Abstract
Cancer neoantigens are antigens that result from somatic mutations present in individual cancers. Neoantigens are considered important targets for cancer immunotherapy because of their immunogenicity and lack of expression in normal tissues. Next-generation sequencing technologies and computational analysis have recently made neoantigen discovery possible. Although neoantigens are important targets of checkpoint blockade therapy, neoantigen vaccines are currently being investigated in preclinical models and early-phase human clinical trials. Preliminary results from these clinical trials demonstrate that dendritic cell, synthetic long peptide, and RNA-based neoantigen vaccines are safe, and capable of inducing both CD8+ and CD4+ neoantigen-specific T-cell responses. We and others are testing neoantigen vaccines in melanoma, breast cancer, non-small-cell lung cancer and other cancer types. Since cancers have evolved mechanisms to escape immune control, it is particularly important to study the efficacy of neoantigen vaccines in combination with other immunotherapies including checkpoint blockade therapy, and immune therapies targeting the immunosuppressive tumor microenvironment.
Keywords: cancer vaccine, neoantigen, immunotherapy, clinical trial
Introduction
Cancer immunotherapy has evolved into one of the most promising cancer treatment modalities. The goal of cancer immunotherapy is to harness the immune system for the selective destruction of cancers while leaving normal tissues unharmed. Two immune checkpoints, cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) and programmed cell death protein 1 (PD-1), have been the focus of most clinical trials. Antibodies targeting CTLA-4 and PD-1/PD-L1 are relatively well-tolerated, and can elicit durable antitumor responses in a subset of patients with melanoma [1–3], non-small-cell lung cancer (NSCLC), bladder carcinoma and other cancers [4]. Although antibodies targeting CTLA-4 and PD-1/PD-L1 represent the greatest success of cancer immunotherapy, substantial efforts are underway to develop additional immunotherapies that can be used alone or in combination with these checkpoint blockade therapies.
A dynamic interplay exists between cancer cells and the immune system, as described in the cancer immunoediting hypothesis [5–7]. It is now understood that the immune system is capable of both suppressing tumor growth, and/or promoting tumor progression. In terms of suppressing tumor growth, the innate and adaptive immune systems can specifically recognize cancers as nonself and mount antitumor immune responses. Such beneficial immune responses can be induced and/or enhanced by checkpoint blockade therapies, adoptive cell therapies or cancer vaccine approaches. In terms of promoting tumor progression, tumors leverage immune regulatory networks to circumvent immune control and promote growth [6, 8]. To maximize efficacy, it is likely that next-generation immunotherapies will successfully integrate multiple strategies targeting elements of this dynamic interplay. The most promising strategies include, but are not limited to (i) agents that target immune checkpoints, (ii) approaches that inhibit immunosuppression in the tumor microenvironment (e.g. targeting regulatory T cells, myeloid-derived suppressor cells, tumor-associated macrophages etc.), (iii) adoptive transfer of ex vivo expanded and/or genetically engineered T-cell receptor (TCR) or chimeric antigen receptor T cells, and (iv) cancer vaccines designed to elicit robust antitumor immune responses. In this review we focus on the development of neoantigen vaccines with an emphasis on both preclinical development and initial clinical experience.
Cancer neoantigens
Traditionally, cancer vaccines have targeted the so-called tumor-associated antigens (TAA). TAA are typically proteins present in normal tissues but overexpressed in cancers. Examples of TAA include HER2, MART-1, MUC1, tyrosinase, MAGE, mammaglobin-A and NY-ESO-1. Cancer vaccines targeting TAA are presumed capable of inducing T-cell responses to these ‘self’ proteins due to one or more of the following reasons (i) incomplete thymic depletion and/or peripheral tolerance of TAA-reactive T cells; (ii) extremely low expression of TAA in the periphery; (iii) low TCR binding affinity of TAA-reactive T cells; or (iv) restricted TAA expression pattern during development. Unfortunately, most clinical trials targeting TAA have failed to demonstrate durable beneficial effects compared with standard of care treatment [9].
In contrast, neoantigens are tumor-specific antigens resulting from somatic DNA alterations [e.g. nonsynonymous point mutations, insertion-deletions (so-called indels), gene fusions and/or frameshift mutations]. Neoantigens typically have a high predicted binding affinity to major histocompatibility complex (MHC) molecules. Cancer vaccines targeting neoantigens have generated great enthusiasm given the potential advantages of targeting protein sequences that are not present in normal tissues including decreased central immune tolerance, and improved safety profile. This enthusiasm for targeting neoantigens has been enhanced recently, as a strong correlation between somatic tumor mutation burden and favorable clinical benefit of checkpoint blockade therapy has been established in melanoma [10, 11], NSCLC [12] and colorectal cancer [13]. In preclinical models, low mutation burden has been shown to result in a lack of immunoediting in the murine KPC pancreatic cancer model, while introduction of a neoantigen (OVA) results in tumor elimination [14]. These observations suggest an important role for neoantigens in the clinical response to checkpoint blockade therapy and the potential value of mutation burden as a predictive biomarker [15]. A clinical trial is ongoing to study the relationship between tumor mutation burden, predicted neoantigen burden, and clinical response in patients with advanced melanoma or bladder cancer treated with nivolumab (α-PD-1) or nivolumab plus ipilimumab (α-CTLA-4) (NCT02553642).
