Abstract
Background
Circulating tumor DNA (ctDNA) is a potential source for tumor genome analysis. We explored the concordance between the mutational status of RAS in tumor tissue and ctDNA in metastatic colorectal cancer (mCRC) patients to establish eligibility for anti-epidermal growth factor receptor (EGFR) therapy.
Patients and methods
A prospective-retrospective cohort study was carried out. Tumor tissue from 146 mCRC patients was tested for RAS status with standard of care (SoC) PCR techniques, and Digital PCR (BEAMing) was used both in plasma and tumor tissue.
Results
ctDNA BEAMing RAS testing showed 89.7% agreement with SoC (Kappa index 0.80; 95% CI 0.71 − 0.90) and BEAMing in tissue showed 90.9% agreement with SoC (Kappa index 0.83; 95% CI 0.74 − 0.92). Fifteen cases (10.3%) showed discordant tissue-plasma results. ctDNA analysis identified nine cases of low frequency RAS mutations that were not detected in tissue, possibly due to technical sensitivity or heterogeneity. In six cases, RAS mutations were not detected in plasma, potentially explained by low tumor burden or ctDNA shedding. Prediction of treatment benefit in patients receiving anti-EGFR plus irinotecan in second- or third-line was equivalent if tested with SoC PCR and ctDNA. Forty-eight percent of the patients showed mutant allele fractions in plasma below 1%.
Conclusions
Plasma RAS determination showed high overall agreement and captured a mCRC population responsive to anti-EGFR therapy with the same predictive level as SoC tissue testing. The feasibility and practicality of ctDNA analysis may translate into an alternative tool for anti-EGFR treatment selection.
Keywords: anti-EGFR therapy, circulating tumor DNA, metastatic colorectal cancer, RAS analysis
Introduction
In metastatic colorectal cancer (mCRC), treatment with anti-epidermal growth factor receptor (EGFR) monoclonal antibodies cetuximab or panitumumab has demonstrated efficacy in wild-type (WT) RAS mutations and it is now considered imperative this determination at the time of diagnosis [1, 2]. Formalin-fixed, paraffin-embedded (FFPE) tumor tissue with PCR analysis is currently used as standard of care (SoC) for RAS testing and is considered the gold standard [3].
Circulating-free DNA (cfDNA) is natural DNA present in the cell-free fraction of blood. Recent studies have suggested that genomic alterations in solid tumors may be characterized by studying the circulating tumor DNA (ctDNA) released from cancer cells into the plasma [4]. In mCRC, ctDNA is detected in almost all patients but the low abundance requires highly sensitive techniques to study mutations present at low frequencies. This approach represents a liquid non-invasive biopsy with a potential for determining RAS status. The main benefits are based on the safety and convenience associated with minimally invasive procedures, accessibility at any time point—that favor dynamic/evolutive evaluation—and is not affected by sample selection bias, although accuracy and concordance with tumor-based techniques has not been fully elucidated in patients from clinical practice [5–7].
Here, we carried out a concordance biomarker analysis of 146 mCRC patients using plasma and tissue-based RAS mutation testing with BEAMing and SoC techniques in both specimens. Discordant results were analyzed in-depth taking into consideration both technical and clinical conditions. We investigated the value of this determination in terms of progression-free survival (PFS) in patients who had received anti-EGFR as well as overall survival (OS) and mutant allele fraction (MAF) analysis.
Materials and methods
Study design
This prospective-retrospective study recruited patients candidate for therapy from three Spanish hospitals as well as from a phase II multicentric TTD ULTRA clinical trial (NCT01704703) for prospective biomarker investigation. It was approved by the ethics committees of each hospital and all patients provided written informed consent. Patients were required to have a diagnosis of mCRC with available tumor tissue for mutational analysis, have not received anti-EGFR agents before plasma collection, and have evidence of measurable disease according to Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1 [8].
Plasma was obtained from 10 ml of blood and all patients had FFPE tissue (either primary tumor or metastasis) with >15% tumor area. Tumor tissue area was evaluated by the pathologist taking into consideration the amount of sample occupied by the tumor in a standardized procedure.
All samples were analyzed blinded to the study endpoints. Full description in supplementary methods, available at Annals of Oncology online.
