Abstract
Background
Genomic changes that occur in breast cancer during the course of disease have been informed by sequencing of primary and metastatic tumor tissue. For patients with relapsed and metastatic disease, evolution of the breast cancer genome highlights the importance of using a recent sample for genomic profiling to guide clinical decision-making. Obtaining a metastatic tissue biopsy can be challenging, and analysis of circulating tumor DNA (ctDNA) from blood may provide a minimally invasive alternative.
Patients and methods
Hybrid capture-based genomic profiling was carried out on ctDNA from 254 female patients with estrogen receptor-positive breast cancer. Peripheral blood samples were submitted by clinicians in the course of routine clinical care between May 2016 and March 2017. Sequencing of 62 genes was carried out to a median unique coverage depth of 7503×. Genomic alterations (GAs) in ctDNA were evaluated and compared with matched tissue samples and genomic datasets of tissue from breast cancer.
Results
At least 1 GA was reported in 78% of samples. Frequently altered genes were TP53 (38%), ESR1 (31%) and PIK3CA (31%). Temporally matched ctDNA and tissue samples were available for 14 patients; 89% of mutations detected in tissue were also detected in ctDNA. Diverse ESR1 GAs including mutation, rearrangement and amplification, were observed. Multiple concurrent ESR1 GAs were observed in 40% of ESR1-altered cases, suggesting polyclonal origin; ESR1 compound mutations were also observed in two cases. ESR1-altered cases harbored co-occurring GAs in PIK3CA (35%), FGFR1 (16%), ERBB2 (8%), BRCA1/2 (5%), and AKT1 (4%).
Conclusions
GAs relevant to relapsed/metastatic breast cancer management were identified, including diverse ESR1 GAs. Genomic profiling of ctDNA demonstrated sensitive detection of mutations found in tissue. Detection of amplifications was associated with ctDNA fraction. Genomic profiling of ctDNA may provide a complementary and possibly alternative approach to tissue-based genomic testing for patients with estrogen receptor-positive metastatic breast cancer.
Keywords: ctDNA, liquid biopsy, genomic profiling, ER, metastatic breast cancer, ESR1
Introduction
Genomic changes that characterize primary breast cancer (BC) have been elucidated by extensive genomic profiling studies [1]. Comparative analyses of estrogen receptor-positive (ER+) metastatic BC (mBC) have demonstrated genomic evolution during metastatic progression, and following treatment, such as the enrichment of HER2 and ESR1 genomic alterations (GAs) [2, 3]. Clonal evolution can arise due to independent primary lesions, expansion of subclonal populations, or acquisition of novel GAs. Genomic changes following therapy are exemplified by acquired activating ESR1 GAs that mediate aromatase inhibitor (AI) resistance [2–4].
Clonal evolution processes highlight the importance of profiling a contemporaneous sample that is representative of disease progression to guide further management. However, limitations in performing repeated prospective biopsies of metastatic lesions over the disease course for a patient can present challenges for clinical genomic analysis [5]. Liquid biopsy and sequencing of circulating tumor DNA (ctDNA) from blood could provide a complementary approach to tissue-based genomic testing for mBC.
Research studies of BC identified genomic changes in ctDNA following therapy, however, limited numbers of ER+ BC have been profiled [6–8]. In larger studies of ctDNA from ER+ mBC, droplet digital PCR (ddPCR) identified select mutations in ESR1 or PIK3CA [9–11]. In phase 3 trials for ER+/HER2-negative (HER2−) BC, prospective ctDNA assessment identified patients with PIK3CA mutation who derived survival benefit from buparlisib [4]. Retrospective analyses of ctDNA in phase 3 trials suggest that ESR1 mutations are associated with resistance to AI but not selective ER down-regulators (SERDs), and can guide therapy selection [11].
In this study, we carried out hybrid capture-based genomic profiling to characterize GAs in ctDNA from 254 patients with ER+ BC during the course of their clinical care.
Methods
Detailed descriptions of patient samples/methods are presented in supplementary methods, available at Annals of Oncology online. Briefly, peripheral blood samples were collected from 254 patients with ER+ BC, plasma was isolated from 20 ml whole blood, ≥20 ng DNA was extracted, and hybrid capture-based genomic profiling of ctDNA was carried out in a CLIA-certified/CAP-accredited laboratory [Foundation Medicine (FM)] to identify substitutions, short insertions/deletions, rearrangements/fusions, and amplifications [12]. Sixty-two genes (supplementary Table S1, available at Annals of Oncology online) were sequenced (Illumina HiSeq 2500 or 4000) to a median unique coverage depth of 7503×. Maximum somatic allele frequency (MSAF) was used to estimate the ctDNA fraction in plasma.