Cancer neoantigen identification
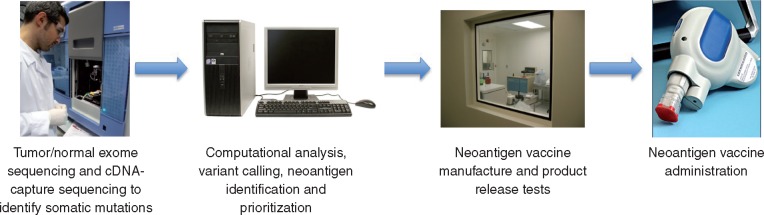
The mutational landscape of cancer is complex as demonstrated by genomic analyses of breast cancer [16], melanoma [17], liver cancer [18] and many other major cancer types [19, 20]. Advances in next-generation sequencing (NGS) technologies have enabled rapid and cost-effective comparisons between tumor and normal sequences, which is the starting point of cancer neoantigen identification [21] (Figure 1). Many somatic mutations detected by DNA sequencing are not expressed (i.e. noncoding mutations, nonsense mutations or monoallelic expression). For those expressed mutations resulting in an altered amino acid sequence, the abnormal amino acid sequences have to be successfully translated and then processed into short peptide fragments and displayed on the cell surface in the context of MHC molecules in order to be recognized by the immune system. Antigen processing and presentation is a complex process involving multiple steps that can impact neoantigen presentation [22]. Antigen processing is different for MHC class I and class II molecules. MHC class I molecules present 8–10 amino acid peptides produced endogenously or acquired by professional antigen presenting cells. MHC class II molecules present longer peptides (11–20 amino acids) derived from exogenous proteins. Therefore, predicting whether a somatic mutation can create a neoantigen depends on several key factors: (i) whether the somatic mutation is expressed at the protein level, (ii) whether the mutant protein can be naturally processed into an appropriate peptide for presentation, (iii) the binding affinity of the mutant peptide to the patient‘s autologous MHC molecules and (iv) the affinity of the mutant peptide/MHC complex to the TCR of responding T cells. It is currently believed that most somatic mutations detected by sequencing do not result in effective neoantigens [23].
Figure 1.
Neoantigen vaccine design and manufacture.
With the large number of somatic mutations, and the intrinsic polymorphism of human MHC molecules [i.e. human leukocyte antigen (HLA) molecules], it remains a challenging task to accurately predict and prioritize neoantigen candidates. A number of computational tools have been developed, which include sequence-based and structure-based algorithms to predict peptide–MHC binding (reviewed in Refs. [24–29]). Structure-based approaches require experimentally acquired crystallographic data for model building as well as significant computing power for simulations. This limits their current application despite an increased accuracy in epitope prediction [28]. On the other hand, in sequence-based predictions, supervised machine learning modules are applied with methods ranging from simple position-specific scoring matrices (PSSM, used by SYFPEITHI [30] and BIMAS [31]), to more sophisticated artificial neural networks (ANN, used by NetMHC [32]), support vector machines or hidden Markov models. These algorithms depend heavily on the size and quality of the training datasets of known MHC binding peptides available, and because of this, the predictive performance of some MHC alleles is still in need of further improvement. Nevertheless the computational methods that predict binding affinity of peptides to MHC class I molecules are the most accurate so far and have been used successfully in studies to identify neoantigens [33–36]. The ideal neoantigen prediction approach would integrate filters based on all of the biologic processes discussed above (i.e. proteasomal cleavage, MHC binding, TCR recognition) in assessing the potential immunogenicity of neoantigen candidates. To this end, the Immune Epitope Database and Analysis Resource (IEDB, www.iedb.org) hosts an array of tools to facilitate peptide–MHC binding prediction with the options of integrating proteasomal processing (NetCHOP and NetCTL) and TAP transport prediction [37]. We have found in our preclinical studies that median affinity is an effective way to predict neoantigens [33] although we are not aware that the median approach and NetMHCpan have been directly compared. In these studies, we calculated a median affinity for each neoantigen using multiple epitope prediction algorithms (NetMHCpan, ANN, SMM and SMMPMBEC). Additional filters were applied to prioritize neoantigen candidates: (i) elimination of hypothetical (Riken) proteins; (ii) use of NetCHOP, an antigen processing algorithm to eliminate epitopes that are not likely to be proteolytically produced by the constitutive- or immune-proteasome and (iii) prioritization of neoantigens where the mutant epitope has a higher predicted binding affinity than the corresponding wildtype sequence.