RAS mutational analysis
RAS status determination was carried out with available plasma and tumor tissue using BEAMing and Real-Time PCR as SoC technique. The DNA extracted from FFPE tissue sections was partitioned and used for both determinations (BEAMing and real-time PCR). The panel of RAS mutations evaluated with BEAMing was identical to that previously validated (supplementary Table S1, available at Annals of Oncology online) [2]. Each plasma and tumor sample was independently processed (using an 8-step workflow, supplementary Figure S1, available at Annals of Oncology online). In discordant cases the historical RAS reports were reviewed and further RAS determinations were carried out when metastases tissue was available, using SoC techniques (supplementary Table S2, available at Annals of Oncology online).
Depending on the specific assay, samples with a detectable mutation rate above 0.02%–0.04% were considered positive using BEAMing in ctDNA and 1% in tumor tissue. CtDNA testing was carried out with the commercially available CE-IVD BEAMing RAS plasma kit with the same thresholds for the specific mutations.
The sensitivity for Real-Time PCR as SoC analysis in tumor tissue is ∼1%–5%. Full description in supplementary methods and Table S3, available at Annals of Oncology online.
Statistics
Full description in supplementary methods, available at Annals of Oncology online.
Results
Patient characteristics
A total of 157 mCRC patients were initially included, 11 of whom were excluded because of specific pre-analytical requirements or lack of tumor tissue availability (supplementary Figure S2, available at Annals of Oncology online).
Patient baseline characteristics, number and location of metastasis, and number and description of previous lines of therapy are summarized in supplementary Table S4, available at Annals of Oncology online.
Overall, 61 (42%) patients were naïve for therapy in the metastatic setting at the time of ctDNA collection, while the remaining 85 (58%) patients had received a range of treatments but all were anti-EGFR therapies naive. The median time from tumor tissue specimen to ctDNA collection was 1.2 months (range 0–34) in therapy-naive patients. The range in previously exposed patients was wide, with a median of 20.2 months (range 0.4–282). A group of 67 (46%) patients received anti-EGFR immediately after ctDNA collection mainly in second and third line (supplementary Table S4, available at Annals of Oncology online). Median PFS and median OS were described in supplementary Table S5, available at Annals of Oncology online.
Correlation between RAS status in tissue and plasma
Using qPCR, we found tumor tissue samples positive for KRAS mutations in 44/146 samples (30%) and NRAS mutations in 10/146 (7%) (Table 1; supplementary Table S6, available at Annals of Oncology online). Using BEAMing in tissue samples, KRAS mutations were detected in 49/130 (38%) available samples and NRAS mutations in 11/130 (8%). For ctDNA analysis, 46/146 (31%) and 11/146 (8%) plasma samples harbored KRAS and NRAS mutations, respectively.
Table 1.
Concordance between tumor-tissue and ctDNA analysis (N = 146)
| ctDNA analysis | Tumor-tissue analysis SoC |
Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |||
|---|---|---|---|---|---|---|---|---|
| KRAS mut | NRAS mut | WT | ||||||
| BEAMing | KRAS mut | 40 | 0 | 6 | 89 | 90 | 84 | 93 |
| NRAS mut | 0 | 8 | 3 | |||||
| WT | 4 | 2 | 83 | |||||
| Total | 44 | 10 | 92 | |||||
| Tumor-tissue analysis BEAMinga | 85 | 91 | 89 | 88 | ||||
| BEAMing | KRAS mut | 42 | 0 | 4 | ||||
| NRAS mut | 0 | 9 | 2 | |||||
| WT | 7 | 2 | 64 | |||||
| Total | 49 | 11 | 70 | |||||
| Tumor-tissue analysis | Tumor-tissue analysis SoC | 94 | 88 | 85 | 96 | |||
| BEAMing | KRAS mut | 42 | 0 | 7 | ||||
| NRAS mut | 0 | 9 | 2 | |||||
| WT | 2 | 1 | 67 | |||||
| Total | 44 | 10 | 76 | |||||
Tumor-tissue analysis with BEAMing was carried out in 130 samples.
WT, wild type; SoC, standard of care; ctDNA, circulating tumor DNA; PPV, positive predictive value; NPV, negative predictive value.
