Abstract
Background
Genetic variations in MicroRNA (miRNA) binding sites may alter structural accessibility of miRNA binding sites to modulate risk of cancer. This large-scale integrative multistage study was aimed to evaluate the interplay of genetic variations in miRNA binding sites of iron regulatory pathway, dietary iron intake and lung cancer (LC) risk.
Patients and methods
The interplay of genetic variant, dietary iron intake and LC risk was assessed in large-scale case–control study. Functional characterization of the validated SNP and analysis of target miRNAs were performed.
Results
We found that the miRNA binding site SNP rs1062980 in 3’ UTR of Iron-Responsive Element Binding protein 2 gene (IREB2) was associated with a 14% reduced LC risk (P value = 4.9×10 − 9). Comparing to AA genotype, GG genotype was associated with a 27% reduced LC risk. This association was evident in males and ever-smokers but not in females and never-smokers. Higher level of dietary iron intake was significantly associated with 39% reduced LC risk (P value = 2.0×10 − 8). This association was only present in individuals with AG + AA genotypes with a 46% reduced risk (P value = 1.0×10 − 10), but not in GG genotype. The eQTL-analysis showed that rs1062980 significantly alters IREB2 expression level. Rs1062980 is predicted to alter a miR-29 binding site on IREB2 and indeed the expression of miR-29 is inversely correlated with IREB2 expression. Further, we found that higher circulating miR-29a level was significantly associated with 78% increased LC risk.
Conclusion
The miRNA binding site SNP rs1062980 in iron regulatory pathway, which may alter the expression of IREB2 potentially through modulating the binding of miR-29a, together with dietary iron intake may modify risk of LC both individually and jointly. These discoveries reveal novel pathway for understanding lung cancer tumorigenesis and risk stratification.
Keywords: microRNA binding site SNP, iron regulatory pathway, circulating miRNA, dietary iron intake, risk of lung cancer
Introduction
The genetic variations in MicroRNAs (miRNA) binding sites of genes may alter structural accessibility of miRNA binding sites and potentially affect the regulatory role of miRNA on gene expression [1]. Studies have shown that miRNA-related single nucleotide polymorphisms (SNPs) affect miRNA processing machinery and target binding sites that modulate a variety of cancers [1], including lung cancer (LC) [2].
As an essential nutrient that facilitates DNA synthesis, cell proliferation, and metabolism, iron is a crucial element that enables the vital function of iron- and/or heme-containing enzymes for normal mammalian cells to survive and replicate [3]. Genes in iron regulatory pathway, such as Iron-Responsive Element Binding Protein 1 and 2 (IREB1 and IREB2), not only regulate iron metabolism but also play important roles in cancer cell reprogramming contributing to malignant growth [3]. Iron also regulates crucial signaling pathways in tumor cell formation including WNT, MYC, and hypoxia-inducible factor (HIF) pathways [4, 5]. Torti et al. [3] highlighted that both beneficial and deleterious aspects of iron and iron regulatory pathway in development of cancer, which has been proved by large-scale epidemiological studies [6, 7]. Although previous LC studies have examined disrupted homeostasis of iron-related signaling pathways in LC cell lines and mice, the evidence of genetic variants of miRNA binding site in intracellular iron regulatory pathways in the risk of LC, especially in the context of dietary iron intake, remains limited and inconclusive. Although genome-wide association studies (GWASs) have identified susceptibility loci on chromosomes 5p15.33, 6p21.33, 15q25, 22q12.1, and 13q12.3 contributing to LC risk in Caucasians [8–10], genetic variants in iron regulatory pathway genes, especially miRNA-binding SNPs, are not well covered in GWAS chips.
Several studies have shown that dietary food intake may modulate the risk of cancer and may play a significant role in miRNA expression in cancer tissues. These findings suggested that miRNA expression may modulate the effect of dietary intake on cancer risk [11]. Iron also participates in potentially harmful reactions such as reactive oxygen species through dietary intake, which results in oxidative damage to DNA and initiates a mutagenic process in host cells [12].