CD4+ T cells, which recognize antigens presented by MHC class II molecules, contribute to antitumor immunity. However, the computational methods available to predict MHC class II epitopes are less informative than the MHC class I algorithms because of the more promiscuous nature of peptide binding to MHC class II molecules [38], and lack of robust training datasets. Unlike MHC class I molecules, whose peptide binding groove tends to be closed at both ends, the ends of the MHC class II peptide binding grove are open, allowing the accommodation of longer peptides [39]. The core binding motif of a set of peptides (with variable lengths) that bind to a particular MHC class II molecule is more difficult to identify. In addition, the proteolytic degradation process of MHC class II-bound peptides is less well characterized. Despite these difficulties, Kreiter et al. [40] successfully prioritized MHC class II neoantigens based solely on expression levels and predicted MHC class II binding affinity using tools available at IEDB. The relevance of these MHC class II neoantigen predictions was confirmed as vaccination with synthetic polyepitope mRNA led to complete rejection of established, aggressively growing syngeneic tumors (B16F10 and CT26) in mice [40]. In addition, recent results from two phase I clinical trials [34, 35] highlight the importance of neoantigen-specific CD4+ T-cell responses following neoantigen vaccination. These trials, which will be discussed in more detail below, investigated personalized synthetic long peptide (SLP) [34] and polyepitope mRNA neoantigen vaccines [35] in patients with advanced melanoma. Both SLP and polyepitope mRNA approaches were able to generate CD8+ and CD4+ neoantigen-specific T-cell responses. This result is notable because the study by Ott et al. [34] did not attempt to identify MHC class II-restricted neoantigens for inclusion in the neoantigen vaccine, and Sahin et al. [35] found that ∼20% of the T-cell responses were induced to neoantigens predicted to bind poorly to HLA class I and II. These results underscore the need to further improve the in silico prediction algorithms for both MHC class I and II epitopes.
Other methods have been used to identify cancer neoantigens. Mass spectrometry analyses of peptides eluted from peptide–MHC complexes have enabled the characterization of the HLA ligandome or immunopeptidome [41, 42]. A series of clinical trials targeting the HLA ligandome have been completed in HLA-A2+ renal-cell carcinoma patients [43]. Another strategy is based on the functional analysis of an individual patient’s peripheral blood mononuclear cells (PBMC) or tumor-infiltrating lymphocytes (TIL). Pre-existing neoantigen-reactive T cells can be stimulated and detected by tetramer/multimer staining (flow cytometry analysis) or cytokine secreting assay (e.g. IFN-γ ELISPOT). Recently, novel platforms that screen a patient’s PBMC against a neoantigen library has been proposed. These platforms have been successfully applied to identify CD4+ and CD8+ T-cell antigens in infectious diseases and cancer [44, 45]. However, these types of biologic assays are currently costly, technically challenging, and may also fail to identify subdominant and/or cryptic neoantigens that do not naturally induce immune responses yet can be activated through vaccination [46].
Preclinical studies
Recent studies have shown that neoantigen vaccine approaches are able to induce robust antitumor responses in mice [33, 40, 47]. In the B16F10 murine melanoma model, Castle et al. [47] vaccinated C57BL/6 mice with SLPs derived from 50 validated mutations. Sixteen of these peptides were immunogenic as determined by IFN-γ ELISPOT assay. Peptide vaccination against two mutant antigens MUT30 (Kif18b K739N) and MUT44 (Cpsf3l D314N) was able to confer a marked in vivo antitumor effect in both preventive and therapeutic settings [47]. In follow-up studies, the same group found that the majority of the immunogenic ‘mutanome’ of B16F10 and CT26 tumors was CD4+ T-cell specific, and mRNA vaccines encoding CD4+ T-cell neoantigens were able to induce potent antitumor immunity [40]. In collaboration with Schreiber et al., we identified two neoantigens responsible for tumor rejection following immune checkpoint blockade with α-CTLA-4 or α-PD-1 antibodies [33] in the murine sarcoma model T3. Vaccination with SLP incorporating these two mutant epitopes, namely Lama4 G1254V and Alg8 A506T, induced antitumor immunity comparable to checkpoint blockade immunotherapy [33]. We have since confirmed and extended these results in the murine 4T1 and E0771 breast tumor models, identifying neoantigens that can be successfully targeted with both SLP and polyepitope DNA neoantigen vaccines (unpublished data). Employing similar strategies, Yadav et al. [36] successfully identified and validated several neoantigens in the MC-38 and TRAMP-C1 tumors.