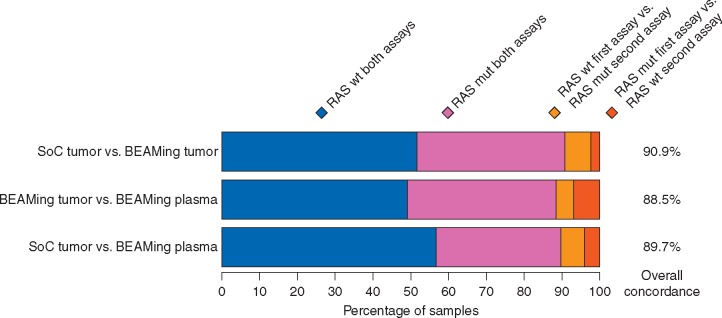
Figure 1 shows concordance of RAS status between the three methods. ctDNA analysis showed a Cohen's Kappa estimate of 0.80 (95% CI 0.71–0.90) compared with tumor tissue evaluated by SoC reflecting almost perfect agreement according to Landis and Koch classification [9]. Results were similar for RAS status in plasma and tissue using BEAMing with a Kappa index of 0.79 (95% CI 0.69–0.89), and in tumor tissue using SoC and BEAMing a Kappa index of 0.83 (95% CI 0.74–0.92).
Figure 1.
Concordance analysis. SoC tumor and BEAMing plasma analysis was carried out in 146 samples, BEAMing tumor was carried out in 130 samples. mut, mutation; SoC, standard of care.
Discordant samples description
In the population of samples with discordance between RAS status according to ctDNA BEAMing and tissue by SoC, two groups were identified, as detailed below (Table 2). To clarify these cases, the historical RAS testing was reviewed and additional RAS determinations were carried out by SoC in metastases whenever tissue was available (supplementary Table S2, available at Annals of Oncology online).
Table 2.
Discordant samples
| ID | qPCR (SoC) tumor | BEAMing plasma | BEAMing tumor | Additional (SoC) tumorc | Historical (SoC) tumorc | Codon | MAF (BEAMing plasma) | adjMAF (BEAMing tumor) | Tissue source | Tissue tumor area (%) | Time tissue- plasma (month) | Previous chemo lines | Previous treatment receivedd | Anti-EGFR after plasma collection and best response | Possible explanation | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group Aa | 1 | WT | MUT | MUT | MUT | WT | NRAS Q61 | 0.43 | 0.072 | Primary | 15 | 8 | 1 | Capox adyuvant | FOLFIRI+ Panitumumab 1L (PR) | SoC sensitivity |
| 2 | WT | MUT | MUT | MUT | WT | KRAS A146 | 0.0065 | 0.25 | Primary | 95 | 1 | 0 | FOLFOX+ Cetuximab 1L (SD) | |||
| 3 | WT | MUT | MUT | NA | NA | KRAS A146 | 0.0061 | 0.29 | Primary | 50 | 7 | 0 | No | |||
| 4 | WT | MUT | MUT | NA | MUT | KRAS G12 | 0.0006 | 0.058 | Primary | 20 | 108 | 2 | 5FU adyuvant, FOLFIRI 1L | No | ||
| 5 | WT | MUT | MUT | NA | MUT | NRAS Q61 | 0.0005 | 0.085 | Primary | 75 | 2 | 0 | No | |||
| 6 | WT | MUT | WT | NA | WT | KRAS G12 | 0.0005 | Metastasis | 45 | 4 | 1 | FOLFIRI+BVZ 1L | FOLFIRI+ Panitumumab 2L (PR) | Molecular heterogeneity | ||
| 7 | WT | MUT | WT | NA | WT | KRAS G12 | 0.0008 | Primary | 70 | 3 | 0 | No | ||||
| 8 | WT | MUT | WT | WT | WT | KRAS Q61 | 0.0015 | Primary | 70 | 1 | 0 | FOLFIRI+ Cetuximab 1L (PR) | ||||
| 9 | WT | MUT | WT | NA | WT | NRAS Q61 | 0.0005 | Primary | 100 | 10 | 1 | FOLFIRI 1L | FOLFIRI+ Panitumumab 2L (PD) | |||
| Group Bb | 10 | MUT | WT | MUT | MUTe | MUT | KRAS G12 | 0,27 | Primary | 50 | 1 | 0 | No | Low tumor burden? | ||
| 11 | MUT | WT | MUT | MUT | MUT | KRAS G12 | 0,14 | Primary | 40 | 33 | 2 | FOLFOX 1L, FOLFIRI 2L | No | |||
| 12 | MUT | WT | MUT | NA | MUT | KRAS G12 | 0,12 | Primary | 95 | 3 | 0 | In course FOLFOX 1L (PR) | No | Chemotherapy effect? | ||
| 13 | NA | WT | MUT | NA | MUT | NRAS G13 | NA | Primary | 70 | 8 | 0 | In course FOLFOX + BVZ 1L (SD) | No | |||
| 14 | NA | WT | MUT | NA | MUT | NRAS Q61 | NA | Primary | 70 | 4 | 0 | In course FOLFOX 1L (PD) | No | |||
| 15 | MUT | WT | WT | NA | MUTf | KRAS Q61 | Primary | 60 | 3 | 0 | No | Technical issues? |
Group A: mutations detected in plasma but not in tissue by SoC.