Results
Patient characteristics
This study of hybrid capture-based sequencing of ctDNA in blood included consecutive genomic profiling results from 254 female patients with an initial diagnosis of ER+ BC, determined by routine IHC. Patient characteristics are described in Table 1 and supplementary Table S2, available at Annals of Oncology online. For patients with available clinical information, 94% were stage IV, 88% had received prior chemotherapy, and 88% had received prior AI in the adjuvant and/or metastatic setting.
Table 1.
Patient characteristics
| All ER + | ER+/HER2− | ER+/HER2+ | ER+/HER2 unk | ||
|---|---|---|---|---|---|
| N | 254 | 197 | 28 | 29 | |
| Median age, years (range) | 58 (32–85) | 58 (33–85) | 57 (33–79) | 62 (32–78) | |
| Stage, N (%) | I | 2 (0.8%) | 2 (1.1%) | 0 (0%) | 0 (0%) |
| II | 5 (2.1%) | 3 (1.6%) | 1 (3.7%) | 1 (3.8%) | |
| III | 7 (2.9%) | 7 (3.7%) | 0 (0%) | 0 (0%) | |
| IV | 226 (94.2%) | 175 (93.6%) | 26 (96.3%) | 25 (96.2%) | |
| Unknown | 14 | 10 | 1 | 3 | |
| Previous chemotherapya | Yes | 120 (88.2%) | 94 (87.0%) | 18 (90.0%) | 8 (100%) |
| [adj/met/unk], N | [24/91/5] | [17/72/5] | [4/14/0] | [3/5/0] | |
| No | 16 (11.8%) | 14 (13.0%) | 2 (10.0%) | 0 (0%) | |
| Unknown | 118 | 89 | 8 | 21 | |
| Previous aromatase inhibitora | Yes | 115 (88.5%) | 95 (92.2%) | 15 (75.0%) | 5 (71.4%) |
| [adj/met/unk], N | [19/91/5] | [15/75/5] | [2/13/0] | [2/3/0] | |
| No | 15 (11.5%) | 8 (7.8%) | 5 (25.0%) | 2 (28.6%) | |
| Unknown | 124 | 94 | 8 | 22 | |
| Previous tamoxifena | Yes | 56 (43.8%) | 41 (40.6%) | 8 (40.0%) | 7 (100%) |
| [adj/met/unk], N | [41/13/2] | [28/11/2] | [7/1/0] | [6/1/0] | |
| No | 72 (56.2%) | 60 (59.4%) | 12 (60.0%) | 0 (0%) | |
| Unknown | 126 | 96 | 8 | 22 | |
| Previous fulvestranta | Yes | 69 (54.3%) | 57 (56.4%) | 7 (36.8%) | 5 (71.4%) |
| No | 58 (45.7%) | 44 (43.6%) | 12 (63.2%) | 2 (28.6%) | |
| Unknown | 127 | 96 | 9 | 22 | |
See supplementary Table S2, available at Annals of Oncology online, for detailed descriptions of treatments in adjuvant/metastatic settings, and for treatment/response status at the time of sample collection.
adj, adjuvant only; met, metastatic or metastatic and adjuvant; unk, unknown.
GAs identified in ctDNA from ER+ BC
At least 1 GA was detected in 78% of cases with an average of 2.5 GA/sample (range 0–27). MSAF was calculated for each case and provided a median estimated ctDNA fraction of 1.7% (interquartile range 0.3%–9.2%). Eighty-four percent of cases have evidence of ctDNA in the blood (MSAF > 0). There was no evidence of ctDNA in the blood (MSAF = 0) in 15% (33/226) of stage IV cases and 43% (6/14) of stage I–III cases. The most frequently altered genes in ER+ BC were TP53 (38%), ESR1 (31%), PIK3CA (31%), CDH1 (10%), and ERBB2 (8%) (supplementary Figure S1, available at Annals of Oncology online).
