Abstract
Background
Cell-free DNA (cfDNA) from plasma offers easily obtainable material for KRAS mutation analysis. Novel, multiplex, and accurate diagnostic systems using small amounts of DNA are needed to further the use of plasma cfDNA testing in personalized therapy.
Patients and methods
Samples of 16 ng of unamplified plasma cfDNA from 121 patients with diverse progressing advanced cancers were tested with a KRASG12/G13 multiplex assay to detect the seven most common mutations in the hotspot of exon 2 using droplet digital polymerase chain reaction (ddPCR). The results were retrospectively compared to mutation analysis of archival primary or metastatic tumor tissue obtained at different points of clinical care.
Results
Eighty-eight patients (73%) had KRASG12/G13 mutations in archival tumor specimens collected on average 18.5 months before plasma analysis, and 78 patients (64%) had KRASG12/G13 mutations in plasma cfDNA samples. The two methods had initial overall agreement in 103 (85%) patients (kappa, 0.66; ddPCR sensitivity, 84%; ddPCR specificity, 88%). Of the 18 discordant cases, 12 (67%) were resolved by increasing the amount of cfDNA, using mutation-specific probes, or re-testing the tumor tissue, yielding overall agreement in 115 patients (95%; kappa 0.87; ddPCR sensitivity, 96%; ddPCR specificity, 94%). The presence of ≥ 6.2% of KRASG12/G13 cfDNA in the wild-type background was associated with shorter survival (P = 0.001).
Conclusion(s)
Multiplex detection of KRASG12/G13 mutations in a small amount of unamplified plasma cfDNA using ddPCR has good sensitivity and specificity and good concordance with conventional clinical mutation testing of archival specimens. A higher percentage of mutant KRASG12/G13 in cfDNA corresponded with shorter survival.
Keywords: cell-free DNA, droplet digital PCR, KRAS, multiplex
Introduction
Mutations in codons 12 and 13 of the KRAS gene (KRASG12/G13 mutations) are prevalent in colorectal cancer, non-small cell lung cancer (NSCLC), and others and can be associated with less favorable prognosis or a lack of benefit from anti-epidermal growth factor receptor antibodies [1–6]. Furthermore, preclinical and early clinical data suggest that KRAS mutations can predict the response of advanced low-grade serous ovarian cancer to combinations of PI3K and MEK inhibitors [7]. Therefore, the accurate assessment of KRAS mutation status is critical to therapeutic decisions.
Current practice prescribes KRAS mutation testing of archival formalin-fixed, paraffin-embedded (FFPE) tumor tissue and a lack of adequate samples can preclude mutation analysis in at least 10% of patients with advanced cancers [8]. Also, mutation status can change over time, and discrepancies between the genomic profiles of primary and metastatic tumors may occur [9–11]. Thus, archival FFPE tumor samples, which may be many years old, might not necessarily reflect the pertinent genotype.
Cell-free DNA (cfDNA) is secreted into the circulation by tumor cells and cells in the tumor microenvironment that are undergoing apoptosis or necrosis and can be isolated from plasma as a minimally invasive alternative for determining KRAS mutation status by polymerase chain reaction (PCR) or next-generation-sequencing (NGS) methods [12]. Droplet digital PCR (ddPCR) has better sensitivity than standard quantitative PCR or NGS and simpler workflow than other digital PCR such as BEAMing [12]. The purpose of the present study of patients with advanced cancer was to determine whether the detection and quantification of KRASG12/G13 mutations in unamplified plasma cfDNA by multiplexed ddPCR has an acceptable level of sensitivity, specificity and concordance with conventional clinical testing for KRAS mutations in FFPE tumor samples performed by a Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory. We also sought to determine whether the number of KRASG12/G13 mutant alleles in the wild-type background (mutant allele frequency [MAF] or copy number) is associated with outcomes.
Methods
Patients
The study enrolled patients with progressing advanced cancers and known KRASG12/G13 mutation status from clinical testing of their FFPE specimens (supplementary Methods, available at Annals of Oncology online) who were referred to MD Anderson’s Department of Investigational Cancer Therapeutics for experimental therapies from October 2010 to June 2015. Patients had the option of providing longitudinally collected plasma samples during the course of their therapy (at baseline, then every 3–4 weeks if feasible). This retrospective study was conducted in accordance with MD Anderson’s Institutional Review Board guidelines.
Plasma collection and cfDNA KRASG12/G13 mutation testing
Whole blood was collected in EDTA-containing tubes and centrifuged and spun twice within 2 h to yield plasma. The QIAamp Circulating Nucleic Acid kit (Qiagen, Valencia, CA) was used to isolate cfDNA according to the manufacturer’s instructions, and 16 ng of unamplified cfDNA was tested with a multiplex ddPCR KRASG12/G13 Screening Kit (Bio-Rad, Pleasanton, CA) to distinguish the wild-type allele from the seven most common mutations in the exon 2 (G12A, G12C, G12D, G12R, G12S, G12V, and G13D) using the QX200 Droplet Digital PCR platform (Bio-Rad) according to the manufacturer’s standard protocol (supplementary Table S1, available at Annals of Oncology online). Mutation-specific assays (e.g. G12V, G12D) or more cfDNA (24–100 ng) were used for re-testing for patients whose FFPE specimens but not plasma cfDNA showed a KRASG12/G13 mutation. The investigators performing the mutation analysis of the cfDNA samples were blinded to the results of the FFPE specimens and used appropriate positive and negative controls. The lower limit of detection is approximately 0.2% MAF for the multiplexed screening assay and <0.1% MAF per single well for the mutation-specific assays.