These facts suggests that an integrated investigation is need to comprehensively evaluate the relationship, underline mechanism, and interplay of miRNA-related genetic variants, dietary iron intake, expression of genes/miRNAs and LC risk. In this study, we examined 1) whether miRNA binding site SNPs in genes from iron regulatory pathways may modify LC risk; 2) functional characterization of the identified SNPs and its correlation with predicted miRNAs; 3) the circulating miRNA which were predicted to regulate IREB2 and its association with LC risk; 4) the relationship between dietary iron intake and LC risk; and 5) whether the association was modified by the genetic variants. To our knowledge, this is the first large-scale integrated investigation of miRNA binding site polymorphisms of iron regulatory pathway together with dietary iron intake in modulating the risk of LC.
Methods
Detailed description of methods can be found in supplementary Methods, available at Annals of Oncology online.
Study design and data collection
The schematic of study design was displayed in supplementary Figure S1, available at Annals of Oncology online. This study was approved by MD Anderson's Institutional Review Board, and written informed consent to participate in the study was obtained from each participant before data and biospecimens were collected.
SNP selection and genotyping
Based on previous literatures of iron regulatory pathways [3] and Integrated Pathway Analysis (IPA) software, we identified 80 major candidate genes involved in regulating intracellular iron level and cancer development (supplementary Table S1, available at Annals of Oncology online). Genomic DNA isolation and genotyping were performed as previously described [13].
Dietary iron level analysis
We used a previously validated semi-quantitative food frequency questionnaire (FFQ), a modified version of the National Cancer Institute’s Health Habits and History Questionnaire [14] to assess dietary intake for the year prior to diagnosis or study enrollment.
Functional characterization and serum miR-29a expression
We used public database to perform eQTL analysis. Predicted miRNAs that alter IREB2 expression were identified using miRdSNP database [15]. The association of IREB2 and miRNAs expression was analyzed in TCGA dataset of 554 LC tumor tissues. Serum miR-29a expression was analyzed in 150 stage I–II LC patients from MD Anderson Cancer Center and 172 controls.
Statistical analysis
The association of each SNP with the risk of LC was estimated using the odds ratio (OR) and 95% confidence interval (CI) for the additive models of inheritance. Multivariable unconditional logistic regression was used to adjust for age, gender and smoking status in the discovery population and for age, gender, study, and significant principal components for population stratification within studies (two for PLCO, and one each for EAGLE and ATBC [16]) in the validation population. Meta-analysis was used to estimate the OR and 95% CI for the pooled discovery and validation population. Multivariable unconditional logistic regression was used to estimate the association of dietary iron intake and the risk of LC while adjusting for age, gender, smoking status and total energy intake. Interactions between dietary iron intake and each variable on LC risk were assessed by the likelihood ratio test.
Results
Genetic variants as LC susceptibility loci
The discovery population included 1,656 LC patients from MD Anderson Cancer Center and 1,486 controls. Of 160 SNPs examined, we found eight SNPs in six genes significantly associated with LC risk (P < 0.05). Two SNPs in strong linkage disequilibrium (r2 = 0.9) were found in the same IREB2 that were associated with LC risk: rs4887057 (OR = 0.85, 95%Cl = 0.76–0.95, P = 0.005) and rs1062980 (OR = 0.85, 95%Cl = 0.76–0.95, P = 0.006). The remaining six SNPs in iron regulatory pathway that were significantly associated with LC risk were in Table 1.
Table 1.
Significant associations of miRNA binding site SNPs in iron regulatory pathway with lung cancer risk
| Gene: SNP | Chromosome | Minor allele | Case | Wild-type wild-type/wild-type variant/variant variant |
||
|---|---|---|---|---|---|---|
| Control | ORa (95% Cl) | P value | ||||
| Discovery phaseb | ||||||
| XPD: rs3916874 | 19 | C | 831\689\136 | 820\549\117 | 1.19 (1.06–1.34) | 4.27E-03 |
| BMP2: rs235769 | 20 | A | 619\764\273 | 613\685\188 | 1.17 (1.05–1.31) | 5.34E-03 |
| IREB2: rs1062980 | 15 | G | 690\756\197 | 554\685\234 | 0.85 (0.76–0.95) | 5.57E-03 |
| STEAP2: rs17621350 | 7 | A | 971\577\105 | 916\507\63 | 1.17 (1.03–1.34) | 0.01 |
| PFAS: rs12951103 | 17 | A | 1059\530\67 | 885\517\84 | 0.83 (0.74–1.03) | 5.78E-03 |
| PFAS: rs1132554 | 17 | C | 523\788\334 | 397\733\342 | 0.87 (0.71–1.01) | 8.90E-03 |
| EPAS1: rs7579899 | 2 | A | 622\779\255 | 519\712\255 | 0.89 (0.79–0.99) | 0.03 |
| Validation phase | ||||||
| IREB2: rs1062980 | 15 | G | 2272\2679\745 | 2098\2809\903 | 0.87 (0.82–0.91) | 2.51E-07 |
| Meta-analysis | ||||||
| IREB2: rs1062980 | 15 | G | 2962\3435\942 | 2652\3494\1137 | 0.86 (0.82–0.91) | 4.87E-09 |
Adjusted for age, gender and smoking status in discovery and age, gender in validation.