Group B: mutation detected in tissue by SoC but not in plasma.
Supplementary Table S2, available at Annals of Oncology online.
In those patients with plasma extraction during chemotherapy immediate response after extraction is reported between brackets.
Codon NRAS A59.
Codon KRAS G13.
SoC, standard of care; MAF, mutant allele fraction; adjMAF, adjusted mutant allele fraction; Chemo, chemotherapy (adyuvant and/or metastatic setting); MUT, mutation; NA, not available; PR, partial response; SD, stable disease; PD, progression disease; 1L, frontline metastatic therapy; 2L, second line metastatic therapy; Capox, Capecitabine + oxaliplatin; 5FU, 5-fluorouracil; BVZ, Bevacizumab; FOLFIRI, 5FU + leucovorin + irinotecan; FOLFOX, 5FU + leucovorin + oxaliplatin.
Group A includes patients with evidence of mutations detected in plasma but not in tissue by SoC techniques. In the first five cases the SoC tissue technique failed to detect mutations that were detected in the same tumor sample by BEAMing (Table 2).
Interestingly, in cases 1 and 2, SoC analysis of additional metastatic samples showed the same mutations as those found in plasma supporting the concept that plasma can be used to capture tumor heterogeneity. Likewise, in cases 4 and 5 the historical reports showed identical mutated as plasma BEAMing but the new qPCR result was WT.
On the remaining four cases (ID 6–9) of this group the mutation detected by plasma BEAMing could not be identified by any other tumor sampling test. These cases appeared not to have specific clinicopathologic features or differential tissue sampling timing.
In group B, mutations were detected in tissue but not in plasma in six patients (Table 2). In this group, we also reviewed the CT scan carried out closest to the blood extraction to calculate tumor burden. Patient 10 had small hepatic lesions (<1.5 cm) and patient 11 had only three peritoneal lesions, both of which reflect low tumor burden. For three patients (ID 12–14), plasma extraction was carried out during the course of chemotherapy, which may have altered ctDNA detection. The immediate RECIST 1.1 response after plasma extraction was also reviewed. The last case (ID 15) had discordant results between tissue BEAMing and SoC evaluations even though the DNA for this analysis originated from the same tumoral tissue block. Again, these cases did not have any other particular clinic-pathologic features or differential time to tumor sampling.
MAF analysis: distribution and median values
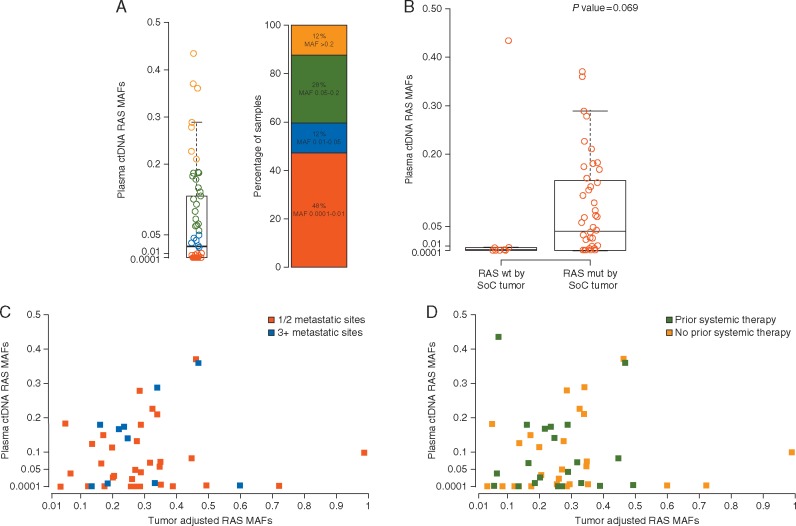
RAS MAFs had a median of 0.02 (range 0.0001–0.43) in plasma and were found in a wide distribution, 48% showed <1% (MAF <0.01) mutant alleles in their cfDNA (Figure 2A). RAS-adjusted MAFs had a median of 0.25 (range 0.03–0.99) in tumor tissue.