Statistical analysis
Concordance between the mutation analyses of FFPE specimens and plasma cfDNA was calculated using a kappa coefficient. Overall survival (OS) was defined as the time from the date of study entry to the date of death or last follow-up. Time to treatment failure (TTF) was defined as the time from the date of systemic therapy initiation to the date of removal from the treatment. The Kaplan–Meier method was used to estimate OS and TTF, and a log-rank test was used to compare OS and TTF among patient subgroups. Cox proportional hazards regression models were fit to assess the association between patient characteristics and OS or TTF. The Spearman rank coefficient was used to assess correlations. All tests were two-sided, and P values <0.05 were considered statistically significant. All statistical analyses were performed with the GraphPad (GraphPad Software, Inc., La Jolla, CA) or SPSS 23 (SPSS, Chicago, IL) software programs.
Results
Patients
The study enrolled 121 patients with diverse advanced cancers and known KRASG12/G13 mutation statuses of archival FFPE specimens (Table 1). The patients’ median age was 56 years (range, 20–84 years). Most patients were white (N = 86; 71%) and male (N = 68; 56%). The most common tumor type was colorectal cancer (N = 71; 59%), NSCLC (N = 14; 12%), and melanoma (N = 10; 8%). The median time from tissue to blood sampling was 18.5 months (range, 1.1–134.4 months). The median amount of cfDNA isolated per 1 ml of plasma was 42 ng (range, 8–3093 ng).
Table 1.
Characteristics of 121 patients with advanced cancers
| Characteristic | Total No. of patients | No. of patients with KRASG12/G13 mutation in FFPE tumor (%) | No. of patients with KRASG12/G13 mutation in plasma cfDNA* (%) |
|---|---|---|---|
| All | 121 | 88 (73) | 78 (64) |
| Sex | |||
| Male | 68 | 53 (78) | 47 (69) |
| Female | 53 | 35 (66) | 31 (58) |
| Race | |||
| Caucasian | 86 | 60 (70) | 56 (65) |
| African American | 21 | 18 (86) | 13 (62) |
| Hispanic | 13 | 10 (77) | 9 (69) |
| Asian | 1 | 0 (0) | 0 (0) |
| Disease | |||
| Colorectal cancer | 71 | 59 (83) | 53 (75) |
| Non-small cell lung cancer | 14 | 10 (71) | 7 (50) |
| Melanoma | 10 | 1 (10) | 2 (20) |
| Appendiceal cancer | 6 | 6 (100) | 4 (67) |
| Pancreatic cancer | 5 | 5 (100) | 5 (100) |
| Ovarian cancer | 5 | 3 (60) | 3 (60) |
| Uterine cancer | 3 | 2 (67) | 2 (67) |
| Breast cancer | 3 | 1 (33) | 2 (67) |
| Duodenal cancer | 1 | 1 (100) | 0 (0) |
| Squamous head and neck cancer | 1 | 0 (0) | 0 (0) |
| Papillary thyroid cancer | 1 | 0 (0) | 0 (0) |
| Erdheim–Chester histiocytosis | 1 | 0 (0) | 0 (0) |
| Method of tumor KRASG12/G13 testing | |||
| PCR | 58 | 42 (72) | 37 (64) |
| NGS | 50 | 36 (72) | 33 (66) |
| MassARRAY | 13 | 10 (77) | 8 (62) |
FFPE, formalin-fixed, paraffin-embedded; cfDNA, cell-free DNA; PCR, polymerase chain reaction; NGS, next-generation sequencing.
Mutations were detected by testing 16 ng of cfDNA using the KRASG12/G13 multiplex probe.
KRAS G12/G13 mutations in FFPE specimens and plasma cfDNA
Of the 121 patients, 88 (73%) had KRASG12/G13 mutations in FFPE specimens, and 78 (64%) had KRASG12/G13 mutations detectable by multiplex ddPCR in 16 ng of unamplified cfDNA. There was overall agreement between cfDNA and FFPE specimens in 103 cases (85%; kappa, 0.66; standard error [SE], 0.07; 95% confidence interval [CI], 0.52–0.80). The cfDNA test had a sensitivity of 84% (95% CI, 0.75–0.91), specificity of 88% (95% CI, 0.72–0.97), positive predictive value (PPV) of 95% (95% CI, 0.87–0.99), and negative predictive value (NPV) of 67% (95% CI, 0.51–0.81; Table 2). Results were similar irrespective of the method used by the CLIA laboratory for tissue KRAS testing (supplementary Table S2 and File S1, available at Annals of Oncology online).
Table 2.
