Abstract
Tissue biopsy is the standard diagnostic procedure for cancer. Biopsy may also provide material for genotyping, which can assist in the diagnosis and selection of targeted therapies but may fall short in cases of inadequate sampling, particularly from highly heterogeneous tumors. Traditional tissue biopsy suffers greater limitations in its prognostic capability over the course of disease, most obviously as an invasive procedure with potential complications, but also with respect to probable tumor clonal evolution and metastasis over time from initial biopsy evaluation. Recent work highlights circulating tumor DNA (ctDNA) present in the blood as a supplemental, or perhaps an alternative, source of DNA to identify the clinically relevant cancer mutational landscape. Indeed, this noninvasive approach may facilitate repeated monitoring of disease progression and treatment response, serving as a means to guide targeted therapies based on detected actionable mutations in patients with advanced or metastatic solid tumors. Notably, ctDNA is heralding a revolution in the range of genomic profiling and molecular mechanisms to be utilized in the battle against cancer. This review will discuss the biology of ctDNA, current methods of detection and potential applications of this information in tumor diagnosis, treatment, and disease prognosis. Conventional classification of tumors to describe cancer stage follow the TNM notation system, heavily weighting local tumor extent (T), lymph node invasion (N), and detectable metastasis (M). With recent advancements in genomics and bioinformatics, it is conceivable that routine analysis of ctDNA from liquid biopsy (B) may make cancer diagnosis, treatment, and prognosis more accurate for individual patients. We put forward the futuristic concept of TNMB tumor classification, opening a new horizon for precision medicine with the hope of creating better outcomes for cancer patients.
Keywords: liquid biopsy, noninvasive, circulating tumor DNA, cancer, cancer staging
Key Message
Ongoing discoveries in the analysis of cell-free circulating tumor DNA (ctDNA) in blood show benefits to cancer patients. This noninvasive, liquid biopsy provides a reservoir of DNA information about cancer status to improve cancer diagnosis, prognosis, and actionable treatment. We propose implementing routine analysis of ctDNA as a means to improve cancer staging and outcome.
Introduction
Malignant tumors are highly heterogeneous at multiple levels [1, 2]. Histologically, tumor tissues may exhibit remarkable variation in morphology and cellular composition within different regions of the same tumor, as well as among different tumors from the same primary site [3, 4]. In the case of metastatic cancer, metastases to regional lymph nodes and at distant sites present further divergence [5]. These heterogeneities may not be fully represented in morphology-based pathological classifications from biopsy of the primary tumor site at diagnosis. More recently, genomic analyses along with molecular characterization of cancers have helped reveal the foundation for these differences [6–8]. Complex relationships with the local tumor environment, particularly immune cells, may alter disease progression. Indeed, there is little doubt that cancer displays dynamic evolution during disease progression. Both intrinsic and extrinsic forces act to drive cancer cells to move—to metastasize—transforming cancer to a systemic disease. While initial biopsy diagnosis and primary tumor site offer critical information for cancer treatment, new methodologies offering diagnostic sensitivity and longitudinal assessment are needed.
Cancer management relies on staging at initial cancer diagnosis. Conventional staging of malignant tumors follows the TNM notation system, encompassing tumor extent (T), lymph node invasion (N), and detectable metastasis (M) [9]. New understanding of genetic and molecular drivers for cancer has led to development of drugs targeting these mutational events [10], and targeted therapy has provided increasing numbers of success stories [11, 12]. Too often, however, these responses are short-lived, and it is understood that a major limitation is the heterogeneous, fundamentally dynamic, and inherently systemic nature of cancer. Considering these barriers, by improving disease characterization—both initially and over cancer progression—the more precisely targeted therapy can be applied. Insight into the systemic nature of cancer has emerged from the study of circulating tumor cells (CTCs), recognizing that cancer cells and their by-products can be detected circulating in blood [13]. Interest in human genomics has driven rapid advances in DNA and RNA sequencing technologies. Streamlined and highly sensitive next-generation sequencing (NGS) has facilitated analysis of cancer mutations in blood [14, 15]. This expanding capacity to detect cancer-specific mutations in circulation, particularly using a noninvasive procedure that enables resampling over time, provides tremendous potential for cancer diagnosis, prognosis, and actionable treatment [16]. Improved characterization of CTCs [17] and cancer-derived DNA [18], RNA [19], and protein-based [20] markers offers additional targets for development of therapeutics. Although not a new concept, a shift toward routine implementation of liquid biopsy as a cancer diagnostic and prognostic tool stands to benefit patients by providing a noninvasive means to detect clinically actionable genetic alterations, and importantly, to monitor disease progression and treatment resistance in real time (Figure 1).
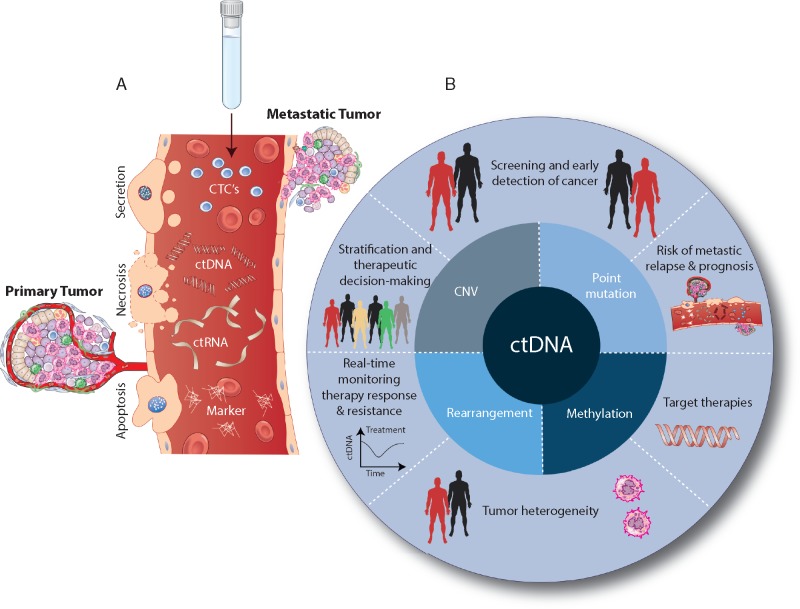
Figure 1.
Biological characteristics and clinical applications of circulating tumor DNA (ctDNA). (A) Circulating tumor cells (CTCs), ctDNA, circulating tumor RNA (ctRNA), and circulating tumor protein as complementary blood-based biomarkers. Tumor cells release ctDNA into the bloodstream through apoptosis, necrosis, and secretion. A subset of aggressive ctDNA enters the bloodstream from the primary tumor or metastatic lesions. (B) Central circle illustrates ctDNA as having multiple classes of genetic and epigenetic alterations. Outer circle presents the potential clinical benefits of ctDNA monitoring in cancer management. CNV, Copy Number Variation.
The clinical applications of circulating tumor DNA (ctDNA) as a ‘liquid biopsy’ have been actively investigated in recent years. Advancements in NGS technologies are making DNA-based liquid biopsy feasible to gain understanding of cancer mechanisms and to predict drug responses [21, 22]. Recent studies support the role of ctDNA as a clinically valuable readout for prognostic staging at diagnosis and monitoring over time [23]. In this new era of precision medicine, we propose that there is diagnostic, therapeutic, and prognostic value to implementing blood-based ctDNA testing concurrent with updating the current TNM system of cancer staging to include this information in a modified system, we suggest as ‘TNMB’ staging.
The history of ctDNA and its utility in cancer
The first evidence of cell-free DNA (cfDNA) in blood dates back to 1948 when Mandel and Métais observed circulating DNA and RNA in human plasma [24]. The implications of this discovery remained obscure for decades, until Leon et al. reported increased concentrations of cfDNA in the circulation of cancer patients in 1977 [25]. After another decade Stroun et al. [26] provided conclusive evidence of neoplastic ctDNA in the serum of cancer patients. The definitive link came in 1994, when Sorenson et al. [27] detected mutated KRAS oncogene sequences in plasma cfDNA by allele-specific polymerase chain reaction (PCR) and convincingly linked the mutant DNA fragments to the original patient tumor. These investigations, over the course of nearly half a century, opened a door to the potential utility of analyses of ctDNA in blood for cancer diagnosis, prognosis, and treatment.
The process by which tumor DNA enters into circulation, consequently, has been of interest, and studies [28–30] have revealed multiple mechanisms (Figure 1A). Migrating tumor cells can enter the bloodstream directly as CTCs (not a topic for this review, see [31]) and may contribute as a source of ctDNA. Actively growing tumors also experience periods of heightened apoptosis or necrosis, processes demonstrated to release DNA into circulation [32]. Tumor cells closely interact with vascular cells and are shown to shed DNA into circulation. These ctDNAs maintain tumor-specific genetic and epigenetic aberrations; including point mutations in tumor suppressors and oncogenes [22, 33], copy number variants [34, 35], DNA methylation patterns, and chromosomal rearrangements [36]. The advantages of noninvasive blood collection over surgery, or even needle biopsy, to identify the genetic and molecular defects reflective of the tumor mass(es) have produced tremendous interest in liquid biopsy. It is envisioned that blood testing could be applied to cancer screening, early detection, evaluation of tumor heterogeneity, observation of dynamic changes, identification of genetic/epigenetic alterations for targeted therapy, and assessment of drug resistance (Figure 1B). Utilization of ctDNA, through implementation of liquid biopsy can provide a new era of comprehensive genomic profiling during the full course of disease, from initial diagnosis through treatment and progression.