Genotyping data for rs4887057 was not shown here due to its strong linkage with rs1062980.
In validation phase from dbGaP including a total of 5,699 LC patients and 5,815 control subjects, genotyping data were identified for two SNPs (rs7579899 and rs1062980) and three tag SNPs (rs235769, rs17621350, and rs12951103). Combining all subjects (7,352 cases, 7,301 controls), the minor G allele of rs1062980 was associated with a 14% reduced LC odds with an OR of 0.86 (95%CI = 0.82–0.91) and overall P value of 4.9×10 − 9, which is more than genome-wide significance level (Table 1).
As shown in supplementary Table S2, available at Annals of Oncology online, in the combined analysis, compared to the common AA genotype, the heterozygous AG genotype was associated with significantly reduced LC odds with an OR of 0.88 (95%CI = 0.82–0.95, P = 9.6×10 − 4); and the variant GG genotype had strongest association with 27% reduced LC odds (OR = 0.73; 95%CI = 0.66–0.81; P = 4.4×10 − 9). This association was more evident in male subjects, younger subjects and ever-smokers.
Dietary iron intake and susceptible loci in risk of LC
Among the MD Anderson study population, 1,340 cases and 1,317 controls had dietary information available that was used for analysis of dietary iron intake. A higher dietary iron intake level was significantly associated with 39% reduced odds of LC (OR = 0.61, 95%CI = 0.52–0.73, P = 2.0×10 − 8). Similar results of the association were also observed as dichotomized by tertiles or continuous/ordinal analyses (supplementary Table S3, available at Annals of Oncology online) and adjusted for BMI at diagnosis or BMI 3 years before diagnosis (supplementary Table S4, available at Annals of Oncology online). Bootstrap analysis goodness of fit showed excellent agreement between predicted and observed events (supplementary Table S5, available at Annals of Oncology online). The association was more substantial in males, older subjects, and ever-smokers (Table 2).
Table 2.
Association between dietary iron intake and lung cancer risk
| Dietary iron intake | No. of cases | No. of controls | ORa (95% Cl) | P value | P for interaction |
|---|---|---|---|---|---|
| All | All | ||||
| Low | 659 | 860 | 1(reference) | ||
| High | 658 | 480 | 0.61 (0.52–0.73) | 2.04E-08 | |
| Gender | Male | 1.34E-03 | |||
| Low | 482 | 403 | 1(reference) | ||
| High | 202 | 402 | 0.50 (0.39–0.64) | 1.58E-08 | |
| Female | |||||
| Low | 378 | 256 | 1(reference) | ||
| High | 278 | 256 | 0.79 (0.61–1.01) | 0.06 | |
| Age | <60 years | 0.60 | |||
| Low | 344 | 266 | 1(reference) | ||
| High | 141 | 193 | 0.67 (0.50–0.89) | 6.89E-03 | |
| ≥60 years | |||||
| Low | 516 | 393 | 1(reference) | ||
| High | 339 | 465 | 0.59 (0.47–0.72) | 7.85E-07 | |
| Smoking status | Ever-smoker | 3.60E-04 | |||
| Low | 749 | 350 | 1(reference) | ||
| High | 364 | 326 | 0.52 (0.42–0.63) | 2.36E-10 | |
| Never-smoker | |||||
| Low | 111 | 309 | 1(reference) | ||
| High | 116 | 332 | 0.94 (0.69–1.29) | 0.71 | |
Adjusted for age, gender, smoking status and total energy intake.