Concordance assessment of KRASG12/G13 mutations in formalin-fixed, paraffin-embedded (FFPE) tumor tissue and plasma cell-free DNA (cfDNA) from 121 patients with advanced cancers
| KRASG12/G13 mutation in tumor | KRASG12/G13 wild-type in tumor | |
|---|---|---|
| Concordance for plasma samples (16 ng of cfDNA) collected before systemic experimental therapy tested with KRASG12/G13 multiplex probe versus FFPE tumor samples tested in the CLIA-certified laboratory | ||
| KRASG12/G13 mutation in cfDNA, no. of patients | 74 | 4 |
| KRASG12/G13 wild-type in cfDNA, no. of patients | 14 | 29 |
| Observed agreements | 103 (85%); kappa, 0.66; SE, 0.07; 95% CI, 0.52–0.80 | |
| Sensitivity | 84% (95% CI, 0.75–0.91) | |
| Specificity | 88% (95% CI, 0.72–0.97) | |
| Positive predictive value | 95% (95% CI, 0.87–0.99) | |
| Negative predictive value | 67% (95% CI, 0.51–0.81) | |
| Concordance for plasma samples (16–100 ng of cfDNA) collected before systemic experimental therapy tested with KRASG12/G13 multiplex and/or mutation specific probes versus FFPE tumor samples tested in the CLIA-certified laboratory | ||
| KRASG12/G13 mutation in cfDNA, no. of patients | 84 | 4 |
| KRASG12/G13 wild-type in cfDNA, no. of patients | 4 | 29 |
| Observed agreements | 113 (93%); kappa, 0.83; SE, 0.06; 95% CI, 0.72–0.95 | |
| Sensitivity | 95% (95% CI, 0.89–0.99) | |
| Specificity | 88% (95% CI, 0.72–0.97) | |
| Positive predictive value | 95% (95% CI, 0.89–0.99) | |
| Negative predictive value | 88% (95% CI, 0.72–0.97) | |
| Concordance for plasma samples (16–100 ng of cfDNA) collected before systemic experimental therapy tested with KRASG12/G13 multiplex and/or mutation specific probes versus FFPE tumor samples tested in the CLIA-certified laboratory or with droplet digital PCR | ||
| KRASG12/G13 mutation in cfDNA, no. of patients | 86 | 2 |
| KRASG12/G13 wild-type in cfDNA, no. of patients | 4 | 29 |
| Observed agreements | 115 (95%); kappa, 0.87; SE, 0.5; 95% CI, 0.77–0.97 | |
| Sensitivity | 96% (95% CI, 0.89–0.99) | |
| Specificity | 94% (95% CI, 0.79–0.99) | |
| Positive predictive value | 98% (95% CI, 0.92–1.00) | |
| Negative predictive value | 88% (95% CI, 0.72–0.97) | |
SE, standard error; CI, confidence interval.
Sixteen nanograms of cfDNA contain only about 5000 genomic equivalents, which enabled us to reliably detect KRASG12/G13 mutations in samples with a MAF of ≥0.2%. In the 14 cases with known KRASG12/G13 mutations in FFPE specimens but wild-type KRAS in cfDNA, we were able to detect KRASG12/G13 mutations in the cfDNA in seven cases by increasing the amount of cfDNA (median, 74 ng; range, 24–100 ng) and in an additional three cases using mutation-specific probes (G12V, G12D) and 49–100 ng of cfDNA. This yielded overall agreement between cfDNA and FFPE specimens in 113 cases (93%; kappa, 0.83; SE, 0.06; 95% CI, 0.72–0.95), and the cfDNA test had a sensitivity of 95% (95% CI, 0.89–0.99), specificity of 88% (95% CI, 0.72–0.97), PPV of 95% (95% CI, 0.89–0.99), and NPV of 88% (95% CI, 0.72–0.97; Table 2). Furthermore, we were able to retrieve archival FFPE specimens for 2 of 4 cases with KRASG12/G13 mutations in cfDNA but wild-type KRAS in FFPE specimens and using ddPCR identified low-frequency KRASG12/G13 mutations in both of them (0.08% and 0.14%, respectively). This yielded overall agreement between cfDNA and FFPE specimens in 115 cases (95%; kappa, 0.87; SE, 0.05; 95% CI, 0.77–0.97), and the cfDNA test had a sensitivity of 96% (95% CI, 0.89–0.99), specificity of 94% (95% CI, 0.79–0.99), PPV of 98% (95% CI, 0.92–1.00), and NPV of 88% (95% CI, 0.72–0.97; Table 2). Of interest, 1 of 4 patients who had KRASG12/G13 mutations in cfDNA but wild-type KRAS in FFPE specimens had advanced colorectal cancer and received chemotherapy with cetuximab, which resulted in accelerated disease progression (supplementary Figure S1, available at Annals of Oncology online).