The immunodeficient NOD.scid.gamma (NSG) [48, 49] and related mouse models have made it possible to study human cancer cell lines and patient-derived xenografts (PDX) in vivo. We have successfully established PDX by injecting human breast cancer cells into NSG mice [50]. Two neoantigens, ROBO3 A1265V and PALB2 H198D were identified in the WHIM30 PDX and parental tumor by computational analysis and in vitro studies. Adoptive transfer of autologous PBMCs stimulated in vitro with mutant ROBO3 and PALB2 peptides resulted in decreased tumor growth [50]. Integrating PDX models into the neoantigen discovery pipeline offers great opportunities. Recently the TRON Cell Line Portal (TCLP) [51, 52] has been assembled which, among others, catalog the HLA type, expression and neoepitope candidates of 1082 human cancer cell lines. TCLP (available at celllines.tron-mainz.de) is the product of data-mining and re-analyzing the public databases generated by the Catalogue of Somatic Mutations In Cancer (COSMIC) [53, 54], the Cancer Cell Line Encyclopedia (CCLE) [55] and Klijn et al. [56]. This valuable resource will help researchers select cancer cell lines based on the HLA type and expression, as well as provide therapeutic target for the development of cancer immunotherapy.
Current clinical trials
The presence of neoantigen-specific CD8+ and CD4+ T cells in TILs from melanoma patients responding to checkpoint blockade therapy [57–60], and promising results from preclinical studies have generated significant interest in the clinical development of neoantigen vaccines (Table 1). Results from several phase I clinical trials [34, 35, 46] in patients with advanced melanoma are quite encouraging, even though the number of patients treated in these studies is small. Main characteristics of current neoantigen vaccine platforms are summarized in Table 2. Carreno et al. [46] were the first to report that neoantigen-pulsed DC can induce neoantigen-specific T-cell responses in melanoma patients (NCT00683670). The initial report details the results of vaccinating three patients. Seven neoantigens with the highest binding scores to HLA-A*02:01 were prioritized in each patient. DC vaccination augmented pre-existing immunity to neoantigens, and induced neoantigen-specific T-cell responses. In addition, the frequency of most existing pre-vaccine TCR-β clonotypes was increased and previously undetected clonotypes were revealed, indicating that vaccination promotes a more diverse T-cell repertoire [46]. However, clinical presentation and responses to neoantigen-pulsed DC vaccine were not reported for these melanoma patients.
Table 1.
Selected clinical trials targeting cancer neoantigens
| ClinicalTrial.gov identifier | Phase | Enrollment statusa | Cancer type | Formulation | Additional intervention |
|---|---|---|---|---|---|
| Polyepitope plasmid DNA | |||||
| NCT02348320 | I | Recruiting | TNBC | Electroporation | Neoadjuvant chemotherapy |
| NCT03122106 | I | Not yet recruiting | Pancreatic cancer | Electroporation | Adjuvant chemotherapy |
| Polyepitope coding RNA | |||||
| NCT02035956 | I | Not recruiting | Melanoma | Intranodal injection | Pembrolizumabb [33] |
| NCT02316457 | I | Recruiting | TNBC | Intranodal injection | |
| Synthetic peptide | |||||
| NCT01970358 | I | Recruiting | Melanoma | Poly-ICLC (NeoVax) | Pembrolizumabb [32] |
| NCT02427581 | I | Recruiting | TNBC | Poly-ICLC | Neoadjuvant chemotherapy |
| NCT02600949 | II | By invitation only | PDA and CRC | IFA, topical imiquimod | Chemotherapy |
| NCT02721043 | II | Recruiting | Solid tumors | Poly-ICLC | Lenalidomide |
| NCT02510950 | I | Recruiting | Glioblastoma | Poly-ICLC | Temozolomide |
| NCT02992977 | I | Recruiting | Advanced cancer | AutoSynVax™c | |
| NCT02933073 | I | Recruiting | Ovarian cancer | OncoImmunome | Chemotherapy |
| NCT02897765 | I | Recruiting | Multiple | Poly-ICLC (Neo-PV-01) | Nivolumab |
As of 15 August 2017.
Only to patients with disease recurrence.
HSP70 conjugated short peptides.
TNBC, triple-negative breast cancer; PDA, pancreatic ductal adenocarcinoma; CRC, colorectal cancer; IFA, incomplete Freund‘s adjuvant.
Table 2.
