower risk could select less aggressive treatment to reduce the side-effects. mRNA expression profiles, but not mutation profiles, have been studied intensively for lung cancer prognosis, so the focus of this study is to evaluate the published mRNA expression prognosis signatures for NSCLC prognosis.
This study focused only on the gene signatures derived from genome-wide microarray studies and did not evaluate genes or biomarkers derived from other assays. The main reason is that the training and testing datasets used in this study were all from microarray studies. The dynamic range and sensitivity of genome-wide microarray platforms may not be good enough to validate some gene signatures that were originally derived from higher resolution technologies, such as qPCR technology. Therefore, we should note that besides the signatures discussed in this study, there are other promising prognostic signatures that have been derived from other platforms and could be considered for further evaluation in prospective clinical studies. In addition to mRNA expression signature, it is also important to compare and integrate with other types of gene signatures, such as protein signatures and gene mutations, for lung cancer prognosis.
Most of published gene signatures were derived from fresh frozen samples which has limited clinical impact. Therefore, for future clinical usage of the gene signatures, validation and application in using CLIA-certifiable assays in FFPE samples are important. In order to apply an mRNA gene signature in clinical practice, the mRNA expression levels of the genes in the signature need to be measured by a CLIA-certifiable assay, such as qPCR or nCounter platform from Nanostring, which usually requires the number of genes to be less than a hundred. When the number of genes in a signature is large, it is hard to translate the signature into a clinical device, so it is important to reduce the number of genes from a molecular signature derived from the genome-wide molecular profiling experiments. Several studies have shown that an appropriate dimension reduction step will great improve the prediction performance [20–22]. We also provide the number of genes for each signature in Table 1 to facilitate the selection of these signatures for clinical use.
In conclusion, we conducted a comprehensive review and meta-analysis of the 42 published mRNA expression prognostic signatures in lung cancer. We found that 17 signatures carried out significantly better than random gene sets and could provide additional prediction power, in addition to complementing known clinical variables for ADC. Of those 42, 8 such signatures were identified for SCC. These signatures provide the following: (i) a statistical basis for developing CLIA-certified tests to help guide patient care and (ii) a biological basis for exploring what are the underlying molecular differences in NSCLCs, which otherwise appear similar, that explain differences in clinical behavior.
Funding
This work was supported by the National Institutes of Health [5R01CA152301, P50CA70907, 5P30CA142543, 1R01GM115473, and 1R01CA172211]; and the Cancer Prevention and Research Institute of Texas [RP120732].
Disclosure
The authors have declared no conflicts of interest.
Supplementary Material
References
- 1.National Comprehensive Cancer Network I. In NCCN Clinical Practice Guidelines in Oncology. Non-small Cell Lung Cancer 2016 (Version 4.2016).
- 2. Tanoue LT. Staging of non-small cell lung cancer. Semin Respir Crit Care Med 2008; 29: 248–260. [DOI] [PubMed] [Google Scholar]
- 3. Subramanian J, Simon R.. Gene expression-based prognostic signatures in lung cancer: Ready for clinical use? J Natl Cancer Inst 2010; 102: 464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Subramanian J, Simon R.. What should physicians look for in evaluating prognostic gene-expression signatures? Nat Rev Clin Oncol 2010; 7: 327–334. [DOI] [PubMed] [Google Scholar]
- 5. Bair E, Tibshirani R.. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol 2004; 2: E108.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhu CQ, Pintilie M, John T. et al. Understanding prognostic gene expression signatures in lung cancer. Clin Lung Cancer 2009; 10: 331–340. [DOI] [PubMed] [Google Scholar]
- 7. Collett D. Modelling Survival Data in Medical Research. Raton, Florida, USA: Chapman & Hall/CRC; 2003. [Google Scholar]
- 8. Zheng Y, Cai T, Feng Z.. Application of the time-dependent ROC curves for prognostic accuracy with multiple biomarkers. Biometrics 2006; 62: 279–287. [DOI] [PubMed] [Google Scholar]
- 9. Chen DT, Hsu YL, Fulp WJ. et al. Prognostic and predictive value of a malignancy-risk gene signature in early-stage non-small cell lung cancer. J Natl Cancer Inst 2011; 103: 1859–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shedden K, Taylor JM, Enkemann SA. et al. Gene expression-based survival prediction in lung adenocarcinoma: A multi-site, blinded validation study. Nat Med 2008; 14: 822–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tomida S, Takeuchi T, Shimada Y. et al. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J Clin Oncol 2009; 27: 2793–2799. [DOI] [PubMed] [Google Scholar]
- 12. Roepman P, Jassem J, Smit EF. et al. An immune response enriched 72-gene prognostic profile for early-stage non-small-cell lung cancer. Clin Cancer Res 2009; 15: 284–290. [DOI] [PubMed] [Google Scholar]
- 13. Herbst RS, Heymach JV, Lippman SM.. Lung cancer. N Engl J Med 2008; 359: 1367–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Venet D, Dumont JE, Detours V.. Most random gene expression signatures are significantly associated with breast cancer outcome. PLoS Comput Biol 2011; 7: e1002240.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lynch TJ, Bell DW, Sordella R. et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004; 350: 2129–2139. [DOI] [PubMed] [Google Scholar]
- 16. Bell DW, Lynch TJ, Haserlat SM. et al. Epidermal growth factor receptor mutations and gene amplification in non-small-cell lung cancer: Molecular analysis of the ideal/intact gefitinib trials. J Clin Oncol 2005; 23: 8081–8092. [DOI] [PubMed] [Google Scholar]
- 17. Mok TS, Wu YL, Thongprasert S. et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009; 361: 947–957. [DOI] [PubMed] [Google Scholar]
- 18. Zhu CQ, da Cunha Santos G, Ding K. et al. Role of KRAS and EGFR as biomarkers of response to erlotinib in National Cancer Institute of Canada Clinical Trials Group Study BR.21. J Clin Oncol 2008; 26: 4268–4275. [DOI] [PubMed] [Google Scholar]
- 19. Byers LA, Diao L, Wang J. et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res 2013; 19: 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao Q, Shi X, Xie Y. et al. Combining multidimensional genomic measurements for predicting cancer prognosis: observations from TCGA. Brief Bioinform 2015; 16: 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma S, Dai Y.. Principal component analysis based methods in bioinformatics studies. Brief Bioinform 2011; 12: 714–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ma S, Kosorok MR, Fine JP.. Additive risk models for survival data with high-dimensional covariates. Biometrics 2006; 62: 202–210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.