KRAS G12/G10 mutations in cfDNA and survival
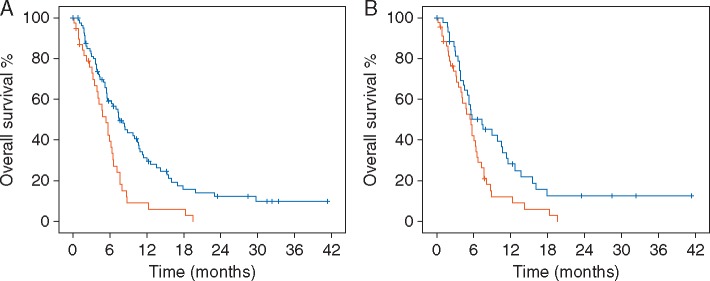
To determine whether the baseline MAF of KRASG12/G13-mutated cfDNA was associated with OS, we divided the 121 patients into two groups according to the percentage of KRASG12/G13-mutated cfDNA (MAF <6.2% versus MAF ≥6.2%). These thresholds were selected based on a 5% trimmed mean value of KRASG12/G13-mutated cfDNA. This was deemed to be representative, as the median percentage of KRASG12/G13-mutant cfDNA was only 0.5% because 33 of 121 patients had no KRASG12/G13 mutations in cfDNA. The median OS of the 82 patients with a KRASG12/G13-mutant cfDNA MAF <6.2% (7.5 months; 95% CI, 5.6–9.4 months) was significantly longer than that of the 39 patients with a KRASG12/G13-mutant cfDNA MAF ≥6.2% (5.4 months; 95% CI, 3.7–7.1 months; P = 0.001; Figure 1A). Similarly, when we used a median percentage of KRASG12/G13-mutant cfDNA (<0.5% versus ≥0.5%) as the cut-off value, the median OS of the 60 patients with a KRASG12/G13-mutant cfDNA MAF <0.5% (8.4 months; 95% CI, 5.5–11.3 months) was significantly longer than that of the 61 patients with a KRASG12/G13-mutant cfDNA MAF ≥0.5% (5.5 months; 95% CI, 4.7–6.3 months; P = 0.004; supplementary Figure S2A, available at Annals of Oncology online). We also performed a separate analysis for patients with colorectal cancer (N = 71, the most frequent cancer type) and found that the median OS of the 41 patients with a KRASG12/G13-mutant cfDNA MAF <6.2% (8.4 months; 95% CI, 5.1–11.7 months) was significantly longer than that of the 30 patients with a KRASG12/G13-mutant cfDNA MAF ≥6.2% (5.7 months; 95% CI, 3.8–7.6 months; P = 0.007; supplementary Figure S2B, available at Annals of Oncology online). In contrast, neither the amount of total cfDNA in plasma (ng/ml) nor the cfDNA concentration (ng/µl) was associated with OS (supplementary Figure S2C and D, available at Annals of Oncology online). Finally, in a separate analysis of patients with KRASG12/G13 mutations in FFPE specimens, the median OS of the 44 patients with a MAF <4% (7.3 months; 95% CI, 3.6–11.0 months) was significantly longer than that of the 44 patients with a MAF ≥4% (5.5 months; 95% CI, 4.3–6.7 months; P = 0.017; Figure 1B).
Figure 1.
(A) Among the 121 patients whose cfDNA samples were tested for KRASG12/G13 mutations, the median overall survival (OS) duration of the 82 patients with a KRASG12/G13-mutant cfDNA percentage of <6.2% (7.5 months; 95% confidence interval [CI], 5.6–9.4 months; blue) was significantly longer than that of the 39 patients with a KRASG12/G13-mutant cfDNA percentage of ≥6.2% (5.4 months; 95% CI, 3.7–7.1; red; P = 0.001). (B) In a separate analysis that included only the 88 patients with KRASG12/G13 mutations in formalin-fixed, paraffin-embedded tumor samples, the median OS duration of the 44 patients with a KRASG12/G13-mutant cfDNA percentage of <4.0% (7.3 months; 95% CI, 3.6–11.0; blue) was significantly longer than that of the 44 patients with a KRASG12/G13-mutant cfDNA percentage of ≥4.0% (5.5 months; 95% CI, 4.3–6.7; red; P = 0.017).
Next, we analyzed the prognostic impact of the MAF of KRASG12/G13-mutant cfDNA on OS using a multi-variable analysis, which included the Royal Marsden Hospital (RMH) prognostic score [13]. The RMH score, a prospectively validated tool used to predict OS in patients with advanced cancers who are referred for early phase clinical trials, is calculated on the basis of lactate dehydrogenase levels (greater than the upper limit of normal versus normal), albumin levels (<3.5 g/ml versus ≥3.5 g/ml), and the number of metastatic sites (>2 sites versus ≤2 sites). Scores of 0 or 1 are associated with longer OS than are scores of 2 or 3. The median OS duration of the 70 patients with an RMH score of 0 or 1 (8.4 months; 95% CI, 5.7–11.1 months) was significantly longer than that of the 51 patients with an RMH score of 2 or 3 (4.4 months; 95% CI, 2.4–6.4 months; P < 0.001). A multi-variable analysis including all 121 patients revealed that, compared to an RMH score of 2 or 3, an RMH score of 0 or 1 was associated with longer OS (hazard ratio [HR], 0.57; 95% CI, 0.36–0.90; P = 0.015; Table 3) and that, compared to a KRAS-mutant cfDNA MAF ≥6.2%, a KRASG12/G13-mutant cfDNA MAF <6.2% demonstrated a trend towards longer OS (HR, 0.63; 95% CI, 0.39–1.02; P = 0.06; Table 3). An additional multi-variable analysis including 88 patients with KRASG12/G13 mutations in FFPE (patients with wild-type KRAS in cfDNA were excluded), which included RMH score (0 or 1 versus 2 or 3) and KRASG12/G13-mutant percentage of cfDNA (≥4.0% versus <4.0%), demonstrated that a KRASG12/G13-mutant cfDNA percentage <4.0% (HR, 0.64; 95% CI, 0.39–1.06; P = 0.08) but not RMH score was associated with a trend towards longer OS (HR, 0.68; 95% CI, 0.42–1.13; P = 0.14; Table 3).
Table 3.