Main characteristics of current neoantigen vaccine platforms
| Vaccine platform | Advantages | Disadvantages | Clinical trials |
|---|---|---|---|
| Synthetic long peptide vaccine |
|
|
|
| RNA vaccine |
|
|
|
| Dendritic cell vaccine |
|
|
|
| DNA vaccine |
|
|
Two additional papers co-published in Nature by Ott et al. [34] and Sahin et al. [35] confirm the potential of neoantigen vaccines in treating melanoma patients. These two studies employed similar strategies to identify neoantigens based on NGS data from cancers and normal cells. Computational algorithms were used to predict the ability of neoantigens to bind MHC class I molecules and to prioritize candidate neoantigens. Ott et al. vaccinated six patients with SLP (up to 20 total peptides in 4 pools for each patient) after surgical resection of the tumors (NCT01970358). Four of the six patients vaccinated showed no disease recurrence during follow-up of 20–32 months after vaccination. The remaining two participants had disease recurrence but both achieved a complete response after treatment with anti-PD-1 antibody. Sahin et al. [61] created synthetic RNA vaccines, each encoding five 27-mer neoantigens (NCT02035956). Such RNA molecules were previously shown to be readily taken up by lymph node resident DCs. Up to 10 mutations were targeted in each patient’s tumor (two RNA vaccines). Of the 13 patients vaccinated via intranodal injection of the RNA vaccine, 8 remained tumor free throughout the follow-up period. The other five participants had tumor relapse. However, after PD-1 blockade therapy, tumor regression occurred in one of these patients. Another patient was noted to have outgrowth of β2-microglobulin deficient cancer cells, indicating MHC loss as an acquired immune escape mechanism. ‘Off-the-shelf’ RNA vaccines targeting shared TAAs were also administered in patients with NY-ESO-1+ and/or tyrosinase+ melanoma, but their contribution to the antitumor immunity was not studied. Immune monitoring analyses of patients’ PBMCs (IFN-γ ELISPOT, intracellular cytokine staining, multimer staining) in both studies revealed that SLP and RNA vaccines can (i) enhance pre-existing but weak neoantigen-specific T-cell responses and (ii) generate de novo neoantigen-specific T-cell responses [34, 35]. As noted above, the majority of the ex vivo IFN-γ responses were generated by CD4+ T cells. Both studies also found that vaccination resulted in an expansion of the neoantigen-specific T-cell repertoire. Taken together, these studies provide strong rationale for further clinical development and testing of neoantigen vaccines.
We and others have initiated clinical trials of neoantigen vaccines in breast and other cancer types (Table 1). For instance, we have initiated trials and are currently recruiting patients with triple-negative breast cancer (TNBC) who do not have a pathologic complete response after neoadjuvant chemotherapy. These patients typically have no gross evidence of disease following standard of care therapy (neoadjuvant chemotherapy, surgery and radiation therapy) but are at high-risk for disease recurrence. Targeting this patient population provides a window-of-opportunity to identify neoantigens and manufacture personalized cancer vaccines, maximizing the potential benefit from the vaccine as the regulatory networks associated with metastatic disease are not present. One trial (NCT02427581) is designed to vaccinate TNBC patients with SLP admixed with poly-ICLC as adjuvant. In a companion trial (NCT02348320), we synthesize polyepitope neoantigen DNA vaccines and vaccinate patients intramuscularly using an electroporation device. Exome and RNA sequencing, neoantigen prediction and vaccine production has been completed for 12 patients, and 11 subjects have completed all scheduled vaccinations. The primary objective of these trials is to assess the safety of personalized neoantigen vaccines. Meanwhile, pre- and post-vaccine PBMC have been collected at various time points from vaccinated patients, and cryopreserved. Neoantigen-specific T-cell responses will be assessed using IFN-γ ELISPOT assay, multiparametric flow cytometry analysis, and related techniques.
Challenges
Despite recent advances, many challenges remain in the development of neoantigen vaccines. First, both the cost and time to manufacture neoantigen vaccines have to be reduced. Although the cost of DNA/RNA sequencing has decreased significantly [21], it remains costly and time-consuming to identify and validate candidate neoantigens. Manufacture of neoantigen vaccines under good manufacturing practice conditions is also very expensive. Currently the time from tissue acquisition to vaccine delivery ranges from 3 to 5 months [34, 35]. This will need to be improved in order to benefit patients with metastatic disease. Second, neoantigen prediction algorithms require further optimization. These include strategies to better predict MHC class I and II neoantigens, as well as potential neoantigens resulting from genetical alterations other than missense mutations, such as gene fusions and indels. Given the recent findings that CD4+ T-cell responses to neoantigen vaccines were more common than CD8+ T-cell responses [34, 35] even when the neoantigens included in the vaccines were prioritized based on predicted MHC class I binding, it is clear that neoantigen prediction algorithms can be improved.
The clinical response to cancer immunotherapies has distinct kinetics compared with the response to cytotoxic or small molecule therapies. Cancer immunotherapies are frequently evaluated using immune-related response criteria (irRC) [62, 63]. Neoantigen vaccines will also leverage these immune-related response criteria, but will also rely on effective immune monitoring to assess vaccine-induced immune responses before clinical end points are reached. Unfortunately, there is still a lack of reliable immune response biomarkers that are predictive of antitumor immunity and, ultimately a survival benefit. Further investigation is needed to identify relevant immune response biomarkers using a systematic approach. From a technical point-of-view, immunological assays currently available (e.g. ELISPOT, flow cytometry-based multimer staining and intracellular cytokine staining) have a reputation of inconsistency among different laboratories. For instance, one study found the inter-laboratory variation of ELISPOT can be as high as 50% [64]. Clearly, standardized and harmonized procedures from specimen banking, assay validation, to result reporting are warranted for successful clinical development.