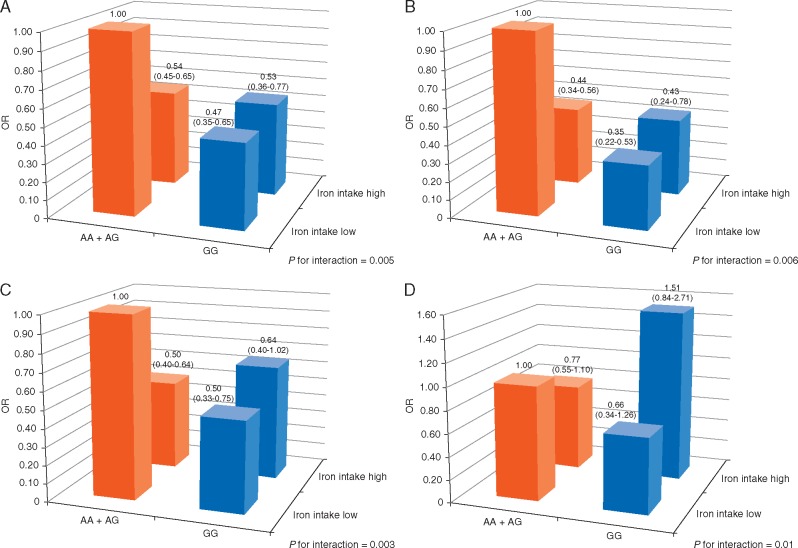
The association of LC and dietary iron intake was also modulated by the genotypes of rs1062980. We found a synergistic interaction between rs1062980 and iron intake in modulating LC risk (P for interaction = 0.005). The protective effect of higher dietary iron intake was evident in individuals carrying common allele genotypes (AA + AG: OR = 0.54, 95%CI = 0.45–0.65, P = 1.0×10 − 10), but not in individuals carrying the rare homozygous genotype (GG: OR = 0.97, P = 0.89). This interaction remained significant in males, older age, and never-smokers with P for interaction of 0.006, 0.003, and 0.01, respectively (Figure 1). The result of additive joint-effects between genotypes of rs1062980 and dietary iron intake was shown in supplementary Table S6, available at Annals of Oncology online.
Figure 1.
Interaction of rs1062980 polymorphism with dietary iron intake in LC risk. The analysis was done for overall population (A), males (B), age ≥ 60 (C), never-smokers (D). Low and high are defined by the median cutoff in controls. OR adjusted for age, gender, smoking status and total energy intake.
Functional characterizing of the SNP
We used eQTL-analysis to examine whether SNP rs1062980 regulates gene expression in three independent public databases: MuTHER, GRASP QTL and GTEx. We observed significant associations of rs1062980 with IREB2 expression in MuTHER, GRASP QTL (supplementary Figure S2A, available at Annals of Oncology online), and GTEx (supplementary Figure S2B, available at Annals of Oncology online). The results indicated that individuals with the high-risk A allele at rs1062980 have significantly lower IREB2 expression level in multiple tissues compared with individuals with lower-risk G allele at rs1062980 with a dose response relationship. All of linked SNPs (LP, r2 > 0.8) of rs1062980 and their imputed function were checked by the HaploReg database (supplementary Table S7, available at Annals of Oncology online) and >95% of them were in or close to the IREB2. The majority of linked SNPs were associated with changed motifs and/or promoter/enhancer marks during protein expression.
Predicted miRNAs that bind to the regions encompassing rs1062980
Since rs1062980 is predicted to be a miRNA binding site SNP, it could alter IREB2 expression by affecting the miRNA binding activity. By using the miRdSNP database, we found 10 predicted miRNAs alter IREB2 expression by binding to the region encompassing rs1062980 at IREB2 (supplementary Table S8, available at Annals of Oncology online).