Multi-variable cox regression models evaluating KRASG12/G13-mutant cell-free DNA (cfDNA) percentage and Royal Marsden Hospital (RMH) score with respect to overall survival (OS)
| Variable | HR | 95% CI | P value |
|---|---|---|---|
| OS, all 121 patients | |||
| KRASG12/G13 cfDNA (<6.2% versus ≥6.2%) | 0.63 | 0.39–1.02 | 0.060 |
| RMH score (0 or 1 versus 2 or 3) | 0.57 | 0.36–0.90 | 0.015 |
| OS, 88 patients with KRASG12/G13 in FFPE | |||
| KRASG12/G13 cfDNA (<4.0% versus ≥4.0%) | 0.64 | 0.39–1.06 | 0.080 |
| RMH score (0 or 1 versus 2 or 3) | 0.68 | 0.42–1.13 | 0.140 |
HR, hazard ratio; CI, confidence interval.
Longitudinal testing for KRASG12/G13 mutations in plasma cfDNA
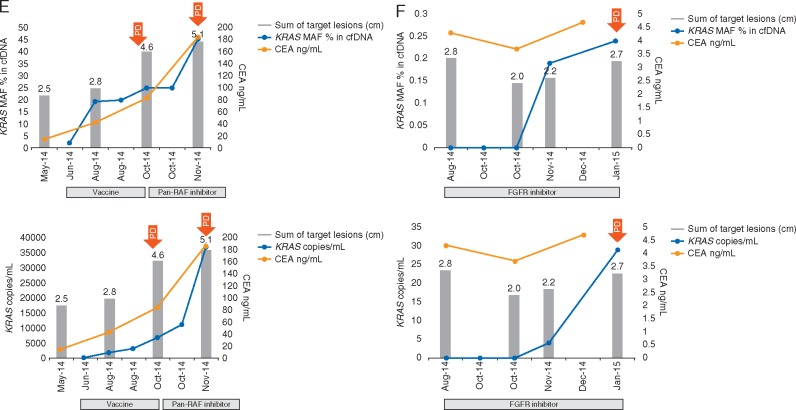
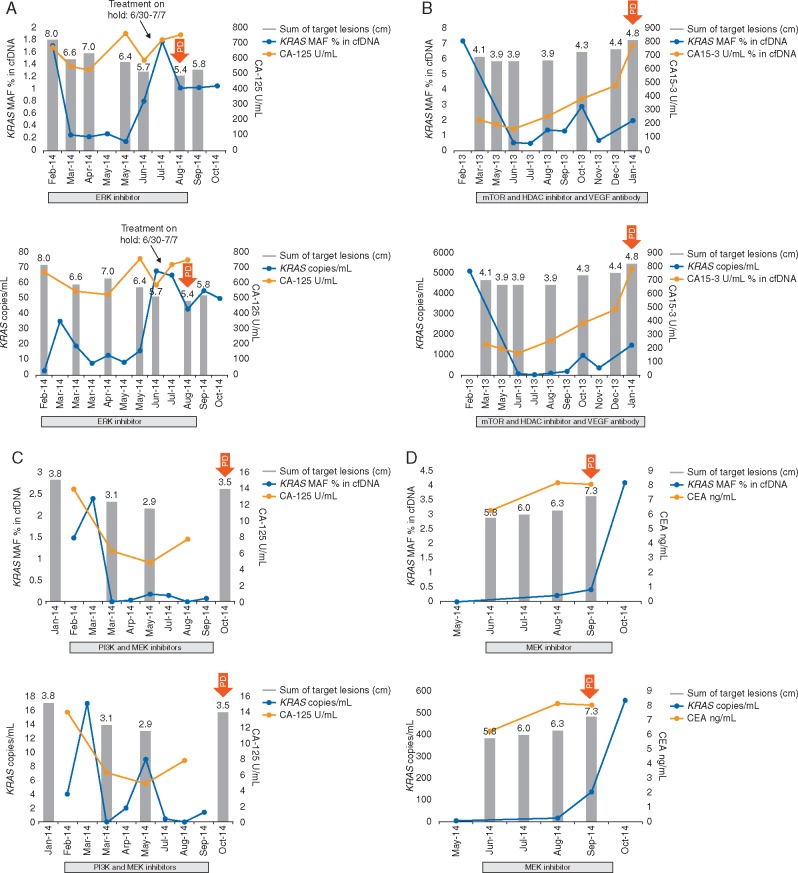
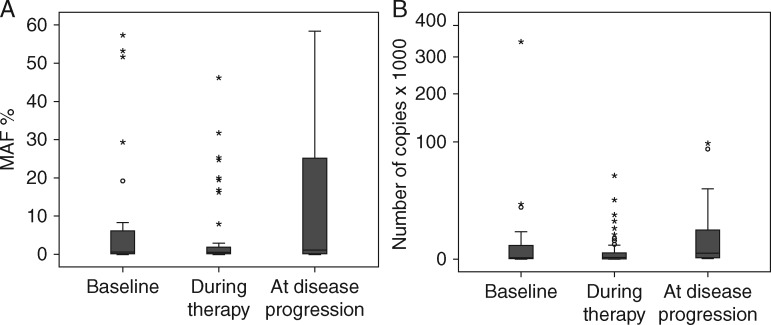
At least two (median, 4; range, 2–12) longitudinal serial plasma collections were obtained before and during systemic therapy from 23 patients with KRASG12/G13 mutations in FFPE specimens who underwent 26 diverse experimental systemic therapies (Figure 2 and supplementary Figure S3, available at Annals of Oncology online). In all 23 patients, KRASG12/G13 mutations were detected in plasma cfDNA at ≥1 time points. Because MAF might be influenced by the number of wild-type copies, which can vary for reasons other than cancer, we also analyzed changes in the number of KRASG12/G13 copies per 1 ml of plasma, which did not take into account wild-type copies [12]. The median KRASG12/G13 MAFs at baseline (0.50%), during therapy (0.22%), and at disease progression (1.13%) differed significantly (P = 0.04), as did the median numbers of KRASG12/G13 copies at baseline (14), during therapy (17) and at disease progression (273; P = 0.03; Figure 3). The best response to therapy on imaging per Response Evaluation Criteria in Solid Tumors (RECIST) was not correlated with the best change in MAF (r = 0.26; P = 0.21) but was correlated with the best change in the number of KRASG12/G13 copies (r = 0.42; P = 0.04) [14, 15]. Of the 26 diverse systemic cancer therapies, 12 decreased the KRASG12/G13 MAF, 14 caused no change or increased the KRASG12/G13 MAF, 11 decreased the number of KRASG12/G13 copies, and 15 caused no change or increased the number of KRASG12/G13 copies. The median TTFs of patients with a decrease in KRASG12/G13 MAF (2.5 months) and those with no change or an increase in KRASG12/G13 MAF (2.8 months) did not differ significantly (P = 0.72); however, there was a trend towards longer TTF for decrease in number of KRASG12/G13 copies compared to no change or increase (3.4 months versus 2.7 months; P = 0.11; supplementary Figure S4, available at Annals of Oncology online). Nevertheless, decrease in size of target tumor lesions on imaging per RECIST vs. no change or increase was the only variable significantly associated with prolonged median TTF (8.2 months versus 2.5 months; P = 0.001; supplementary Figure S4, available at Annals of Oncology online).
Figure 2.
(E) a patient with colorectal cancer who received an intratumoral autologous dendritic cell vaccine; and (F) a patient with non–small cell lung cancer who received targeted therapy with a FGFR inhibitor.
Figure 2.
Dynamic changes in KRASG12/G13-mutated cfDNA in various patients with heavily pretreated KRASG12/G13-mutated advanced cancers. In each panel, the top graph depicts the KRASG12/G13 mutation allelic frequency (MAF) in the wild-type background, and the bottom graph depicts the number of KRASG12/G13 copies per 1 ml of plasma. Red arrows indicate the onset of progressive disease. Shown are sequential measurements of KRASG12/G13 (blue); cancer antigen 125 (CA125) levels (panels A and C), cancer antigen 15-3 (CA15-3) levels (panel B), carcinoembryonic antigen (CEA) levels (panels D–F; orange); and the sum (cm) of target lesions on imaging (grey bars) in (A) a patient with metastatic ovarian carcinoma who received targeted therapy with an ERK inhibitor; (B) a patient with metastatic breast carcinoma who received therapy with an mTOR inhibitor, HDAC inhibitor, and anti-VEGF antibody; (C) a patient with metastatic ovarian carcinoma who received targeted therapy with PI3K and MEK inhibitors; (D) a patient with metastatic ampullary carcinoma who received targeted therapy with a MEK inhibitor.