Figure 2.
Mutant allele fraction analysis. (A) RAS mutant allele fractions in ctDNA BEAMing, a MAF of 0.01 corresponds to a percentage of mutant alleles of 1%. (B) Comparison of RAS mutant allele fractions in ctDNA and positivity for RAS mut tumor by SoC testing. (C) Correlation of RAS mutant allele fractions with BEAMing carried out in tumor (adjusted for purity) and ctDNA, according to prior systemic therapy exposure. Samples with RAS wild-type by SoC were excluded. (D) Correlation of RAS mutant allele fractions with BEAMing carried out in tumor (adjusted for purity) and ctDNA, according to number of metastatic sites. Samples with RAS wild-type by SoC were excluded. mut, mutation SoC, standard of care.
In the group of patients with concordant mutant samples in ctDNA and tissue by SoC (N = 48), median MAF in plasma was 0.04 (range 0.0001–0.37) (Figure 2B). In the discordant cases (n = 9) median MAF was 0.0008 (range 0.0004–0.43) (P = 0.069, Kruskal test).
In concordant samples by BEAMing tested in both tumor and plasma (N = 48), median adjusted MAF was 0.26 (95% CI 0.04–0.99) in tumor and 0.14 (95% CI 0.05–0.99) (P = 0.16, Kruskal test) in discordant samples (N = 7). Overall, there was a tendency for lower MAFs both in tumor and plasma for the samples with discordant results.
The median MAF in ctDNA was also described according to prior chemotherapy exposure and number of metastatic sites. In the first case, median MAF was 0.07 (95% CI 0.002–0.16) and 0.04 (95% CI 0.006–0.15) in those with no prior therapy and those exposed, respectively (P = 0.69, Kruskal test). In the second case, median MAF was 0.05 (95% CI 0.002–0.13) in those with one or two metastatic sites and 0.15 (95% CI 0.009–0.18) in those with three or more (P = 0.24, Kruskal test).
Correlation of MAF in concordant mutant samples in plasma and tissue
We carried out a RAS-adjusted MAF correlation analysis with BEAMing carried out in tumor and ctDNA in the same patient according to prior systemic therapy exposure or number of metastatic sites (Figure 2C and D). Mutational load showed very high heterogeneity and poor correlation, with a Pearson correlation coefficient in the overall population (N = 43) of 0.10 (95% CI −0.21 to 0.39, P = 0.54).
RAS status and correlation with anti-EGFR treatment benefit
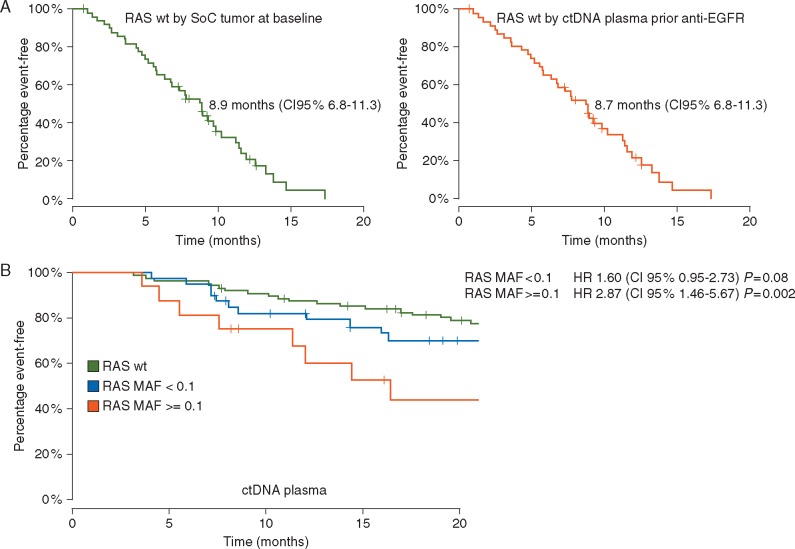
The predictive value of RAS WT status from plasma and tumor determination was analyzed in the subset of patients who received anti-EGFR plus the irinotecan backbone in second- or third-line therapy (N = 52). RAS WT patients detected by SoC (N = 50) had a median PFS of 8.9 months (95% CI 6.8–11.3). RAS WT patients detected by ctDNA (N = 47) showed a median PFS of 8.7 months (95% CI 6.8–11.3) (Figure 3A).