The correlation of IREB2 and the predicted miRNAs expression were analyzed in 554 LC tumor tissues. We found that miR-29a and 29b were the most strongly inversely correlated with IREB2 expression (rho=−0.25 and −0.26, P = 2.0×10 − 9 and 1.1×10 − 9, respectively). Subsequently, we measured the expression of serum miR-29a in 150 early-stage LC patients and 172 healthy controls and found that a higher miR-29a level was significantly associated with 78% increased odds of LC (OR = 1.78; 95%CI = 1.06–3.00, P = 0.03, supplementary Table S9, available at Annals of Oncology online). The association was more evident in females than that in males.
Discussion
To the best of our knowledge, this is the first large-scale integrative analysis of iron regulatory pathway with LC risk that included data on genotyping, gene expression, miRNA expression, and dietary iron intake levels. In this study, we performed a comprehensive analysis of miRNA binding site SNPs in iron regulatory pathway with LC risk and identified a functional SNP rs1062980 in IREB2 as a LC susceptibility loci. Additionally, we showed significant association of dietary iron intake with LC risk and modulating effect of rs1062980 on the association between dietary iron intake and LC risk. Functional characterization using eQTL-analysis showed a strong correlation of genotypes with IREB2 expression level. Further, evaluation of the predicted miRNAs that target the binding site demonstrated a significant correlation of mir-29a and IREB2 expression level and the significant association between circulating mir-29a and LC risk.
Genetic variations in miRNA binding sites may alter structural accessibility of miRNA binding sites, affect gene expression, and modulate cancer risk. IREB2/IREB1 are master regulators in iron homeostasis by increasing or decreasing protein expression level to regulate intracellular iron [3]. On one hand, mice knocked out IREB2 depleted functional cellular iron and developed microcytic anemia by low TfR1 and high ferritin expression [17]. The depleted functional intracellular iron may further negatively affect the proliferation of host cells such as immune cells. On the other hand, IREB2 expression was 2–10 times higher in cancer cell lines than in somatic cells [18] suggesting that iron regulated by IREB2 is necessary for cancer cell growth and proliferation. IREB2 is located in the 15q25 region which is a well-known GWAS identified LC susceptibility region. However, the molecular function of 15q25 locus is little known. Tekpli et al. [19] showed that polymorphisms of nicotinic receptor genes (CHRNA family) are associated with mRNA level of CHRNA5. However, no GWAS identified SNPs have been shown to associate with IREB2 expression in eQTL-analysis. In this study, we used three independent databases to perform eQTL-analysis and found consistent association of rs1062980 with IREB2 expression. Although rs1062980 was not in strong linkage disequilibrium to any previous published GWAS LC SNPs in 15q25 (data not shown), after adjusting the GWAS identified SNPs, the effect disappeared. However, in contrast to GWAS identified SNPs [20], rs1062980 was not associated with smoking behavior (data not shown) or with nicotinic receptor genes in this study, suggesting the involvement of different signaling pathways. Since smoking is the predominant risk factor of LC, the effect of rs1062980 may be masked by the GWAS identified SNPs related to nicotinic receptor genes which represent mixed signals from smoking behavior, nicotine addiction, acetylcholine receptor activity, and iron regulation. Taken together, our study provides biological plausibility for the rs1062980 polymorphism associated modification of IREB2 expression, which altered LC risk.
In our study population with higher percentage of males than females, the daily dietary iron intake of males in control subjects (mean: 17.9 mg) was comparable to the values of adult male reported by the National Health and Nutrition Examination Survey (NHANES), 1999–2000, for the US population [21], which are 17.2 mg. Consistent with NHANES, the iron intake of female population of our study was lower than male. However, the overall iron intake of adult female in our study (mean: 16.8 mg) was higher than NHANES (mean: 13.4 mg), which may explain the divergent result in stratified analysis of male (P = 1.6×10 − 8) versus female (P = 0.06). As intake for dietary trace metals may vary by region, our female subjects, the majority of whom reside in Houston metropolitan area, might consume more iron compared with the national average leading to the observed results.