Concluding remarks
Neoantigen vaccines have shown encouraging results in terms of inducing neoantigen-specific T-cell responses [34, 35]. RNA, SLP, dendritic cell and DNA neoantigen vaccines are being rigorously tested in phase I clinical trials. NGS technologies and computational algorithms demonstrate great promise but still need to be optimized to most effectively prioritize candidate neoantigens. Given that cancers can escape immune control through various mechanisms, including some that are not fully understood, it will be important to explore the efficacy of neoantigen vaccines in combination with other immunotherapies including checkpoint blockade therapy, and emerging therapies targeting the immunosuppressive tumor microenvironment.
Acknowledgement
We would like to thank Enid McIntosh for administrative assistance.
Funding
This project was supported by grants from Susan G. Komen for the Cure (KG111025), the Alvin J. Siteman Cancer Center (Siteman Investment Program grant 4035); the National Cancer Institute at the National Institute of Health (Cancer Center Support Grant P30-CA091842, and SPORE in Pancreatic Cancer P50-CA196510); and the Foundation for Barnes-Jewish Hospital (to SPG).
This supplement was sponsored by F. Hoffmann-La Roche.
Disclosure
The authors have declared no conflicts of interest.
References
- 1. Hodi FS, O’Day SJ, McDermott DF. et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010; 363(8): 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Postow MA, Chesney J, Pavlick AC. et al. Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N Engl J Med 2015; 372(21): 2006–2017.http://dx.doi.org/10.1056/NEJMoa1414428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Robert C, Schachter J, Long GV. et al. Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med 2015; 372(26): 2521–2532.http://dx.doi.org/10.1056/NEJMoa1503093 [DOI] [PubMed] [Google Scholar]
- 4. Zou W, Wolchok JD, Chen L.. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci Transl Med 2016; 8(328): 328rv324.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dunn GP, Old LJ, Schreiber RD.. The three Es of cancer immunoediting. Annu Rev Immunol 2004; 22: 329–360.http://dx.doi.org/10.1146/annurev.immunol.22.012703.104803 [DOI] [PubMed] [Google Scholar]
- 6. Schreiber RD, Old LJ, Smyth MJ.. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science 2011; 331(6024): 1565–1570.http://dx.doi.org/10.1126/science.1203486 [DOI] [PubMed] [Google Scholar]
- 7. Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ.. Natural innate and adaptive immunity to cancer. Annu Rev Immunol 2011; 29: 235–271.http://dx.doi.org/10.1146/annurev-immunol-031210-101324 [DOI] [PubMed] [Google Scholar]
- 8. Chen DS, Mellman I.. Elements of cancer immunity and the cancer-immune set point. Nature 2017; 541(7637): 321–330.http://dx.doi.org/10.1038/nature21349 [DOI] [PubMed] [Google Scholar]
- 9. Melero I, Gaudernack G, Gerritsen W. et al. Therapeutic vaccines for cancer: an overview of clinical trials. Nat Rev Clin Oncol 2014; 11(9): 509–524.http://dx.doi.org/10.1038/nrclinonc.2014.111 [DOI] [PubMed] [Google Scholar]
- 10. Snyder A, Makarov V, Merghoub T. et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014; 371(23): 2189–2199.http://dx.doi.org/10.1056/NEJMoa1406498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Van Allen EM, Miao D, Schilling B. et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015; 350(6257): 207–211.http://dx.doi.org/10.1126/science.aad0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rizvi NA, Hellmann MD, Snyder A. et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015; 348(6230): 124–128.http://dx.doi.org/10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Le DT, Uram JN, Wang H. et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015; 372(26): 2509–2520.http://dx.doi.org/10.1056/NEJMoa1500596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Evans RA, Diamond MS, Rech AJ. et al. Lack of immunoediting in murine pancreatic cancer reversed with neoantigen. JCI Insight 2016; 1(14): e88328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gubin MM, Schreiber RD.. CANCER. The odds of immunotherapy success. Science 2015; 350(6257): 158–159.http://dx.doi.org/10.1126/science.aad4140 [DOI] [PubMed] [Google Scholar]
- 16. Stephens PJ, Tarpey PS, Davies H. et al. The landscape of cancer genes and mutational processes in breast cancer. Nature 2012; 486(7403): 400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hodis E, Watson IR, Kryukov GV. et al. A landscape of driver mutations in melanoma. Cell 2012; 150(2): 251–263.http://dx.doi.org/10.1016/j.cell.2012.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fujimoto A, Furuta M, Totoki Y. et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet 2016; 48(5): 500–509.http://dx.doi.org/10.1038/ng.3547 [DOI] [PubMed] [Google Scholar]