Recent growth in literature attests to the value of ctDNA in cancer diagnosis, prognosis, and monitoring of disease progression and therapy response (Figure 2 and Table 1). In 2005, methylation in ctDNA was shown useful for early detection [37] and much later as a diagnostic tool for monitoring cancer dynamics [38]. In 2006, Kimura et al. [39] demonstrated the role of EGFR mutations in predicting response to treatment with molecularly targeted inhibitors; a similar line of study was reported by Kuang et al. [40]. In 2008, ctDNA dynamics measured in patients undergoing treatment for colorectal cancer (CRC) reflected tumor responses and progression, and ctDNA detection after surgery indicated residual disease [41]. In 2013, Murtaza et al. [42] described utility of exome sequencing of cfDNA from serial plasma samples to study clonal evolution and to track ctDNA dynamics in high-burden disease. Additional examples follow.
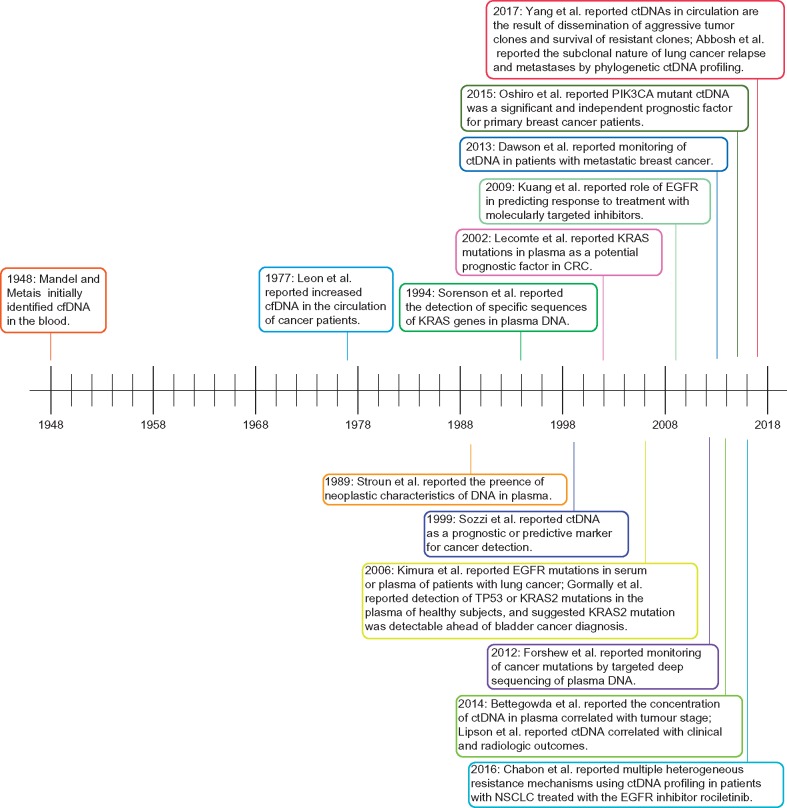
Figure 2.
ctDNA discovery and concomitant developments in cancer: a timeline of genomics advances impacting cancer research over the past 70 years. Landmark discoveries are indicated on the timeline; the numbers of which have increased dramatically in the past 10 years owing in large part to next-generation sequencing capabilities.
Table 1.
Trends in ctDNA literature by cancer type
| Cancer types | 1989–1999 | 2000–2004 | 2005–2009 | 2010–2014 | 2015– |
|---|---|---|---|---|---|
| NSCLC | 1 [104] | 6 [39, 40, 105–108] | 22 [49, 50, 56, 76, 79, 82, 84, 89, 90, 109–121] | 21 [10, 14, 16, 47, 52, 57, 64, 8 3, 85, 122–133] | |
| SCLC | 1 [134] | ||||
| Lung cancer | 1 [78] | 1 [135] | |||
| CRC | 1 [77] | 2 [41, 136] | 12 [48, 98, 137–146] | 9 [15, 62, 87, 147–152] | |
| Colon cancer | 1 [73] | ||||
| Breast cancer | 4 [66, 69, 153, 154] | 7 [8, 61, 67, 99, 155–157] | |||
| Ovarian cancer | 1 [158] | 1 [159] | 2 [160, 161] | ||
| HCC | 2 [34, 162] | 2 [163, 164] | |||
| Brain tumor | 1 [165] | ||||
| Pancreatic adenocarcinoma | 2 [27, 166] | 1 | |||
| Bladder cancer | 1 [167] | ||||
| Prostate cancer | 1 [35] | 2 [53, 168] | |||
| Renal cell carcinoma | 1 [1] | ||||
| Melanoma | 1 [169] | 5 [88, 170–173] | |||
| Lymphoma | 1 [174] | ||||
| Nasopharyngeal carcinoma | 1 [175] | 1 [176] | |||
| Various type | 1 [26] | 1 [32] | 2 [177, 178] | 4 [36, 42, 71, 179] | 9 [21, 22, 97, 180–185] |
Note: The number of publications followed by the references in the bracket are shown in the table.
Detection and quantification of ctDNA: technological development
Clinical applications for liquid biopsy are largely driven by technology development from genomics research. Applications include real-time PCR (qPCR) [43, 44]; the Scorpion Amplification-Refractory Mutation System [45]; Beads, Emulsion, Amplification and Magnetics [46]; digital PCR (dPCR) [47, 48]; and NGS [49, 50]. Being most broadly applicable, dPCR and NGS are briefly described.
dPCR to identify targetable mutations and track targeted therapy responses
The dPCR approach expands quantification of gene expression beyond traditional qPCR by directly quantifying the exact number of target molecules. In this manner, dPCR allows detection of a single copy of mutated ctDNA, even in complex mixtures, rendering it highly sensitive [51]. Importantly, dPCR has a relatively simple workflow facilitating implementation in the clinical laboratory.
NGS to screen and detect disease burden
NGS refers to high-throughput genome sequencing using one of several available platforms. Through parallel sequencing of millions of DNA templates, NGS reveals a large portion of the genome. The richness of NGS is realized in the diversity of genomic input material (whole-genome, exome, de novo, targeted, RNA, ChIP, methylation, etc.) and analysis pipelines. Currently, multiple applications are used in oncology, such as targeted sequencing (gene panels) and whole-exome or whole-genome sequencing [14, 21, 50, 52, 53].
Each analysis method has its own diagnostic niche. dPCR is rapid, relatively inexpensive, and allows quantitation of mutant alleles, particularly at very low concentrations. A significant constraint, however, is required a priori knowledge of specific mutations for analysis, with limited detection of rearrangements, and analysis of many mutations presenting a challenge. NGS allows multiplex analysis of thousands of genomic positions and readily detects rearrangements and copy number variation. There is a growing consensus that NGS will be the test of the future but the current downside is high cost of the deep sequencing necessary for high sensitivity. With technological demand leading to innovation, however, the detection capabilities should logically lead toward cost-effective gene coverage. Of note, GRAIL—a new company with a mission to facilitate early detection of cancer—is pioneering in this direction by development of higher coverage and deeper sequencing depth that is poised to significantly circumvent these limitations [54, 55]. Our recent study [22] showed that even a single mutation in blood from cancer patients was diagnostic of decreased survival. Moreover, mutation rates in plasma were cancer stage-independent, supporting wide-ranging early diagnostic relevancy of a sensitive NGS-based approach. Newman et al. introduced an economical, ultrasensitive modification for quantifying ctDNA: Cancer Personalized Profiling by deep Sequencing (CAPP-Seq) to expand ctDNA detection for broad clinical applicability [56]. The use of CAPP-Seq in non-small-cell lung cancer (NSCLC) revealed emergence of drug-specific mutational patterns associated with resistance to EGFR-targeted therapies [57]. In a majority of patients analyzed, post-treatment residual disease detected at the molecular level by CAPP-Seq preceded cancer progression detected by radiographic measures by a median of 5.2 months [58]. MSKCC researchers using the Grail platform for NGS, reported detection of mutations in circulation in 89% of 151 metastatic cancer patients using ultra broad coverage (508 genes) and ultra-deep sequencing (60 000×) [59]. Accumulating evidence suggests that ctDNA detection techniques are quantitative and that changes in ctDNA levels during chemotherapy are associated with tumor response or progression in several tumor types [41, 60–62]. These studies begin to demonstrate the clinical utility of ctDNA detection at all stages of disease.
New insight into cancer hallmarks through ctDNA
Our current understanding of cancer is derived primarily from investigation by cancer types and their tissues of origin. The systems for staging and for designing cancer therapeutics also rely on this information. The unprecedented ability to interrogate cancer as a systemic disease by examining ctDNA has led to a number of new perspectives.