Prior studies of serum iron level and cancer risk suggested a J-shape model, both low and high end of iron associated with cancer risk [6], supporting the notion that iron regulatory pathway both beneficially and deleteriously impacts cancer risk [3]. Mahabir et al. [22] found that increased dietary iron intake was associated with decreased LC risk and that the population with the lowest iron and suboptimal DNA repair capacity was at the highest risk of developing LC. However, Zhou et al. [23] suggested that higher LC risk was associated with higher total iron intake, whereas lower cancer risk was associated with higher heme iron. In a large scale meta-analysis study, no significant association of iron intake and LC risk was observed [7]. These discrepancies and inconsistencies might be due to distinct questionnaires and the formulas used to calculate the iron intake and/or the definition for iron/heme iron intake or genetic, gender, age, smoking specific effects as shown in our study. In our study, although we found significant association between high dietary iron intake and LC risk, this association was modulated by the genotype of rs1062980 in IREB2. Higher dietary iron intake was associated with reduced LC risk among carriers of common high-risk A allele containing genotypes of rs1062980; however, higher dietary iron intake was not associated with LC risk among carriers of minor homozygozite low-risk G allele genotype. As shown by the eQTL-analysis, the common high-risk allele was associated with lower IREB2 expression indicating lower intracellular iron activity. Therefore, higher iron intake is needed to boost the intracellular iron activity. The effect of interaction appeared stronger in males, older age, or none smokers possibly suggest the effect underlines the LC initiation and development besides smoking associated LC. The association of miR-29a and cancer has been extensively studied in multiple cancer sites including LC. Consistent with us, at 10 months after diagnosis of LC, the death rate for patients with a high level of miR-29a expression was 6.4 times higher than for patients with a low level of miR-29a expression [24].
There were several strengths in this investigation. The large sample size provided enough power for us to assess the LC risk associated with genetic variants and to stratify all possible confounders to comprehensively evaluate the contribution of LC risk. The validation phrase was conducted to avoid the possibility of false-positive findings. The large population used in this study provided genetic and dietary data as well as complete demographic information, which allowed us to test our hypothesis integratively. The sensitivity analyses, including Bootstrap analysis and consistency of iron intake of general US population and our population, suggested robustness of the findings. We used commonly used FFQ methods to assess dietary intake in this study. While nutrition data collected one-year prior to diagnoses might be a concern, longitudinal studies indicated that a single FFQ measurement could characterize dietary habits for a period of at least 5–10 years [25], and dietary patterns assessed with a FFQ were stable over time [26]. However, these intriguing joint associations between the rs1062980 genotype and dietary iron intake need to be confirmed in a prospective cohort study.
In summary, this is the first large-scale investigation of germline miRNA-related genetic variants in genes of the iron regulatory pathway, miRNA expression, dietary iron intake level, and LC risk. The miRNA binding site SNP rs1062980 in iron regulatory pathway, which may alter the expression of IREB2 potentially through modulating the binding of miR-29a, and together with dietary iron intake may modify the risk of LC both individually and jointly. These results reveal a novel pathway for understanding LC tumorigenesis and risk stratification.
Key Message
Genetic variations in miRNA binding sites may alter structural accessibility of miRNA to modulate cancer risk. The identified miRNA binding site SNP, which may alter the expression of IREB2 through miR-29a, together with iron intake may modify lung cancer risk both individually and jointly. These discoveries reveal novel pathway for understanding lung cancer tumorigenesis and risk stratification.
Funding
This work was supported in part by grants from the Cancer Prevention and Research Institute of Texas (RP1300502) and National Cancer Institute (P50 CA070907, R01 CA176568). Additional funding was provided by MD Anderson institutional support for the Center for Translational and Public Health Genomics, Duncan Family Institute for Cancer Prevention and Risk Assessment (no grant).
Disclosure
The authors have declared no conflicts of interest.