Figure 3.
(A) KRASG12/G13 mutant allele frequencies (MAFs) at baseline (median, 0.50%), during therapy (median, 0.22%), and at disease progression (median, 1.13%) differed significantly (P = 0.04). (B) KRASG12/G13 copy numbers at baseline (median, 14), during therapy (median, 17), and at disease progression (median, 273) differed significantly (P = 0.03).
Discussion
Our findings demonstrate that the ddPCR KRAS multiplex assay of a small amount of unamplified, plasma cfDNA from patients with advanced cancers can detect KRASG12/G13 mutations and has acceptable concordance (85%), sensitivity (84%), and specificity (88%) compared with the CLIA-certified laboratory-based testing of FFPE tumor tissue obtained at different times during routine care. The addition of more cfDNA with or without the use of mutation-specific probes, as well as retesting discrepant FFPE specimens with ddPCR, resolved the majority of discrepancies between cfDNA and tumor tissue and increased concordance, sensitivity, and specificity to 95%, 96%, and 94%, respectively. We recently demonstrated in a similar patient population that the testing of plasma cfDNA for KRASG12/G13 mutations with BEAMing PCR is concordant with the standard-of-care mutation analysis of FFPE primary or metastatic tumor in 83% of cases, which is similar to the results obtained with the ddPCR KRAS multiplex assay in the present study [16]. A certain level of discordance can be anticipated if the tumor tissue and plasma are not obtained at the same time. Higgins et al. [17] found 100% agreement between PIK3CA mutation testing of plasma cfDNA with BEAMing PCR and tumor tissue collected in a cohort of patients with advanced breast cancer and simultaneous collection. However, the concordance between the methods decreased to 79% in a cohort of patients whose tumor and plasma cfDNA samples were obtained at different times, which is consistent with our results. In addition, Tabernero et al. [18], using BEAMing PCR, showed concordant KRAS mutation status between plasma-derived cfDNA and archival tumor samples in 76% of tested patients with advanced colorectal cancer who had participated in a phase III randomized trial of regorafenib or placebo. Finally, Thierry et al. [19] demonstrated a 96% concordance for combined KRAS and BRAF mutation testing using allele-specific quantitative PCR of plasma cfDNA and mutation detection in primary or metastatic tissue. Our concordance results for KRASG12/G13 mutation compare favorably to most of these previously published studies despite the fact that we used a very low amount of cfDNA, which in most cases was isolated from much less than 0.5 ml of plasma [17–21]. Collectively, there is increasing evidence that the mutation analysis results for cfDNA are highly concordant with those for archival tumor tissue for concordantly, but not discordantly, collected samples, which may be explained by tumor biology, including heterogeneity and evolution over time [10, 21]. In addition, our KRASG12/G13 multiplex assay detects seven of the most frequent KRASG12/G13 mutations in one reaction; in other PCR approaches, this would require seven separate tests. At the same time, the KRASG12/G13 multiplex ddPCR assay, unlike next-generation sequencing, does not compromise on sensitivity [12].