Tumor heterogeneity and clonal evolution
Cancer is a heterogeneous disease with respect to the molecular mechanisms underlying its development. Solid tumors also exhibit temporal heterogeneity, evolving spontaneously over time and shaped by responses to selection-pressures, such as the immune system and treatment. Almost all cancers treated with anticancer agents have the capacity for resistance as a function of tumor heterogeneity, clonal evolution, and selection [63].
Jamal-Hanjani et al. [64] prospectively investigated intratumor heterogeneity in relation to measures of clinical outcome, clonal nature of driver events, and evolutionary processes in early-stage NSCLC. They reported that driver mutations of EGFR, MET, BRAF, and TP53 were almost always clonal. In addition, heterogeneous driver alterations occurred later in evolution in more than 75% of tumors, commonly in PIK3CA and NF1 and genes involved in chromatin modification and DNA damage response and repair. Gerlinger et al. [1] observed intratumor heterogeneity in renal carcinoma, detecting a minority of the total genetic burden in any one biopsy. Of note, among compartmentalized sites within the same tumor, they report convergent evolution events among several tumor suppressor genes leading to loss of function. This complex level of mutational heterogeneity highlights consequences for patients if treatment decisions depend on results from an initial, prototypical tumor biopsy. Consequently, relevant mutations might be overlooked.
Tumor subclones may arise during disease progression, altering the proportion and pattern of specific aberrations between the primary tumor and metastases [65]. The analysis of ctDNA addresses this issue, because ctDNA released from multiple tumor regions may reflect both intratumoral heterogeneity [66] and clonal evolution [16]. Evidence demonstrates that ctDNA from plasma reveals this clonal tumor hierarchy in cancer. Murtaza et al. [67] extensively analyzed sequential tumor tissue biopsies and plasma ctDNA samples in a ER+/HER2+ breast cancer patient over 3 years, including multiple metastatic deposits, and determined that ctDNA reflected the dynamics of clonal evolution over disease progression. CAPP-seq screening of ctDNA by Chabon et al. [57] identified multiple heterogeneous resistance mechanisms after EGFR inhibitor treatment in patients with NSCLC. They described a novel EGFR L798I mutation and found that EGFR-C797S, which arises in ∼33% of patients after osimertinib treatment, occurred in <3% after rociletinib. Increased MET copy number was the most frequent rociletinib resistance mechanism and patients with multiple preexisting mechanisms (EGFR-T790M and MET) had inferior responses. Interestingly, Abbosh et al. [16] demonstrated the subclonal nature of lung cancer relapse and metastases by a new tumor-specific phylogenetic approach using ctDNA profiling. They showed that mean plasma variant allele frequency of clonal SNVs was higher than that of subclonal SNVs, supporting use of clonal alterations as a more sensitive method of ctDNA detection than subclonal alterations. They also demonstrate the feasibility of using ctDNA platforms to guide drug development, identify residual disease, and target emerging subclones before clinical recurrence in NSCLC. Yang et al. [22] demonstrates a key goal of tumor heterogeneity investigations is to identify clinically aggressive or therapy-resistant clones. Analysis of DNA mutations in lung tumor tissue compared with liquid biopsy from the same patients revealed that mutations in TP53, EGFR, BRAF, CTNNB1, ARID1A, ERBB2, and PDGFRA present in minor tumor clones were detectable in plasma. This study points to ctDNA in circulation as a meaningful indication of dissemination of aggressive tumor clones and survival of resistant clones. Thus, ctDNA analyses have provided direct evidence of spatial and temporal intratumor heterogeneity and show that the range of subclonal heterogeneity is variable among cancers.
ctDNA improves early detection of metastasis
The metastatic process is a complex evolutionary progression of cellular events whereby malignant cells from the primary tumor become migratory and move into the circulation, either directly via a blood vessel or indirectly via a lymphatic vessel, to finally inhabit distant sites as metastases [5]. Since tumor biopsy from one site may not completely reveal the genomic landscape of a patient’s disease burden, blood ctDNA analysis to characterize cancer subclones would help guide treatment decisions. Deryugina et al. [68] illustrated that metastasis can occur throughout tumor progression, even early stage. Dawson et al. [69] compared radiographic imaging of tumor progression to assays detecting ctDNA, cancer antigen 15-3, and CTCs in 30 women receiving systemic therapy for metastatic breast cancer. Of these biomarkers, ctDNA levels showed the greatest detection sensitivity, dynamic range and correlation to disease burden, importantly providing an average 5-month advantage over CT imaging in detecting disease progression in 53% of patients. The utility of ctDNA detection in late stage monitoring extends applicability to early detection since ctDNA alterations are identified in blood from patients with early stage disease [70, 71]. Higher ctDNA levels in early stage cancers may predict more rapid progression to late stage disease. Notably, Naxerova et al. [72] recently illustrated that lymph node metastases may not always be the source of cancer’s spread to other organs; in 65% of cases lymphatic and distant metastases in CRC arose from independent subclones from the primary tumor. An ultrasensitive approach recently developed at Johns Hopkins demonstrated that ctDNA detects early-stage tumors and that ctDNA levels are associated with disease recurrence and decreased overall survival [23]. Tie et al. also used NGS-based assays to evaluate ctDNA in 1046 plasma samples from a prospective cohort of 230 patients with resected stage II colon cancer; they demonstrated ctDNA detection after resection provides direct evidence of residual disease and identifies patients at very high risk of recurrence [73].
Early detection of ctDNA may identify driver mutations and metastatic markers during tumor progression. Although patients with early stage or minimal residual disease usually have lower levels of ctDNA, deep sequencing may detect specific alterations to allow therapeutic intervention with the goal of preventing metastatic progression [73]. Hence, ctDNA could offer comprehensive insight into a patient’s disease, even at an early stage, long before clinical manifestations of disease progression [74].
Prognostic and predictive implications
ctDNA detection may more accurately estimate patient prognosis. Previous studies have shown that plasma-based testing and detection of molecular heterogeneity can predict patient outcome [75, 76]. In a retrospective study of patients with stages I–III CRC, detection of ctDNA implied a higher risk of recurrence or shorter overall survival in patients treated with surgery, chemotherapy, radiotherapy, or targeted therapy [77]. High levels of KRAS mutated plasma DNA have been reported as an indicator of poor outcome in lung cancer patients [78, 79]. Sirera et al. [80] found that high pretreatment levels of circulating DNA acted as an independent prognostic marker for shorter survival. Moreover, KRAS status in plasma ctDNA was associated with poor tumor response to EGFR tyrosine kinase inhibitors (TKIs) in NSCLC patients and served as a predictive marker for selecting appropriate treatments [78, 81–84]. In a retrospective analysis [85], patients with NSCLC who were positive for EGFR-T790M in plasma showed outcomes with osimertinib equivalent to patients classified as positive by a tissue-based assay, supporting plasma analysis to avoid invasive tumor biopsies in such patients. A systematic review including 23 studies reported that the presence of ctDNA in blood is associated with worse survival in patients with solid tumors [86]. Taken together, these studies propose that ctDNA measures provide insight for patient prognosis that could further inform clinical decision-making.
Monitoring of treatment responses
Detection of molecular aberrations in ctDNA provides a powerful tool to monitor response to therapy and emergence of secondary mutations associated with therapy resistance [22, 61, 76, 87, 88]. Given the dynamic nature of cancer, ctDNA investigation at multiple time points during cancer treatment and progression may provide crucial information for patient management. Studies have reported that first-line treatment of patients harboring EGFR activating mutations with EGFR TKIs gefitinib, erlotinib, or afatinib results in superior overall response rates, progression-free survival and quality of life compared with chemotherapy [40, 83, 84, 89, 90]. Detection of EGFR-T790M mutation at follow-up facilitates treatment with the third generation EGFR TKI Osimertinib [59]. Thress et al. [10] reported, however, that serial ctDNA monitoring of lung cancer patients treated with Osimertinib revealed a diversity of therapy resistance mechanisms. Cabel et al. [91] demonstrated that quantitative ctDNA monitoring was a valuable tool to assess tumor response in five metastatic melanoma patients treated with anti-PD1 drugs [91]. Furthermore, longitudinal assessment of ctDNA in metastatic melanoma patients who were treated with PD1 inhibitors was an accurate predictor of tumor response and overall survival [92]. Vidal et al. determined baseline RAS at diagnosis and monitored of the emergence of RAS mutations as a mechanism of resistance to anti-EGFR therapy [93]. In advanced prostate cancer, ctDNA can detect aberrations in the androgen receptor and may help to predict for response or resistance to androgen directed therapies [94]. Thus, in such an aggressive disease targeted therapy success is entwined with vigilant monitoring of treatment response. Critically, ctDNA genomic alterations over disease progression provide real-time therapeutic guidance, predict prognosis and assess for therapy resistance ahead of imaging studies.
ctDNA early detection for staging and auxiliary diagnostic screening
The amount of ctDNA detected in blood is correlated with cancer stage and tumor aggressiveness [95]. Comparing late to early stage disease, ctDNA was detected in 100% of patients with stages II–IV NSCLC and in 50% of patients with stage I NSCLC [96]. ctDNA detection shows some variation among tumor types. A high proportion of patients with advanced primary pancreatic, ovarian, colorectal, breast, bladder, esophageal, melanoma, and hepatocellular carcinoma have measurable ctDNA while detection falls under 50% for patients with brain, renal, prostate, and thyroid cancers [71], though this will likely vary with the technology used for ctDNA detection. The same group reports detecting ctDNA from about 50% or more of patients with localized disease (colorectal, esophageal, pancreatic, and breast adenocarcinoma). Moreover, a study with 95% of patients having advanced or metastatic disease reported that 58% of patients had at least one detectable alteration, which increased to 65% when glioblastoma was excluded [97]. In their comprehensive quantitative analysis, Bettegowda et al. [71] reported varying levels of ctDNA across patients with distinct cancer types and provided expected ranges of ctDNA levels across the stages of disease.