Supplementary Material
References
- 1. Ryan BM, Robles AI, Harris CC.. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer 2010; 10: 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hu Z, Chen J, Tian T. et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest 2008; 118: 2600–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Torti SV, Torti FM.. Iron and cancer: more ore to be mined. Nat Rev Cancer 2013; 13: 342–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu KJ, Polack A, Dalla-Favera R.. Coordinated regulation of iron-controlling genes, H-ferritin and IRP2, by c-MYC. Science 1999; 283: 676–679. [DOI] [PubMed] [Google Scholar]
- 5. Brookes MJ, Boult J, Roberts K. et al. A role for iron in Wnt signalling. Oncogene 2008; 27: 966–975. [DOI] [PubMed] [Google Scholar]
- 6. Wen CP, Lee JH, Tai YP. et al. High serum iron is associated with increased cancer risk. Cancer Res 2014; 74: 6589–6597. [DOI] [PubMed] [Google Scholar]
- 7. Fonseca-Nunes A, Jakszyn P, Agudo A.. Iron and cancer risk–a systematic review and meta-analysis of the epidemiological evidence. Cancer Epidemiol Biomarkers Prev 2014; 23: 12–31. [DOI] [PubMed] [Google Scholar]
- 8. Amos CI, Wu X, Broderick P. et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet 2008; 40: 616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang Y, Broderick P, Webb E. et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet 2008; 40: 1407–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu Z, Wu C, Shi Y. et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat Genet 2011; 43: 792–796. [DOI] [PubMed] [Google Scholar]
- 11. Ross SA, Davis CD.. MicroRNA, nutrition, and cancer prevention. Adv Nutr 2011; 2: 472–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dizdaroglu M, Jaruga P.. Mechanisms of free radical-induced damage to DNA. Free Radic Res 2012; 46: 382–419. [DOI] [PubMed] [Google Scholar]
- 13. Liang D, Meyer L, Chang DW. et al. Genetic variants in MicroRNA biosynthesis pathways and binding sites modify ovarian cancer risk, survival, and treatment response. Cancer Res 2010; 70: 9765–9776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Block G, Coyle LM, Hartman AM, Scoppa SM.. Revision of dietary analysis software for the Health Habits and History Questionnaire. Am J Epidemiol 1994; 139: 1190–1196. [DOI] [PubMed] [Google Scholar]
- 15. Bruno AE, Li L, Kalabus JL. et al. miRdSNP: a database of disease-associated SNPs and microRNA target sites on 3'UTRs of human genes. BMC Genomics 2012; 13: 44.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Landi MT, Chatterjee N, Yu K. et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet 2009; 85: 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ghosh MC, Tong WH, Zhang D. et al. Tempol-mediated activation of latent iron regulatory protein activity prevents symptoms of neurodegenerative disease in IRP2 knockout mice. Proc Natl Acad Sci USA 2008; 105: 12028–12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang W, Deng Z, Hatcher H. et al. IRP2 regulates breast tumor growth. Cancer Res 2014; 74: 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tekpli X, Landvik NE, Skaug V. et al. Functional effect of polymorphisms in 15q25 locus on CHRNA5 mRNA, bulky DNA adducts and TP53 mutations. Int J Cancer 2013; 132: 1811–1820. [DOI] [PubMed] [Google Scholar]
- 20. Broderick P, Wang Y, Vijayakrishnan J. et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res 2009; 69: 6633–6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ervin RB, Wang CY, Wright JD, Kennedy-Stephenson J.. Dietary intake of selected minerals for the United States population: 1999-2000. Adv Data 2004; 341: 1–5. [PubMed] [Google Scholar]
- 22. Mahabir S, Forman MR, Barerra SL. et al. Joint effects of dietary trace metals and DNA repair capacity in lung cancer risk. Cancer Epidemiol Biomarkers Prev 2007; 16: 2756–2762. [DOI] [PubMed] [Google Scholar]
- 23. Zhou W, Park S, Liu G. et al. Dietary iron, zinc, and calcium and the risk of lung cancer. Epidemiology 2005; 16: 772–779. [DOI] [PubMed] [Google Scholar]
- 24. Joerger M, Baty F, Fruh M. et al. Circulating microRNA profiling in patients with advanced non-squamous NSCLC receiving bevacizumab/erlotinib followed by platinum-based chemotherapy at progression (SAKK 19/05). Lung Cancer 2014; 85: 306–313. [DOI] [PubMed] [Google Scholar]
- 25. Goldbohm RA, van 't Veer P, van den Brandt PA. et al. Reproducibility of a food frequency questionnaire and stability of dietary habits determined from five annually repeated measurements. Eur J Clin Nutr 1995; 49: 420–429. [PubMed] [Google Scholar]
- 26. Hu FB, Rimm E, Smith-Warner SA. et al. Reproducibility and validity of dietary patterns assessed with a food-frequency questionnaire. Am J Clin Nutr 1999; 69: 243–249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.