Figure 3.
(A) Progression-free survival after anti-EGFR plus irinotecan-based therapy in the second or third-line setting in RAS wild-type metastatic colorectal cancer patients according to method of RAS mutation detection (SoC tumor tissue at baseline N =50 or ctDNA plasma before therapy N =47). (B) Survival in metastatic setting according to RAS mutant allele fraction by ctDNA plasma. MAF of 0.1 corresponds to a percentage of mutant alleles of 10%. SoC, standard of care.
Potential impact in OS
We describe outcomes for OS according to RAS MAF detection by ctDNA (Figure 3B). In the group of patients with RAS mutant samples with MAF < 0.1 by ctDNA (N = 40), median OS was 27.8 months (95% CI 24.9–47.2), with an HR of 1.60 (95% CI 0.95–2.73; P = 0.08) when compared with RAS WT population. In the group with MAF ≥0.1 (n = 16) median OS was 16.4 months (95% CI 11.4–not reached), and the HR for this group was 2.87 (95% CI 1.46–5.67, P = 0.002) when compared with RAS WT population.
Relevant parameters were included in a multivariable Cox proportional hazards model on the entire cohort: mutation status and MAF in two ranges by ctDNA, tumor location and number of metastatic sites. RAS mutation with MAF ≥0.1 by ctDNA was shown to be a significant prognostic factor with a HR of 2.47 (95% CI 1.2–5.0, P = 0.01) (supplementary Table S7, available at Annals of Oncology online).
Discussion
This is the first clinical series showing the usefulness of detecting RAS point mutations by ctDNA in the largest cohort of patients published so far and carried out locally in a general hospital. Our data revealed a very high overall concordance, close to 90% compared with gold standard tumor tissue analysis techniques. This result is in accordance with previous reports, where RAS mutation detection in cfDNA has been directly compared with tumor tissue in CRC cohorts [4–6]. Siravegna et al. [7] focused on clonal evolution and resistance to EGFR blockade, also described excellent concordance in matched tissue and plasma samples using droplet digital PCR (N = 100). Our results prove the feasibility for implementing this technique in the day-by-day care.
The detailed description of discordant samples reflected in Table 2 confirms the complexity of RAS genotyping in both tumor tissue and plasma samples. Translation of these new technologies to clinical practice reveal not only the technical limitations, but also bring to light conflicting data that provide information about the biological behavior of each tumor. Tumor tissue genotyping has inherent limitations the genomic profiles of primary tumors and metastases are not always concordant owing to the intrinsic molecular tumor heterogeneity [10, 11]. Likewise, several reports have shown differences ranging 3%–20% between different techniques to detect RAS mutations in tissue [12–14]. When analyzing tumor tissue by SoC and BEAMing analysis we detected a 9.1% rate of discordance, mostly justified by differences in sensitivity cut-off.
To account for spatial and temporal changes, the genomic profiles of CRC patients should be evaluated repeatedly during the course of therapy and liquid biopsies could play a role for determinations that are more representative of the specific molecular scenario of a patient at the time of anti-EGFR therapy selection [7, 15]. The possibility of RAS testing at the time of decision-making is one of the strongest points arguing in favor of this minimally invasive technique.
Furthermore, we consider several issues regarding RAS genotyping in plasma need to be highlighted. In our cohort, six patients had mutations in tissue that could not be detected in plasma. Lack of RAS mutations in plasma may be attributed to biological factors that impact ctDNA release and is an important matter that should be investigated. False negative results represent a major issue for RAS mutation testing on plasma because of the possible negative interaction of anti-EGFR agents with oxaliplatin-based regimens in RAS mutant patients.