Quantifying ctDNA levels enables earlier response assessment than standard radiographic approaches. Misale et al. [98] reported serial ctDNA analysis identified KRAS-mutant alleles in the plasma of cetuximab-treated patients 10 months before disease relapse was identified by imaging. They suggest that ctDNA could supplement standard screening or restaging approaches for cancers—such as mammography for breast, low dose spiral CT for lung, colonoscopy for CRC, and PSA for prostate—potentially increasing diagnostic sensitivity and specificity. In a study designed to monitor treatment response, Garcia-Murillas et al. [99] used NGS of tumor biopsies in patients with early stage breast cancer to identify patient-tumor-specific mutations and developed personalized dPCR assays to detect and track ctDNA in 55 patients receiving neoadjuvant chemotherapy followed by surgery: detection of ctDNA after surgery or during serial sampling was a significant predictor of early relapse. Furthermore, ctDNA provided a median lead time of 7.9 months before discovery of clinical relapse. Similarly, Olsson et al. [61] used ctDNA for early detection of metastasis in women who presented with early stage, nonmetastatic breast cancer and received no neoadjuvant therapy; reporting 13 of 14 patients with eventual clinical recurrence showed positive ctDNA levels postoperatively, whereas patients with long-term disease-free survival had no detectable ctDNA. Critically, ctDNA molecular detection of occult metastasis preceded the clinical diagnosis in 12 of 14 patients with an average lead time of 11 months.
These studies indicate that ctDNA detected in the blood of early stage patients is a robust and independent marker of disease progression, which may significantly enhance the current prognostic assessment. Doctors have already begun to order ctDNA and NGS tests as part of precision oncology programs. Because of the noninvasive nature, implementation of ctDNA testing is straightforward but utility must develop from a network encompassing research knowledge, product/test development and clinical validation.
Proposing a modified cancer staging system for solid tumors: TNMB
The Tumor, Node, and Metastasis staging system was devised by Pierre Denoix during 1943–1952 [100] to classify malignant tumors with a goal of standardizing treatment regimens and survival expectations by providing a uniform guide to describe the anatomical extent of disease. Since introduction, the TNM system has gained wide international acceptance for staging solid tumors. Applying TNM taxonomy criterion, cancers are staged into one of four groupings (I, II, III, and IV) for a diagnosis that provides a prediction of treatment course and disease prognosis. Stage IV, exclusively indicating distant metastases, represents late stage disease and generally poor prognosis, thus imploring any mechanism to better identify systemic disease as being highly meaningful. While prognostic biomarker research in cancer has great potential to drive personalized medicine, the complexity of implementation is equally great. A logical first step, quantifying ctDNA levels in blood, offers a practical solution to the quandary.
In light of new genomic technologies that improve assessment of risk for cancer progression and/or metastatic disease, the time has come to consider amending the TNM system [101]. We propose a modified staging system for future development—‘TNMB’. Figure 3 shows how TNMB (B represents blood) complements TNM staging by adding a liquid biopsy ‘B’ to capture prognostic and therapeutic implications gained from ctDNA evaluation. Paralleling the ‘M’ category, initial categorization may be defined as the absence (‘B0’) or presence (‘B1’) of detectable ctDNA. Although in need of standardized criteria, B staging should apply to most cancers independent of site. As literature accumulates, classification could be refined to include ctDNA quantification data with clinically meaningful cut-offs by site [71, 102]. Future incorporation of data such as mutational burden, actionable mutations, and metastasis-related mutations will further enhance the clinical impact of the ‘B’ designation. Foundation Medicine has recently put forward ctDNA mutation load as an end point for their liquid biopsy test because of demonstrated clinical utility [63]. Prospective studies to compare TNM staging to TNMB staging on their accuracy in predicting cancer recurrence and patient survival are critically needed and it is likely that the growing body of literature on ctDNA will help define clinically meaningful categories within the ‘B’ designation for each tumor type.
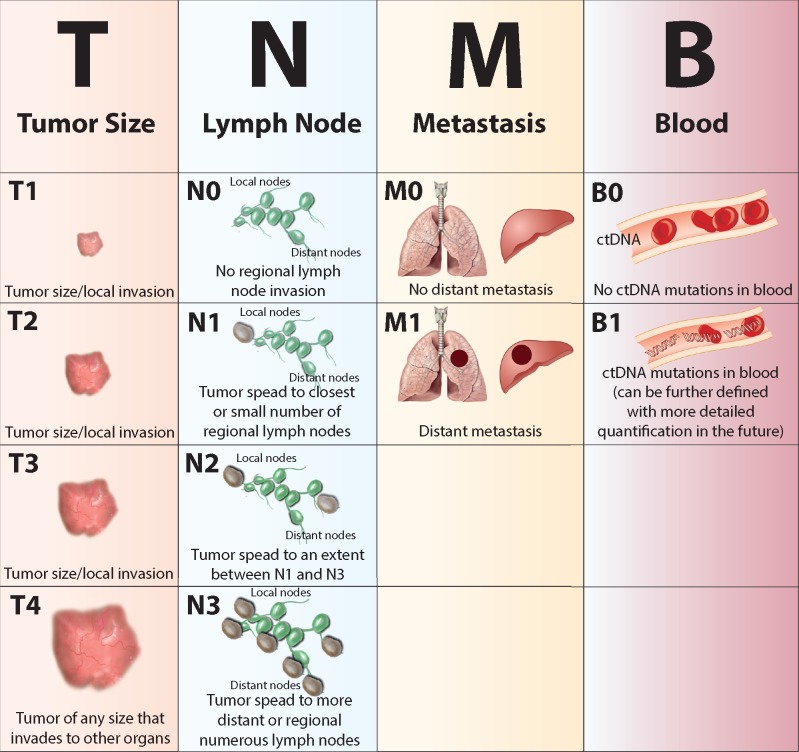
Figure 3.
Generalized overview showing the proposed ctDNA incorporation into the hallmark cancer staging system as TNMB. Each categorical column indicates the anatomical valuation criteria by row for advancing cancer stage, thus reflecting tumor progression. Advancing genomics technologies promise to reveal this previously hidden anatomical indicator—ctDNA— to more accurately stage and treat cancers. Information for TNM staging in this figure derived from the TNM Classification of Malignant Tumours by UICC (7th Edition) [103].
Discussion
Conclusions
Beyond simply reproducing information from tissue biopsies, noninvasive ctDNA analysis offers a comprehensive and integrated view of the systemic evolution of cancer. ctDNA has been shown to be a relevant blood-based biomarker useful as a complementary method for cancer screening and diagnostic tool. By providing material for mutational analysis in the clinical setting, ctDNA facilitates highly sensitive monitoring for the acquisition of treatment resistance. Results of ctDNA detection have been successfully used to guide targeted therapies aiming at key driver events for metastasis. Consequently, we propose that development of a TNMB staging system to include blood ctDNA information to enhance the current TNM cancer staging system. This new component will stimulate further development of specific and sensitive cancer detection technologies for blood biomarkers in the rapidly evolving field of precision oncology. We envision an iterative process is needed where clinical data from population-based cohorts are compiled to evaluate survival associated with staging groups. To initiate use within the TNMB system, large data registries are needed to start capturing ‘B’ so that the next iteration of staging revisions can incorporate the important prognostic information from this variable with the hope to improve outcomes for all cancer patients.
Acknowledgements
The authors would like to thank all the investigators who have contributed to the liquid biopsy project at Wake Forest Baptist Comprehensive Cancer Center that inspired this review article.
Funding
Cancer Center Support Grant from the National Cancer Institute to the Comprehensive Cancer Center of Wake Forest Baptist Medical Center (P30 CA012197). WZ is supported by an endowed Hanes and Willis Professorship in Cancer and a Fellowship by the National Foundation for Cancer Research.
Disclosure
The authors have declared no conflicts of interest.