- 19. Kandoth C, McLellan MD, Vandin F. et al. Mutational landscape and significance across 12 major cancer types. Nature 2013; 502(7471): 333–339.http://dx.doi.org/10.1038/nature12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zehir A, Benayed R, Shah RH. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017; 23(6): 703–713.http://dx.doi.org/10.1038/nm.4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li L, Goedegebuure P, Mardis ER. et al. Cancer genome sequencing and its implications for personalized cancer vaccines. Cancers (Basel) 2011; 3(4): 4191–4211.http://dx.doi.org/10.3390/cancers3044191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Blum JS, Wearsch PA, Cresswell P.. Pathways of antigen processing. Annu Rev Immunol 2013; 31: 443–473.http://dx.doi.org/10.1146/annurev-immunol-032712-095910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schumacher TN, Schreiber RD.. Neoantigens in cancer immunotherapy. Science 2015; 348(6230): 69–74.http://dx.doi.org/10.1126/science.aaa4971 [DOI] [PubMed] [Google Scholar]
- 24. Cherryholmes GA, Stanton SE, Disis ML.. Current methods of epitope identification for cancer vaccine design. Vaccine 2015; 33(51): 7408–7414.http://dx.doi.org/10.1016/j.vaccine.2015.06.116 [DOI] [PubMed] [Google Scholar]
- 25. Desai DV, Kulkarni-Kale U.. T-cell epitope prediction methods: an overview. Methods Mol Biol 2014; 1184: 333–364. [DOI] [PubMed] [Google Scholar]
- 26. Patronov A, Doytchinova I.. T-cell epitope vaccine design by immunoinformatics. Open Biol 2013; 3(1): 120139..http://dx.doi.org/10.1098/rsob.120139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Singh SP, Mishra BN.. Major histocompatibility complex linked databases and prediction tools for designing vaccines. Hum Immunol 2016; 77(3): 295–306.http://dx.doi.org/10.1016/j.humimm.2015.11.012 [DOI] [PubMed] [Google Scholar]
- 28. Backert L, Kohlbacher O.. Immunoinformatics and epitope prediction in the age of genomic medicine. Genome Med 2015; 7: 119..http://dx.doi.org/10.1186/s13073-015-0245-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Soria-Guerra RE, Nieto-Gomez R, Govea-Alonso DO, Rosales-Mendoza S.. An overview of bioinformatics tools for epitope prediction: implications on vaccine development. J Biomed Inform 2015; 53: 405–414. [DOI] [PubMed] [Google Scholar]
- 30. Rammensee H, Bachmann J, Emmerich NP. et al. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 1999; 50(3–4): 213–219. [DOI] [PubMed] [Google Scholar]
- 31. Parker KC, Bednarek MA, Coligan JE.. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J Immunol 1994; 152: 163–175. [PubMed] [Google Scholar]
- 32. Lundegaard C, Lamberth K, Harndahl M. et al. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Res 2008; 36(Suppl 2): W509–W512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gubin MM, Zhang X, Schuster H. et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 2014; 515(7528): 577–581.http://dx.doi.org/10.1038/nature13988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ott PA, Hu Z, Keskin DB. et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 2017; 547(7662): 217–221.http://dx.doi.org/10.1038/nature22991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sahin U, Derhovanessian E, Miller M. et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017; 547(7662): 222–226.http://dx.doi.org/10.1038/nature23003 [DOI] [PubMed] [Google Scholar]
- 36. Yadav M, Jhunjhunwala S, Phung QT. et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 2014; 515(7528): 572–576.http://dx.doi.org/10.1038/nature14001 [DOI] [PubMed] [Google Scholar]
- 37. Vita R, Overton JA, Greenbaum JA. et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res 2015; 43(Database issue): D405–D412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nielsen M, Lund O, Buus S, Lundegaard C.. MHC class II epitope predictive algorithms. Immunology 2010; 130(3): 319–328.http://dx.doi.org/10.1111/j.1365-2567.2010.03268.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Castellino F, Zhong G, Germain RN.. Antigen presentation by MHC class II molecules: invariant chain function, protein trafficking, and the molecular basis of diverse determinant capture. Hum Immunol 1997; 54(2): 159–169.http://dx.doi.org/10.1016/S0198-8859(97)00078-5 [DOI] [PubMed] [Google Scholar]
- 40. Kreiter S, Vormehr M, van de Roemer N. et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 2015; 520(7549): 692–696.http://dx.doi.org/10.1038/nature14426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rammensee HG, Singh-Jasuja H.. HLA ligandome tumor antigen discovery for personalized vaccine approach. Expert Rev Vaccines 2013; 12(10): 1211–1217.http://dx.doi.org/10.1586/14760584.2013.836911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Singh-Jasuja H, Emmerich NP, Rammensee HG.. The Tubingen approach: identification, selection, and validation of tumor-associated HLA peptides for cancer therapy. Cancer Immunol Immunother 2004; 53(3): 187–195.http://dx.doi.org/10.1007/s00262-003-0480-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Walter S, Weinschenk T, Stenzl A. et al. Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat Med 2012; 18(8): 1254–1261.http://dx.doi.org/10.1038/nm.2883 [DOI] [PubMed] [Google Scholar]