Commonly used chemotherapeutic agents as well as targeted drugs can alter the molecular landscape in these tumors. It is widely acknowledged that acquired KRAS mutations are associated with secondary resistance to EGFR blockade [15, 16]. However, the effect on the molecular profile derived from other therapies such as anti-angiogenics or cytostatic agents before anti-EGFR administration is yet to be determined [17, 18]. Patients 6 and 9 (Table 2) may be such cases.
Tie et al. [19] reported changes in ctDNA for mCRC patients during the course of the chemotherapy, with significant reductions in ctDNA levels (median 5.7-fold) observed before cycle 2 in 41 of the 48 patients with concordant mutant samples in ctDNA and tissue by SoC. This could impact RAS status determination in patients exposed to therapy, we hypothesize that this could be the case for three patients in our cohort (ID 12–14 in Table 2), although we could not associate this with a homogeneous pattern of response. Taking this a step further, we detected a lower median MAF in the group of patients exposed to prior therapy.
If ultimately we move towards routine RAS determination in plasma in clinical practice, there will likely be subgroups of patients in whom we should continue to perform determinations in tissue for possible alterations in ctDNA release after a negative liquid biopsy.
Although the cohort size of patients with mutations (N = 48) in our study is a somewhat limiting factor, we nonetheless could draw interesting conclusions from analyzing MAF, providing to our knowledge, the first published data in this field. When considering MAF distribution, a high proportion of patients showed mutant alleles in cfDNA between 0.0001 (0.01%) and 0.01 (1%). This highlights the importance of using an extremely sensitive technique when analyzing plasma samples and must be considered at the time of analysis to translate this into clinical practice. Interestingly, there is a tendency for lower MAFs both in tumor tissue and plasma for samples with discordant results, suggesting that sensitivity for mutation detection in tumor tissue is a real issue that needs to be addressed. We found no correlation of RAS MAF with BEAMing carried out in tumor and ctDNA, regardless of prior systemic therapies. The concept of a cut-off for plasma samples similar to that applied in tissue is complex and in our interpretation should not be equivalent.
Finally, in an exploratory analysis, and as an indirect way of confirming the possibility of selecting patients for anti-EGFR therapy with plasma, a PFS analysis was carried out in the most homogeneous group of our cohort, showing no relevant differences between detection methods. To our knowledge no other concordance studies have reported this, and this type of analysis is relevant to the implementation of liquid biopsies in clinical practice.
We can conclude that ctDNA analysis in plasma can detect RAS mutations to an equivalent level as SoC techniques in tissue, and thus detecting potential mCRC patients who could benefit from anti-EGFR therapies.
Supplementary Material
Acknowledgements
The authors acknowledge the excellent medical editing assistance of Sarah MacKenzie (PhD). We want to acknowledge the Cellex Foundation for providing facilities and equipment as well as the Tumor Biomarkers Research Program of the Banco Bilbao Vizcaya Argentaria (BBVA) Foundation for their financial support to the Cancer Genomics Lab, VHIO.
Funding
This work was supported by Merck, S.L., Madrid, Spain, an affiliate of Merck KGaA, Darmstadt, Germany and partially by the Instituto de Salud Carlos III (Ministerio de Economía y Competitividad) and ‘Fondo Europeo de Desarrollo Regional (FEDER), una manera de hacer Europa’ grants [FIS PI12-01589 to RS] and RETICC Cancer: Grupo Cáncer Digestivo – Instituto de Salud Carlos III.TTD ULTRA study (EC11-050) was supported by the Ministerio de Sanidad y Politica Social [SPI/2885/2011]. To CM grants [PI15/00457 and DTS15/00048].