References
- 1. Gerlinger M, Rowan AJ, Horswell S. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012; 366(10): 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Campbell PJ, Pleasance ED, Stephens PJ. et al. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc Natl Acad Sci USA 2008; 105(35): 13081–13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Bruin EC, McGranahan N, Mitter R. et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 2014; 346(6206): 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang J, Fujimoto J, Zhang J. et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 2014; 346(6206): 256–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Turajlic S, Swanton C.. Metastasis as an evolutionary process. Science 2016; 352(6282): 169–175. [DOI] [PubMed] [Google Scholar]
- 6. Parsons DW, Jones S, Zhang X. et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008; 321(5897): 1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature 2011; 474: 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yates LR, Gerstung M, Knappskog S. et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med 2015; 21(7): 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tazawa K, Tsutsumi Y.. The TNM classification of cancer—from the viewpoint of pathology. Gan to Kagaku Ryoho 1998; 25: 617–622. [PubMed] [Google Scholar]
- 10. Thress KS, Paweletz CP, Felip E. et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med 2015; 21(6): 560–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang JCH, Wu YL, Schuler M. et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol 2015; 16(2): 141–151. [DOI] [PubMed] [Google Scholar]
- 12. Krop IE, Kim SB, Martin AG. et al. Trastuzumab emtansine versus treatment of physician's choice in patients with previously treated HER2-positive metastatic breast cancer (TH3RESA): final overall survival results from a randomised open-label phase 3 trial. Lancet Oncol 2017; 18(6): 743–754. [DOI] [PubMed] [Google Scholar]
- 13. Alix-Panabieres C, Pantel K.. Circulating tumor cells: liquid biopsy of cancer. Clin Chem 2013; 59(1): 110–118. [DOI] [PubMed] [Google Scholar]
- 14. Paweletz CP, Sacher AG, Raymond CK. et al. Bias-corrected targeted next-generation sequencing for rapid, multiplexed detection of actionable alterations in cell-free DNA from advanced lung cancer patients. Clin Cancer Res 2016; 22(4): 915–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Beranek M, Sirak I, Vosmik M. et al. Carrier molecules and extraction of circulating tumor DNA for next generation sequencing in colorectal cancer. Acta Med (Hradec Kralove, Czech Repub) 2016; 59: 54–58. [DOI] [PubMed] [Google Scholar]
- 16. Abbosh C, Birkbak NJ, Wilson GA. et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017; 545(7655): 446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alix-Panabieres C, Pantel K.. Challenges in circulating tumour cell research. Nat Rev Cancer 2014; 14(9): 623–631. [DOI] [PubMed] [Google Scholar]
- 18. Wan JC, Massie C, Garcia-Corbacho J. et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017; 17(4): 223–238. [DOI] [PubMed] [Google Scholar]
- 19. Shen J, Stass SA, Jiang F.. MicroRNAs as potential biomarkers in human solid tumors. Cancer Lett 2013; 329(2): 125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hundt S, Haug U, Brenner H.. Blood markers for early detection of colorectal cancer: a systematic review. Cancer Epidemiol Biomarkers Prev 2007; 16(10): 1935–1953. [DOI] [PubMed] [Google Scholar]
- 21. Frenel JS, Carreira S, Goodall J. et al. Serial next-generation sequencing of circulating cell-free DNA evaluating tumor clone response to molecularly targeted drug administration. Clin Cancer Res 2015; 21(20): 4586–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang M, Topaloglu U, Petty WJ. et al. Circulating mutational portrait of cancer: manifestation of aggressive clonal events in both early and late stages. J Hematol Oncol 2017; 10(1): 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jillian Phallen MSV, Adleff A, Leal C. et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med 2017; 9(403). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mandel P, Metais P.. Les Acides Nucleiques Du Plasma Sanguin Chez Lhomme. C R Seances Soc Biol Fil 1948; 142: 241–243. [PubMed] [Google Scholar]
- 25. Leon SA, Shapiro B, Sklaroff DM, Yaros MJ.. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 1977; 37: 646–650. [PubMed] [Google Scholar]
- 26. Stroun M, Anker P, Maurice P. et al. Neoplastic characteristics of the DNA found in the plasma of cancer-patients. Oncology 1989; 46(5): 318–322. [DOI] [PubMed] [Google Scholar]
- 27. Sorenson GD, Pribish DM, Valone FH. et al. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Biomarkers Prev 1994; 3(1): 67–71. [PubMed] [Google Scholar]
- 28. Stroun M, Lyautey J, Lederrey C. et al. About the possible origin and mechanism of circulating DNA—apoptosis and active DNA release. Clin Chim Acta 2001; 313(1–2): 139–142. [DOI] [PubMed] [Google Scholar]
- 29. Thierry AR, El Messaoudi S, Gahan PB. et al. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev 2016; 35(3): 347–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Diaz LA Jr, Bardelli A.. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014; 32(6): 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thiele JA, Bethel K, Kralickova M, Kuhn P.. Circulating tumor cells: fluid surrogates of solid tumors. Annu Rev Pathol 2017; 12: 419–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jahr S, Hentze H, Englisch S. et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001; 61: 1659–1665. [PubMed] [Google Scholar]
- 33. Wang JY, Hsieh JS, Chang MY. et al. Molecular detection of APC, K-ras, and p53 mutations in the serum of colorectal cancer patients as circulating biomarkers. World J Surg 2004; 28(7): 721–726. [DOI] [PubMed] [Google Scholar]
- 34. Chan KC, Jiang P, Zheng YW. et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem 2013; 59(1): 211–224. [DOI] [PubMed] [Google Scholar]
- 35. Heitzer E, Ulz P, Belic J. et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med 2013; 5(4): 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Leary RJ, Sausen M, Kinde I. et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med 2012; 4(162): 162ra154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fujiwara K, Fujimoto N, Tabata M. et al. Identification of epigenetic aberrant promoter methylation in serum DNA is useful for early detection of lung cancer. Clin Cancer Res 2005; 11: 1219–1225. [PubMed] [Google Scholar]
- 38. Lehmann-Werman RND, Zemmour H, Moss J. et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc Natl Acad Sci USA 2016; 113(13): E1826.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kimura H, Kasahara K, Kawaishi M. et al. Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non-small-cell lung cancer. Clin Cancer Res 2006; 12(13): 3915–3921. [DOI] [PubMed] [Google Scholar]
- 40. Kuang Y, Rogers A, Yeap BY. et al. Noninvasive detection of EGFR T790M in gefitinib or erlotinib resistant non-small cell lung cancer. Clin Cancer Res 2009; 15(8): 2630–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Diehl F, Schmidt K, Choti MA. et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008; 14(9): 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Murtaza M, Dawson SJ, Tsui DW. et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013; 497(7447): 108–112. [DOI] [PubMed] [Google Scholar]
- 43. Li J, Wang L, Mamon H. et al. Replacing PCR with COLD-PCR enriches variant DNA sequences and redefines the sensitivity of genetic testing. Nat Med 2008; 14(5): 579–584. [DOI] [PubMed] [Google Scholar]
- 44. Nagai Y, Miyazawa H, Huqun. et al. Genetic heterogeneity of the epidermal growth factor receptor in non-small cell lung cancer cell lines revealed by a rapid and sensitive detection system, the peptide nucleic acid-locked nucleic acid PCR clamp. Cancer Res 2005; 65: 7276–7282. [DOI] [PubMed] [Google Scholar]
- 45. Duan HL, Lu JL, Lu T. et al. Comparison of EGFR mutation status between plasma and tumor tissue in non-small cell lung cancer using the Scorpion ARMS method and the possible prognostic significance of plasma EGFR mutation status. Int J Clin Exp Pathol 2015; 8: 13136. [PMC free article] [PubMed] [Google Scholar]
- 46. Taniguchi K, Uchida J, Nishino K. et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res 2011; 17(24): 7808–7815. [DOI] [PubMed] [Google Scholar]
- 47. Zhu G, Ye X, Dong Z. et al. Highly sensitive droplet digital PCR method for detection of EGFR-activating mutations in plasma cell-free DNA from patients with advanced non-small cell lung cancer. J Mol Diagn. 2015; 17(3): 265–272. [DOI] [PubMed] [Google Scholar]
- 48. Taly V, Pekin D, Benhaim L. et al. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin Chem 2013; 59(12): 1722–1731. [DOI] [PubMed] [Google Scholar]
- 49. Couraud S, Vaca-Paniagua F, Villar S. et al. Noninvasive diagnosis of actionable mutations by deep sequencing of circulating free DNA in lung cancer from never-smokers: a proof-of-concept study from BioCAST/IFCT-1002. Clin Cancer Res 2014; 20(17): 4613–4624. [DOI] [PubMed] [Google Scholar]