In the present study, we did not use pre-amplification or other methods of enrichment to detect KRASG12/G13-mutant alleles, as we believe that such approaches can skew KRASG12/G13 MAF or copy number values, which can have important prognostic significance. Indeed, we found that patients with a low KRASG12/G13-mutant cfDNA MAF had a significantly longer median OS duration than patients with a high KRASG12/G13-mutant cfDNA MAF did (7.5 months versus 5.4 months; P = 0.001) and confirmed these trend in multivariable analysis, which included prospectively validated RMH score. We previously used BEAMing PCR to assess plasma cfDNA for KRASG12/G13 mutations in patients with advanced cancers and found that a high amount of KRAS-mutant cfDNA was associated with shorter OS (4.8 months versus 7.3 months; P = 0.008)[16]. In another study, using the Idylla system (Biocartis, Mechelen, Belgium) to detect BRAFV600 mutations in plasma-derived cfDNA from patients with diverse advanced cancers, we also found that a higher percentage of BRAFV600-mutant cfDNA was associated with shorter OS (4.4 months versus 10.7 months, P = 0.005) and, in patients treated with BRAF and/or MEK inhibitors, shorter TTF (3.0 versus 7.4 months, P = 0.001) [22]. Similarly, high baseline levels of KRAS-mutant cfDNA were found to be associated with shorter OS in patients with advanced colorectal cancer who were treated in a phase III randomized trial of regorafenib versus placebo [18]. Also, higher amounts of KRAS-mutant cfDNA were associated with shorter progression-free survival and OS in patients with advanced colorectal cancer treated with irinotecan and cetuximab and in patients with advanced NSCLC treated with carboplatin and vinorelbine [23, 24]. Similarly, a BRAFV600E mutation in cfDNA was associated with shorter OS in a combined analysis of clinical trials of BRAF and MEK inhibitors in patients with advanced melanomas [25].
The detection of molecular aberrations can be used to monitor therapy response [26–31]. In the present study, we assessed serially collected plasma cfDNA from patients treated with systemic therapies and found that the KRASG12/G13 MAFs and copy numbers before therapy, during therapy, and at the time of disease progression differed significantly. In addition, the number of KRASG12/G13 copies but not the KRASG12/G13 MAF was correlated with radiographic response, perhaps because KRASG12/G13 MAF is influenced by the amount of wild-type DNA, which can vary for reasons other than cancer (e.g. inflammation, exertion) [12]. We did not find an association between changes in KRASG12/G13 mutations in cfDNA and TTF, which may have been due to the small number of patients in the study and the lack of an effective KRAS inhibitor in clinical testing. Although several previous studies’ findings have supported the concept that changes in cfDNA can predict or at least correspond with treatment outcomes overall the evidence remains conflicting [20, 21, 29–32].
In summary, the molecular analysis of a small amount of unamplified cfDNA for KRASG12/G13 mutations using a ddPCR multiplex assay to detect the most frequent seven hotspot mutations is feasible and has good concordance with standard mutation testing of discordantly collected FFPE tumor tissue. Our results also suggest that the number of KRASG12/G13-mutant alleles in cfDNA is a prognostic biomarker for OS. Our study had several potential limitations. First, we investigated only KRASG12/G13 mutations, which are clinically relevant to only a limited number of patients with certain tumor types. Second, because the study retrospectively analyzed OS data, its findings with regard to these measures need to be validated in future prospective studies. Third, we used archival tumor tissue, which was not collected at same time as plasma samples. Fourth, more than half of the patients had colorectal cancer, which could have influenced our results. Finally, despite the clinical utility of cfDNA mutation testing is increasingly accepted additional prospective clinical trials in which therapeutic interventions are tailored on the basis of patients’ respective cfDNA mutation statuses are needed.
Supplementary Material
Acknowledgement
Authors thank Mr. Joseph Munch for his kind help with editorial assistance.
Funding
Sidney Kimmel Foundation for Cancer Research; Sheikh Khalifa Al Nahyan Ben Zayed Institute for Personalized Cancer Therapy; National Center for Advancing Translational Sciences (grant no. UL1 TR000371); National Institutes of Health through MD Anderson’s Cancer Center Support Grant (P30 CA016672).
Disclosure
FJ has research support from Novartis, Biocartis, and Trovagene and is on the Scientific Advisory Boards of Trovagene and Guardant Health. DNS and GAK-N are employees of Bio-Rad. All remaining authors have declared no conflicts of interest.