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure
JMV has received honoraria for advisory role from Merck, S.L., Madrid, Amgen and investigation from Merck, S.L., Madrid. CL has received honoraria for advisory role and investigation from Merck, S.L., Madrid, Amgen, Roche, Sanofi. EA has received honoraria for advisory role from Amgen, Bayer, Celgene, Merck, S.L., Madrid, Roche, Sanofi. FJ and VS are employees of Sysmex Inostics, Inc. JT has served in a consulting or advisory role for Amgen, Boehringer Ingelheim, Celgene, Chugai, Imclone, Lilly, Merck, S.L., Madrid, Merck Serono, Millennium Pharmaceuticals, Inc., Novartis, Roche, Sanofi, and Taiho. CM has served in a consultant or advisory role for Merck, S.L., Madrid, Roche and Sanofi. RS has served in a consultant or advisory role for Amgen, Merck, S.L., Madrid, Roche Dx and research funding for Roche Dx. AV has served in a consultant or advisory role for Merck, S.L., Madrid, Merck Serono and Sysmex. All remaining authors have declared no conflicts of interest.
References
- 1. Douillard JY, Siena S, Cassidy J. et al. Final results from PRIME: randomized phase III study of panitumumab with FOLFOX4 for first-line treatment of metastatic colorectal cancer. Ann Oncol 2014; doi:10.1093/annonc/mdu141. [DOI] [PubMed] [Google Scholar]
- 2. Van Cutsem E, Lenz HJ, Kohne CH. et al. Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. J Clin Oncol 2015; 33(7): 692–700. [DOI] [PubMed] [Google Scholar]
- 3. Van Krieken JHJM, Rouleau E, Ligtenberg MJL. et al. RAS testing in metastatic colorectal cancer: advances in Europe. Virchows Arch 2016; 468(4): 383–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bettegowda C, Sausen M, Leary RJ. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014; 6(224): 224ra24.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thierry AR, Mouliere F, El Messaoudi S. et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med 2014; 20(4): 430–435. [DOI] [PubMed] [Google Scholar]
- 6. Schmiegel W, Scott RJ, Dooley S. et al. Blood-based detection of RAS mutations to guide anti-EGFR therapy in colorectal cancer patients: concordance of results from circulating tumor DNA and tissue-based RAS testing. Mol Oncol 2017; 11(2): 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Siravegna G, Mussolin B, Buscarino M. et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 2015; 21(7): 795–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eisenhauer EA, Therasse P, Bogaerts J. et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009; 45(2): 228–247. [DOI] [PubMed] [Google Scholar]
- 9. Landis JR, Koch GG.. The measurement of observer agreement for categorical data. Biometrics 1977; 159–174. [PubMed] [Google Scholar]
- 10. Kim M-J, Lee HS, Kim JH. et al. Different metastatic pattern according to the KRAS mutational status and site-specific discordance of KRAS status in patients with colorectal cancer. BMC Cancer 2012; 12(1): 1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Knijn N, Mekenkamp LJM, Klomp M. et al. KRAS mutation analysis: a comparison between primary tumours and matched liver metastases in 305 colorectal cancer patients. Br J Cancer 2011; 104(6): 1020–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Laurent-Puig P, Pekin D, Normand C. et al. Clinical relevance of KRAS-mutated subclones detected with picodroplet digital PCR in advanced colorectal cancer treated with anti-EGFR therapy. Clin Cancer Res 2015; 21(5): 1087–1097. [DOI] [PubMed] [Google Scholar]
- 13. de Castro DG, Angulo B, Gomez B. et al. A comparison of three methods for detecting KRAS mutations in formalin-fixed colorectal cancer specimens. Br J Cancer 2012; 107(2): 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Azuara D, Santos C, Lopez-Doriga A. et al. Nanofluidic digital PCR and extended genotyping of RAS and BRAF for improved selection of metastatic colorectal cancer patients for anti-EGFR therapies. Mol Cancer Ther 2016; 15(5): 1106–1112. [DOI] [PubMed] [Google Scholar]
- 15. Misale S, Yaeger R, Hobor S. et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012; 486(7404): 532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Diaz LA Jr, Williams RT, Wu J. et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012; 486(7404): 537–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Derangère V, Fumet JD, Boidot R. et al. Does bevacizumab impact anti-EGFR therapy efficacy in metastatic colorectal cancer? Oncotarget. 2016; 7(8): 9309–9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tabernero J, Lenz H-J, Siena S. et al. Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: a retrospective, exploratory analysis of the CORRECT trial. Lancet Oncol 2015; 16(8): 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tie J, Kinde I, Wang Y. et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol 2015; doi:10.1093/annonc/mdv177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.