- 50. Narayan A, Carriero NJ, Gettinger SN. et al. Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Cancer Res 2012; 72(14): 3492–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zonta E, Garlan F, Pecuchet N. et al. Multiplex detection of rare mutations by picoliter droplet based digital PCR: sensitivity and specificity considerations. PLoS One 2016; 11(7): e0159094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dietz S, Schirmer U, Merce C. et al. Low input whole-exome sequencing to determine the representation of the tumor exome in circulating DNA of non-small cell lung cancer patients. PLoS One 2016; 11(8): e0161012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ulz P, Belic J, Graf R. et al. Whole-genome plasma sequencing reveals focal amplifications as a driving force in metastatic prostate cancer. Nat Commun 2016; 7: 12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xu T, Kang X, You X. et al. Cross-platform comparison of four leading technologies for detecting EGFR mutations in circulating tumor DNA from non-small cell lung carcinoma patient plasma. Theranostics 2017; 7(6): 1437–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shu YQ, Wu X, Tong XL. et al. Circulating tumor DNA mutation profiling by targeted next generation sequencing provides guidance for personalized treatments in multiple cancer types. Sci Rep 2017; 7: 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Newman AM, Bratman SV, To J. et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014; 20(5): 548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chabon JJ, Simmons AD, Lovejoy AF. et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016; 7: 11815.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chaudhuri AA, Chabon JJ, Lovejoy AF. et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov 2017; 7(12): 1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Razavi P, Li BT, Abida W. et al. Performance of a high-intensity 508-gene circulating-tumor DNA (ctDNA) assay in patients with metastatic breast, lung, and prostate cancer. J Clin Oncol 2017; 35(Suppl 18): LBA11516. [Google Scholar]
- 60. Riva F, Bidard FC, Houy A. et al. Patient-specific circulating tumor DNA detection during neoadjuvant chemotherapy in triple-negative breast cancer. Clin Chem 2017; 63(3): 691–699. [DOI] [PubMed] [Google Scholar]
- 61. Olsson E, Winter C, George A. et al. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol Med 2015; 7(8): 1034–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Siravegna G, Mussolin B, Buscarino M. et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 2015; 21(7): 827. [DOI] [PubMed] [Google Scholar]
- 63. Kytola V, Topaloglu U, Miller LD. et al. Mutational landscapes of smoking-related cancers in caucasians and African Americans: precision oncology perspectives at wake forest baptist comprehensive cancer center. Theranostics 2017; 7: 2914–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jamal-Hanjani M, Wilson GA, Horswell S. et al. Detection of ubiquitous and heterogeneous mutations in cell-free DNA from patients with early-stage non-small-cell lung cancer. Ann Oncol 2016; 27(5): 862–867. [DOI] [PubMed] [Google Scholar]
- 65. Kleppe M, Levine RL.. Tumor heterogeneity confounds and illuminates: assessing the implications. Nat Med 2014; 20(4): 342–344. [DOI] [PubMed] [Google Scholar]
- 66. De Mattos-Arruda L, Weigelt B, Cortes J. et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol 2014; 25(9): 1729–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Murtaza M, Dawson SJ, Pogrebniak K. et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun 2015; 6: 8760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Deryugina EI, Kiosses WB.. Intratumoral cancer cell intravasation can occur independent of invasion into the adjacent stroma. Cell Rep 2017; 19(3): 601–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dawson SJ, Tsui DW, Murtaza M. et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013; 368(13): 1199–1209. [DOI] [PubMed] [Google Scholar]
- 70. Yong E. Cancer biomarkers: written in blood. Nature 2014; 511(7511): 524–526. [DOI] [PubMed] [Google Scholar]
- 71. Bettegowda C, Sausen M, Leary RJ. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014; 6: 224ra224.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Naxerova K, Reiter JG, Brachtel E. et al. Origins of lymphatic and distant metastases in human colorectal cancer. Science 2017; 357(6346): 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tie J, Wang Y, Tomasetti C. et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 2016; 8: 346ra392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kim ST, Banks KC, Lee SH. et al. Prospective feasibility study for using cell-free circulating tumor DNA–guided therapy in refractory metastatic solid cancers: an interim analysis. JCO Precision Oncology 2017; 1–15. [DOI] [PMC free article] [PubMed]
- 75. Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A.. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013; 10(8): 472–484. [DOI] [PubMed] [Google Scholar]
- 76. Sorensen BS, Wu L, Wei W. et al. Monitoring of epidermal growth factor receptor tyrosine kinase inhibitor-sensitizing and resistance mutations in the plasma DNA of patients with advanced non-small cell lung cancer during treatment with erlotinib. Cancer 2014; 120(24): 3896–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lecomte T, Berger A, Zinzindohoue F. et al. Detection of free-circulating tumor-associated DNA in plasma of colorectal cancer patients and its association with prognosis. Int J Cancer 2002; 100(5): 542–548. [DOI] [PubMed] [Google Scholar]
- 78. Gautschi O, Huegli B, Ziegler A. et al. Origin and prognostic value of circulating KRAS mutations in lung cancer patients. Cancer Lett 2007; 254(2): 265–273. [DOI] [PubMed] [Google Scholar]
- 79. Nygaard AD, Garm Spindler KL, Pallisgaard N. et al. The prognostic value of KRAS mutated plasma DNA in advanced non-small cell lung cancer. Lung Cancer 2013; 79(3): 312–317. [DOI] [PubMed] [Google Scholar]
- 80. Sirera R, Bremnes RM, Cabrera A. et al. Circulating DNA is a useful prognostic factor in patients with advanced non-small cell lung cancer. J Thorac Oncol 2011; 6(2): 286–290. [DOI] [PubMed] [Google Scholar]
- 81. Kim ST, Sung JS, Jo UH. et al. Can mutations of EGFR and KRAS in serum be predictive and prognostic markers in patients with advanced non-small cell lung cancer (NSCLC)? Med Oncol 2013; 30(1): 328. [DOI] [PubMed] [Google Scholar]
- 82. Wang S, An T, Wang J. et al. Potential clinical significance of a plasma-based KRAS mutation analysis in patients with advanced non-small cell lung cancer. Clin Cancer Res 2010; 16(4): 1324–1330. [DOI] [PubMed] [Google Scholar]
- 83. Mok T, Wu YL, Lee JS. et al. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin Cancer Res 2015; 21(14): 3196–3203. [DOI] [PubMed] [Google Scholar]
- 84. Douillard JY, Ostoros G, Cobo M. et al. First-line gefitinib in Caucasian EGFR mutation-positive NSCLC patients: a phase-IV, open-label, single-arm study. Br J Cancer 2014; 110(1): 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Oxnard GR, Thress KS, Alden RS. et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol 2016; 34(28): 3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ocana A, Diez-Gonzalez L, Garcia-Olmo DC. et al. Circulating DNA and survival in solid tumors. Cancer Epidemiol Biomarkers Prev 2016; 25(2): 399–406. [DOI] [PubMed] [Google Scholar]
- 87. Reinert T, Scholer LV, Thomsen R. et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016; 65(4): 625–634. [DOI] [PubMed] [Google Scholar]
- 88. Gray ES, Rizos H, Reid AL. et al. Circulating tumor DNA to monitor treatment response and detect acquired resistance in patients with metastatic melanoma. Oncotarget 2015; 6(39): 42008–42018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Douillard JY, Ostoros G, Cobo M. et al. Gefitinib treatment in EGFR mutated caucasian NSCLC circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol 2014; 9(9): 1345–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Goto K, Ichinose Y, Ohe Y. et al. Epidermal growth factor receptor mutation status in circulating free DNA in serum: from IPASS, a phase III study of gefitinib or carboplatin/paclitaxel in non-small cell lung cancer. J Thorac Oncol 2012; 7(1): 115–121. [DOI] [PubMed] [Google Scholar]
- 91. Cabel L, Riva F, Servois V. et al. Circulating tumor DNA changes for early monitoring of anti-PD1 immunotherapy: a proof-of-concept study. Ann Oncol 2017; 28(8): 1996–2001. [DOI] [PubMed] [Google Scholar]
- 92. Lee JH, Long GV, Boyd S. et al. Circulating tumour DNA predicts response to anti-PD1 antibodies in metastatic melanoma. Ann Oncol 2017; 28(5): 1130–1136. [DOI] [PubMed] [Google Scholar]
- 93. Vidal J, Muinelo L, Dalmases A. et al. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann Oncol 2017; 28(6): 1325–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wyatt AW, Azad AA, Volik SV. et al. Genomic alterations in cell-free DNA and enzalutamide resistance in castration-resistant prostate cancer. JAMA Oncol 2016; 2(12): 1598–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Chen KZ, Lou F, Yang F. et al. Circulating tumor DNA detection in early-stage non-small cell lung cancer patients by targeted sequencing. Sci Rep 2016; 6: 31985.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sorber L, Zwaenepoel K, Deschoolmeester V. et al. Circulating cell-free nucleic acids and platelets as a liquid biopsy in the provision of personalized therapy for lung cancer patients. Lung Cancer 2017; 107: 100–107. [DOI] [PubMed] [Google Scholar]