- 44. Long D, Skoberne M, Gierahn TM. et al. Identification of novel virus-specific antigens by CD4(+) and CD8(+) T cells from asymptomatic HSV-2 seropositive and seronegative donors. Virology 2014; 464-465: 296–311. [DOI] [PubMed] [Google Scholar]
- 45. Bentzen AK, Marquard AM, Lyngaa R. et al. Large-scale detection of antigen-specific T cells using peptide–MHC-I multimers labeled with DNA barcodes. Nat Biotechnol 2016; 34(10): 1037–1045.http://dx.doi.org/10.1038/nbt.3662 [DOI] [PubMed] [Google Scholar]
- 46. Carreno BM, Magrini V, Becker-Hapak M. et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 2015; 348(6236): 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Castle JC, Kreiter S, Diekmann J. et al. Exploiting the mutanome for tumor vaccination. Cancer Res 2012; 72(5): 1081–1091.http://dx.doi.org/10.1158/0008-5472.CAN-11-3722 [DOI] [PubMed] [Google Scholar]
- 48. Ishikawa F, Yasukawa M, Lyons B. et al. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor {gamma} chain(null) mice. Blood 2005; 106(5): 1565–1573.http://dx.doi.org/10.1182/blood-2005-02-0516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shultz LD, Lyons BL, Burzenski LM. et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol 2005; 174(10): 6477–6489.http://dx.doi.org/10.4049/jimmunol.174.10.6477 [DOI] [PubMed] [Google Scholar]
- 50. Zhang X, Kim S, Hundal J. et al. Breast cancer neoantigens can induce CD8 T cell responses and antitumor immunity. Cancer Immunol Res 2017; 5(7): 516–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Boegel S, Lower M, Bukur T. et al. A catalog of HLA type, HLA expression, and neo-epitope candidates in human cancer cell lines. Oncoimmunology 2014; 3(8): e954893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Scholtalbers J, Boegel S, Bukur T. et al. TCLP: an online cancer cell line catalogue integrating HLA type, predicted neo-epitopes, virus and gene expression. Genome Med 2015; 7(1): 118.http://dx.doi.org/10.1186/s13073-015-0240-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bamford S, Dawson E, Forbes S. et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer 2004; 91(2): 355–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Forbes SA, Beare D, Boutselakis H. et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res 2017; 45(D1): D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Barretina J, Caponigro G, Stransky N. et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012; 483(7391): 603–607.http://dx.doi.org/10.1038/nature11003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Klijn C, Durinck S, Stawiski EW. et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol 2014; 33(3): 306–312.http://dx.doi.org/10.1038/nbt.3080 [DOI] [PubMed] [Google Scholar]
- 57. Kvistborg P, Philips D, Kelderman S. et al. Anti-CTLA-4 therapy broadens the melanoma-reactive CD8+ T cell response. Sci Transl Med 2014; 6(254): 254ra128. [DOI] [PubMed] [Google Scholar]
- 58. Linnemann C, van Buuren MM, Bies L. et al. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nat Med 2014; 21(1): 81–85. [DOI] [PubMed] [Google Scholar]
- 59. Lu YC, Yao X, Crystal JS. et al. Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin Cancer Res 2014; 20(13): 3401–3410.http://dx.doi.org/10.1158/1078-0432.CCR-14-0433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tran E, Turcotte S, Gros A. et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 2014; 344(6184): 641–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kreiter S, Selmi A, Diken M. et al. Intranodal vaccination with naked antigen-encoding RNA elicits potent prophylactic and therapeutic antitumoral immunity. Cancer Res 2010; 70(22): 9031–9040.http://dx.doi.org/10.1158/0008-5472.CAN-10-0699 [DOI] [PubMed] [Google Scholar]
- 62. Hoos A. Development of immuno-oncology drugs - from CTLA4 to PD1 to the next generations. Nat Rev Drug Discov 2016; 15(4): 235–247.http://dx.doi.org/10.1038/nrd.2015.35 [DOI] [PubMed] [Google Scholar]
- 63. Wolchok JD, Hoos A, O’Day S. et al. Guidelines for the evaluation of immune therapy activity in solid tumors: immune-related response criteria. Clin Cancer Res 2009; 15(23): 7412–7420. [DOI] [PubMed] [Google Scholar]
- 64. Janetzki S, Britten CM, Kalos M. et al. “MIATA”-minimal information about T cell assays. Immunity 2009; 31(4): 527–528. [DOI] [PMC free article] [PubMed] [Google Scholar]