References
- 1. Said R, Ye Y, Falchook GS. et al. Outcomes of patients with advanced cancer and KRAS mutations in phase I clinical trials. Oncotarget 2014; 5: 8937–8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hutchins G, Southward K, Handley K. et al. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol 2011; 29: 1261–1270. [DOI] [PubMed] [Google Scholar]
- 3. Ihle NT, Byers LA, Kim ES. et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst 2012; 104: 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garrido-Laguna I, Hong DS, Janku F. et al. KRASness and PIK3CAness in patients with advanced colorectal cancer: outcome after treatment with early-phase trials with targeted pathway inhibitors. PLoS One 2012; 7: e38033.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Douillard JY, Oliner KS, Siena S. et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 2013; 369: 1023–1034. [DOI] [PubMed] [Google Scholar]
- 6. De Roock W, Claes B, Bernasconi D. et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 2010; 11: 753–762. [DOI] [PubMed] [Google Scholar]
- 7. Bedard PL, Tabernero J, Janku F. et al. A phase Ib dose-escalation study of the oral pan-PI3K inhibitor buparlisib (BKM120) in combination with the oral MEK1/2 inhibitor trametinib (GSK1120212) in patients with selected advanced solid tumors. Clin Cancer Res 2015; 21: 730–738. [DOI] [PubMed] [Google Scholar]
- 8. Tsimberidou AM, Iskander NG, Hong DS. et al. Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res 2012; 18: 6373–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sequist LV, Waltman BA, Dias-Santagata D. et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011; 3: 75ra26.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Meric-Bernstam F, Frampton GM, Ferrer-Lozano J. et al. Concordance of genomic alterations between primary and recurrent breast cancer. Mol Cancer Ther 2014; 13: 1382–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vakiani E, Janakiraman M, Shen R. et al. Comparative genomic analysis of primary versus metastatic colorectal carcinomas. J Clin Oncol 2012; 30: 2956–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Polivka J Jr, Pesta M, Janku F.. Testing for oncogenic molecular aberrations in cell-free DNA-based liquid biopsies in the clinic: are we there yet? Expert Rev Mol Diagn 2015; 15: 1631–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arkenau HT, Barriuso J, Olmos D. et al. Prospective validation of a prognostic score to improve patient selection for oncology phase I trials. J Clin Oncol 2009; 27: 2692–2696. [DOI] [PubMed] [Google Scholar]
- 14. Eisenhauer EA, Therasse P, Bogaerts J. et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009; 45: 228–247. [DOI] [PubMed] [Google Scholar]
- 15. Therasse P, Arbuck SG, Eisenhauer EA. et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 2000; 92: 205–216. [DOI] [PubMed] [Google Scholar]
- 16. Janku F, Angenendt P, Tsimberidou AM. et al. Actionable mutations in plasma cell-free DNA in patients with advanced cancers referred for experimental targeted therapies. Oncotarget 2015; 6: 12809–12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Higgins MJ, Jelovac D, Barnathan E. et al. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin Cancer Res 2012; 18: 3462–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tabernero J, Lenz HJ, Siena S. et al. Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: a retrospective, exploratory analysis of the CORRECT trial. Lancet Oncol 2015; 16: 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thierry AR, Mouliere F, El Messaoudi S. et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med 2014; 20: 430–435. [DOI] [PubMed] [Google Scholar]
- 20. Frenel JS, Carreira S, Goodall J. et al. Serial next-generation sequencing of circulating cell-free DNA evaluating tumor clone response to molecularly targeted drug administration. Clin Cancer Res 2015; 21: 4586–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sacher AG, Paweletz C, Dahlberg SE. et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol 2016; 2: 1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Janku F, Huang HJ, Claes B. et al. BRAF mutation testing in cell-free DNA from the plasma of patients with advanced cancers using a rapid, automated molecular diagnostics system. Mol Cancer Ther 2016; 15: 1397–1404. [DOI] [PubMed] [Google Scholar]
- 23. Nygaard AD, Garm Spindler KL, Pallisgaard N. et al. The prognostic value of KRAS mutated plasma DNA in advanced non-small cell lung cancer. Lung Cancer 2013; 79: 312–317. [DOI] [PubMed] [Google Scholar]
- 24. Spindler KL, Pallisgaard N, Vogelius I, Jakobsen A.. Quantitative cell-free DNA, KRAS, and BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin Cancer Res 2012; 18: 1177–1185. [DOI] [PubMed] [Google Scholar]
- 25. Santiago-Walker A, Gagnon R, Mazumdar J. et al. Correlation of BRAF mutation status in circulating-free DNA and tumor and association with clinical outcome across four BRAFi and MEKi clinical trials. Clin Cancer Res 2016; 22: 567–574. [DOI] [PubMed] [Google Scholar]
- 26. Hyman DM, Diamond EL, Vibat CR. et al. Prospective blinded study of BRAFV600E mutation detection in cell-free DNA of patients with systemic histiocytic disorders. Cancer Discov 2015; 5: 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Misale S, Yaeger R, Hobor S. et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012; 486: 532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bettegowda C, Sausen M, Leary RJ. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014; 6: 224ra224.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dawson SJ, Tsui DW, Murtaza M. et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013; 368: 1199–1209. [DOI] [PubMed] [Google Scholar]
- 30. Forshew T, Murtaza M, Parkinson C. et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 2012; 4: 136ra168.. [DOI] [PubMed] [Google Scholar]
- 31. Murtaza M, Dawson SJ, Tsui DW. et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013; 497: 108–112. [DOI] [PubMed] [Google Scholar]
- 32. Diehl F, Schmidt K, Choti MA. et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008; 14: 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.