- 97. Schwaederle M, Husain H, Fanta PT. et al. Detection rate of actionable mutations in diverse cancers using a biopsy-free (blood) circulating tumor cell DNA assay. Oncotarget 2016; 7(9): 9707–9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Misale S, Yaeger R, Hobor S. et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012; 486(7404): 532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Garcia-Murillas I, Schiavon G, Weigelt B. et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 2015; 7(302): 302ra133. [DOI] [PubMed] [Google Scholar]
- 100. Harmer M, Denoix P, Hamperl H.. TNM-system for classification of tumors. Klin Wochenschr 1968; 46(22): 1181. [Google Scholar]
- 101. Gospodarowicz MK, Miller D, Groome PA. et al. The process for continuous improvement of the TNM classification. Cancer 2004; 100(1): 1–5. [DOI] [PubMed] [Google Scholar]
- 102. Schwaederle M, Chattopadhyay R, Kato S. et al. Genomic alterations in circulating tumor DNA from diverse cancer patients identified by next-generation sequencing. Cancer Res 2017; 77(19): 5419–5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Sobin LH, Gospodarowicz MK, Wittekind C (eds). International Union Against Cancer (UICC). TNM Classification of Malignant Tumours, 7th edition. USA: Wiley-Blackwell 2009. [Google Scholar]
- 104. Sozzi G, Musso K, Ratcliffe C. et al. Detection of microsatellite alterations in plasma DNA of non-small cell lung cancer patients: a prospect for early diagnosis. Clin Cancer Res 1999; 5(10): 2689–2692. [PubMed] [Google Scholar]
- 105. Kimura H, Suminoe M, Kasahara K. et al. Evaluation of epidermal growth factor receptor mutation status in serum DNA as a predictor of response to gefitinib (IRESSA). Br J Cancer 2007; 97(6): 778–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Yung TK, Chan KC, Mok TS. et al. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res 2009; 15(6): 2076–2084. [DOI] [PubMed] [Google Scholar]
- 107. Bai H, Mao L, Wang HS. et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J Clin Oncol 2009; 27: 2653–2659. [DOI] [PubMed] [Google Scholar]
- 108. He C, Liu M, Zhou C. et al. Detection of epidermal growth factor receptor mutations in plasma by mutant-enriched PCR assay for prediction of the response to gefitinib in patients with non-small-cell lung cancer. Int J Cancer 2009; 125(10): 2393–2399. [DOI] [PubMed] [Google Scholar]
- 109. Sriram KB, Tan ME, Savarimuthu SM. et al. Screening for activating EGFR mutations in surgically resected nonsmall cell lung cancer. Eur Respir J 2011; 38(4): 903–910. [DOI] [PubMed] [Google Scholar]
- 110. Jiang B, Liu F, Yang L. et al. Serum detection of epidermal growth factor receptor gene mutations using mutant-enriched sequencing in Chinese patients with advanced non-small cell lung cancer. J Int Med Res 2011; 39(4): 1392–1401. [DOI] [PubMed] [Google Scholar]
- 111. Xu F, Wu JX, Xue C. et al. Comparison of different methods for detecting epidermal growth factor receptor mutations in peripheral blood and tumor tissue of non-small cell lung cancer as a predictor of response to gefitinib. Onco Targets Ther 2012; 5: 439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Hu C, Liu X, Chen Y. et al. Direct serum and tissue assay for EGFR mutation in non-small cell lung cancer by high-resolution melting analysis. Oncol Rep 2012; 28(5): 1815–1821. [DOI] [PubMed] [Google Scholar]
- 113. Zhao X, Han RB, Zhao J. et al. Comparison of epidermal growth factor receptor mutation statuses in tissue and plasma in stage I-IV non-small cell lung cancer patients. Respiration 2013; 85(2): 119–125. [DOI] [PubMed] [Google Scholar]
- 114. Liu X, Lu Y, Zhu G. et al. The diagnostic accuracy of pleural effusion and plasma samples versus tumour tissue for detection of EGFR mutation in patients with advanced non-small cell lung cancer: comparison of methodologies. J Clin Pathol 2013; 66(12): 1065–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Zhang H, Liu DR, Li SQ. et al. Comparison of EGFR signaling pathway somatic DNA mutations derived from peripheral blood and corresponding tumor tissue of patients with advanced non-small-cell lung cancer using liquidchip technology. J Mol Diagn 2013; 15(6): 819–826. [DOI] [PubMed] [Google Scholar]
- 116. Kim HR, Lee SY, Hyun DS. et al. Detection of EGFR mutations in circulating free DNA by PNA-mediated PCR clamping. J Exp Clin Cancer Res 2013; 32(1): 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Oxnard GR, Paweletz CP, Kuang Y. et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res.2014; 20(6): 1698–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Jing CW, Wang Z, Cao HX. et al. High resolution melting analysis for epidermal growth factor receptor mutations in formalin-fixed paraffin-embedded tissue and plasma free DNA from non-small cell lung cancer patients. Asian Pac J Cancer Prev 2013; 14(11): 6619–6623. [DOI] [PubMed] [Google Scholar]
- 119. Wang S, Han X, Hu X. et al. Clinical significance of pretreatment plasma biomarkers in advanced non-small cell lung cancer patients. Clin Chim Acta 2014; 430: 63–70. [DOI] [PubMed] [Google Scholar]
- 120. Li X, Ren R, Ren S. et al. Peripheral blood for epidermal growth factor receptor mutation detection in non-small cell lung cancer patients. Transl Oncol 2014; 7(3): 341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Weber B, Meldgaard P, Hager H. et al. Detection of EGFR mutations in plasma and biopsies from non-small cell lung cancer patients by allele-specific PCR assays. BMC Cancer 2014; 14: 294.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Karachaliou N, Mayo-de las Casas C, Queralt C. et al. Association of EGFR L858R mutation in circulating free DNA with survival in the EURTAC trial. JAMA Oncol 2015; 1(2): 149–157. [DOI] [PubMed] [Google Scholar]
- 123. Thress KS, Brant R, Carr TH. et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: a cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015; 90(3): 509–515. [DOI] [PubMed] [Google Scholar]
- 124. Marchetti A, Palma JF, Felicioni L. et al. Early prediction of response to tyrosine kinase inhibitors by quantification of EGFR mutations in plasma of NSCLC patients. J Thorac Oncol 2015; 10(10): 1437–1443. [DOI] [PubMed] [Google Scholar]
- 125. Uchida J, Kato K, Kukita Y. et al. Diagnostic accuracy of noninvasive genotyping of EGFR in lung cancer patients by deep sequencing of plasma cell-free DNA. Clin Chem 2015; 61(9): 1191–1196. [DOI] [PubMed] [Google Scholar]
- 126. Lam DC, Tam TC, Lau KM. et al. Plasma EGFR mutation detection associated with survival outcomes in advanced-stage lung cancer. Clin Lung Cancer 2015; 16(6): 507–513. [DOI] [PubMed] [Google Scholar]
- 127. Reckamp KL, Melnikova VO, Karlovich C. et al. A highly sensitive and quantitative test platform for detection of NSCLC EGFR mutations in urine and plasma. J Thorac Oncol 2016; 11(10): 1690–1700. [DOI] [PubMed] [Google Scholar]
- 128. Sacher AG, Paweletz C, Dahlberg SE. et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol 2016; 2(8): 1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Newman AM, Lovejoy AF, Klass DM. et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016; 34(5): 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Gale D, Plagnol V, Lawson A. et al. Analytical performance and validation of an enhanced TAm-Seq circulating tumor DNA sequencing assay. Cancer Res 2016; 76(Suppl 14): 3639. [Google Scholar]
- 131. Reck M, Hagiwara K, Han B. et al. ctDNA determination of EGFR mutation status in European and Japanese patients with advanced NSCLC: the ASSESS study. J Thorac Oncol 2016; 11(10): 1682–1689. [DOI] [PubMed] [Google Scholar]
- 132. Sherwood JL, Corcoran C, Brown H. et al. Optimised pre-analytical methods improve KRAS mutation detection in circulating tumour DNA (ctDNA) from patients with non-small cell lung cancer (NSCLC). PLoS One 2016; 11(2): e0150197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Remon J, Caramella C, Jovelet C. et al. Osimertinib benefit in EGFR-mutant NSCLC patients with T790M-mutation detected by circulating tumour DNA. Ann Oncol 2017; 28(4): 784–790. [DOI] [PubMed] [Google Scholar]
- 134. Fernandez-Cuesta L, Perdomo S, Avogbe PH. et al. Identification of circulating tumor DNA for the early detection of small-cell lung cancer. EBioMedicine 2016; 10: 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Underhill HR, Kitzman JO, Hellwig S. et al. Fragment length of circulating tumor DNA. PLoS Genet 2016; 12(7): e1006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Diehl F, Li M, Dressman D. et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci USA 2005; 102(45): 16368–16373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Thierry AR, Mouliere F, Gongora C. et al. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res 2010; 38(18): 6159–6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Garcia-Olmo DC, Dominguez C, Garcia-Arranz M. et al. Cell-free nucleic acids circulating in the plasma of colorectal cancer patients induce the oncogenic transformation of susceptible cultured cells. Cancer Res 2010; 70(2): 560–567. [DOI] [PubMed] [Google Scholar]
- 139. Mouliere F, Robert B, Arnau Peyrotte E. et al. High fragmentation characterizes tumour-derived circulating DNA. PLoS One 2011; 6(9): e23418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Mouliere F, El Messaoudi S, Gongora C. et al. Circulating cell-free DNA from colorectal cancer patients may reveal high KRAS or BRAF mutation load. Transl Oncol 2013; 6(3): 319–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Heitzer E, Auer M, Hoffmann EM. et al. Establishment of tumor-specific copy number alterations from plasma DNA of patients with cancer. Int J Cancer 2013; 133(2): 346–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Mouliere F, El Messaoudi S, Pang D. et al. Multi-marker analysis of circulating cell-free DNA toward personalized medicine for colorectal cancer. Mol Oncol 2014; 8(5): 927–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Thierry AR, Mouliere F, El Messaoudi S. et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med 2014; 20(4): 430–435. [DOI] [PubMed] [Google Scholar]
- 144. Kuo YB, Chen JS, Fan CW. et al. Comparison of KRAS mutation analysis of primary tumors and matched circulating cell-free DNA in plasmas of patients with colorectal cancer. Clin Chim Acta 2014; 433: 284–289. [DOI] [PubMed] [Google Scholar]
- 145. Mohan S, Heitzer E, Ulz P. et al. Changes in colorectal carcinoma genomes under anti-EGFR therapy identified by whole-genome plasma DNA sequencing. PLoS Genet 2014; 10: e1004271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Spindler KL, Pallisgaard N, Andersen RF, Jakobsen A.. Changes in mutational status during third-line treatment for metastatic colorectal cancer—results of consecutive measurement of cell free DNA, KRAS and BRAF in the plasma. Int J Cancer 2014; 135(9): 2215–2222. [DOI] [PubMed] [Google Scholar]
- 147. Tie J, Kinde I, Wang Y. et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol 2015; 26(8): 1715–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Russo M, Siravegna G, Blaszkowsky LS. et al. Tumor heterogeneity and lesion-specific response to targeted therapy in colorectal cancer. Cancer Discov 2016; 6(2): 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Morelli MP, Overman MJ, Dasari A. et al. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol 2015; 26(4): 731–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Kidess E, Heirich K, Wiggin M. et al. Mutation profiling of tumor DNA from plasma and tumor tissue of colorectal cancer patients with a novel, high-sensitivity multiplexed mutation detection platform. Oncotarget 2015; 6(4): 2549–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Russo M, Misale S, Wei G. et al. Acquired resistance to the TRK inhibitor entrectinib in colorectal cancer. Cancer Discov 2016; 6(1): 36–44. [DOI] [PubMed] [Google Scholar]
- 152. El Messaoudi S, Mouliere F, Du Manoir S. et al. Circulating DNA as a strong multimarker prognostic tool for metastatic colorectal cancer patient management care. Clin Cancer Res 2016; 22(12): 3067–3077. [DOI] [PubMed] [Google Scholar]
- 153. McBride DJ, Orpana AK, Sotiriou C. et al. Use of cancer-specific genomic rearrangements to quantify disease burden in plasma from patients with solid tumors. Genes Chromosom Cancer 2010; 49: 1062–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Beaver JA, Jelovac D, Balukrishna S. et al. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin Cancer Res 2014; 20(10): 2643–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Schiavon G, Hrebien S, Garcia-Murillas I. et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med 2015; 7(313): 313ra182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Kirkizlar E, Zimmermann B, Constantin T. et al. Detection of clonal and subclonal copy-number variants in cell-free DNA from patients with breast cancer using a massively multiplexed PCR methodology. Transl Oncol 2015; 8(5): 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Oshiro C, Kagara N, Naoi Y. et al. PIK3CA mutations in serum DNA are predictive of recurrence in primary breast cancer patients. Breast Cancer Res Treat 2015; 150(2): 299–307. [DOI] [PubMed] [Google Scholar]
- 158. Swisher EM, Wollan M, Mahtani SM. et al. Tumor-specific p53 sequences in blood and peritoneal fluid of women with epithelial ovarian cancer. Am J Obstet Gynecol 2005; 193(3): 662–667. [DOI] [PubMed] [Google Scholar]
- 159. Forshew T, Murtaza M, Parkinson C. et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 2012; 4(136): 136ra168. [DOI] [PubMed] [Google Scholar]
- 160. Parkinson CA, Gale D, Piskorz AM. et al. Exploratory analysis of TP53 mutations in circulating tumour DNA as biomarkers of treatment response for patients with relapsed high-grade serous ovarian carcinoma: a retrospective study. PLoS Med 2016; 13(12): e1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Cohen PA, Flowers N, Tong S. et al. Abnormal plasma DNA profiles in early ovarian cancer using a non-invasive prenatal testing platform: implications for cancer screening. BMC Med 2016; 14(1): 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Chan KC, Jiang P, Chan CW. et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci USA 2013; 110(47): 18761–18768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Sun K, Jiang P, Chan KC. et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci USA 2015; 112(40): E5503–E5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Jiang P, Chan CW, Chan KC. et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci USA 2015; 112(11): E1317–E1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. De Mattos-Arruda L, Mayor R, Ng CKY. et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun 2015; 6: 8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Giacona MB, Ruben GC, Iczkowski KA. et al. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas 1998; 17(1): 89–97. [DOI] [PubMed] [Google Scholar]
- 167. Birkenkamp-Demtroder K, Nordentoft I, Christensen E. et al. Genomic alterations in liquid biopsies from patients with bladder cancer. Eur Urol 2016; 70(1): 75–82. [DOI] [PubMed] [Google Scholar]
- 168. Romanel A, Tandefelt DG, Conteduca V. et al. Plasma AR and abiraterone-resistant prostate cancer. Sci Transl Med 2015; 7(312): 312re310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Lipson EJ, Velculescu VE, Pritchard TS. et al. Circulating tumor DNA analysis as a real-time method for monitoring tumor burden in melanoma patients undergoing treatment with immune checkpoint blockade. J Immunother Cancer 2014; 2(1): 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Sullivan RJ, O'Neill VJ, Brinkmann K. et al. Plasma-based monitoring of BRAF mutations during therapy for malignant melanoma (MM) using combined exosomal RNA and cell-free DNA analysis. J Clin Oncol 2015; 33(Suppl 15): 9017–9017. [Google Scholar]
- 171. Santiago-Walker A, Gagnon R, Mazumdar J. et al. Correlation of BRAF mutation status in circulating-free DNA and tumor and association with clinical outcome across four BRAFi and MEKi clinical trials. Clin Cancer Res 2016; 22(3): 567–574. [DOI] [PubMed] [Google Scholar]
- 172. Schreuer M, Meersseman G, Van Den Herrewegen S. et al. Quantitative assessment of BRAF V600 mutant circulating cell-free tumor DNA as a tool for therapeutic monitoring in metastatic melanoma patients treated with BRAF/MEK inhibitors. J Transl Med 2016; 14: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Xi LQ, Pham THT, Payabyab EC. et al. Circulating tumor DNA as an early indicator of response to T-cell transfer immunotherapy in metastatic melanoma. Clin Cancer Res 2016; 22(22): 5480–5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Scherer F, Kurtz DM, Newman AM. et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci Transl Med 2016; 8(364): 364ra155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. To EW, Chan KC, Leung SF. et al. Rapid clearance of plasma Epstein-Barr virus DNA after surgical treatment of nasopharyngeal carcinoma. Clin Cancer Res 2003; 9: 3254–3259. [PubMed] [Google Scholar]
- 176. Chan KC, Hung EC, Woo JK. et al. Early detection of nasopharyngeal carcinoma by plasma Epstein-Barr virus DNA analysis in a surveillance program. Cancer 2013; 119(10): 1838–1844. [DOI] [PubMed] [Google Scholar]
- 177. Gormally E, Vineis P, Matullo G. et al. TP53 and KRAS2 mutations in plasma DNA of healthy subjects and subsequent cancer occurrence: a prospective study. Cancer Res 2006; 66(13): 6871–6876. [DOI] [PubMed] [Google Scholar]
- 178. Umetani N, Hiramatsu S, Hoon DS.. Higher amount of free circulating DNA in serum than in plasma is not mainly caused by contaminated extraneous DNA during separation. Ann N Y Acad Sci 2006; 1075: 299–307. [DOI] [PubMed] [Google Scholar]
- 179. Leary RJ, Kinde I, Diehl F. et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med 2010; 2(20): 20ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Lanman RB, Mortimer SA, Zill OA. et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS One 2015; 10(10): e0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Lebofsky R, Decraene C, Bernard V. et al. Circulating tumor DNA as a non-invasive substitute to metastasis biopsy for tumor genotyping and personalized medicine in a prospective trial across all tumor types. Mol Oncol 2015; 9(4): 783–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Belic J, Koch M, Ulz P. et al. Rapid identification of plasma DNA samples with increased ctDNA levels by a modified FAST-SeqS approach. Clin Chem 2015; 61(6): 838–849. [DOI] [PubMed] [Google Scholar]
- 183. Schwaederle M, Husain H, Fanta PT. et al. Use of liquid biopsies in clinical oncology: pilot experience in 168 patients. Clin Cancer Res 2016; 22(22): 5497–5505. [DOI] [PubMed] [Google Scholar]
- 184. Ulz P, Thallinger GG, Auer M. et al. Inferring expressed genes by whole-genome sequencing of plasma DNA. Nat Genet 2016; 48(10): 1273–1278. [DOI] [PubMed] [Google Scholar]
- 185. Janku F, Huang HJ, Claes B. et al. BRAF mutation testing in cell-free DNA from the plasma of patients with advanced cancers using a rapid, automated molecular diagnostics system. Mol Cancer Ther 2016; 15(6): 1397–1404. [DOI] [PubMed] [Google Scholar]