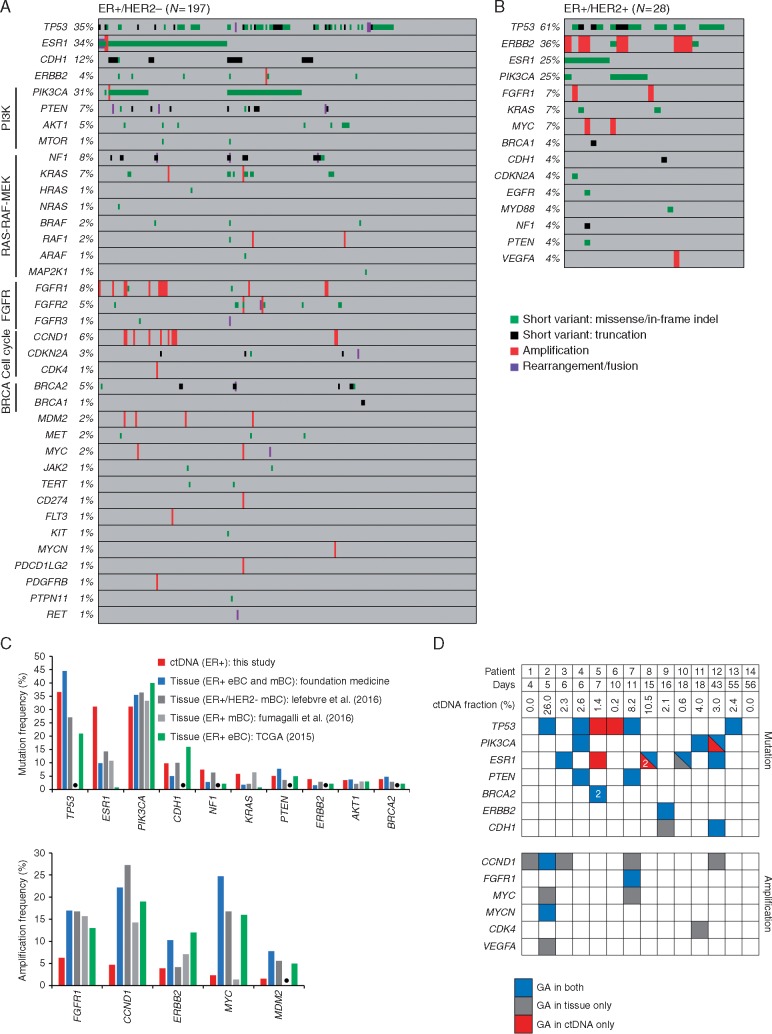
For ER+/HER2− BC, the most frequently altered genes were TP53 (35%), ESR1 (34%), PIK3CA (31%), and CDH1 (12%) (Figure 1A). ERBB2 activating mutations were identified in 3% of cases. ERBB2 amplification was identified in one patient initially diagnosed with ER+/HER2− BC (IHC on a breast biopsy); gain of HER2 was subsequently confirmed by IHC (3+) on a metastatic biopsy. Activating kinase fusions (FGFR2-INA, FGFR3-TACC3, NCOA4-RET) were observed in three cases (2%). Frequently altered pathways included PI3K-AKT-mTOR (38%), RAS-RAF-MEK (15%), FGFR (14%), cell cycle (8%), and BRCA (6%).
Figure 1.
Genomic alterations in ctDNA from patients with ER+ breast cancer and comparisons with tissue. (A) GAs identified in 197 cases of ER+/HER2− BC. Percent of cases altered is indicated. (B) GAs identified in 28 cases of ER+/HER2+ BC. (C) Comparison of the most frequently mutated (top panel) or amplified (bottom panel) genes observed in ctDNA in this study (N = 254) with tissue-based genomic profiling of ER+ BC. Datasets from ER+ BC tissue used for comparison were from the FM database (N = 851) and published studies of tissue from early BC (eBC, TCGA [1]: N = 594) and mBC (Lefebvre et al. [3]: N = 143; Fumagalli et al. [2]: N = 182). Data from [1, 3] were extracted from cBioPortal. Black dots represent genes that were not assessed in [2]. (D) Concordance between GAs found in ctDNA and matched tumor tissue from 14 patients. Days between ctDNA and tissue collection are shown. The ctDNA fraction was estimated using MSAF. Concordant/shared GAs are in blue, GAs found in tissue only are in grey, and GAs found in ctDNA only are in red. For samples with multiple unique mutations in a gene (patient-5 and patient-8), the number of mutations is shown.
For ER+/HER2+ BC, the most frequently altered genes were TP53 (61%), ERBB2 (36%), PIK3CA (25%), and ESR1 (25%) (Figure 1B). ERBB2 amplification was observed in 29% (8/28) of HER2+ cases, consistent with next-generation sequencing (NGS) studies of ctDNA in HER2+ BC [7, 13]. The estimated ctDNA fraction was significantly higher for HER2+ cases with ERBB2 amplification compared with cases without ERBB2 amplification detected (supplementary Figure S2A, available at Annals of Oncology online), suggesting that the ability to detect ERBB2 amplification was associated with the quantity of ctDNA in the blood.
Comparison of GAs in ctDNA and tissue
We compared frequently altered genes in ctDNA with ER+ BC tissue samples from the FM database and published studies [1–3] (Figure 1C). For the majority of genes, mutation frequencies in ctDNA were similar to the range observed in tissue; ESR1 was mutated at a higher frequency (greater than twofold) compared with tissue, as expected from our study population comprising mostly patients who had received or were receiving AI treatment (supplementary Table S2, available at Annals of Oncology online) [9]. Amplifications were observed at lower frequencies in ctDNA compared with tissue, consistent with other studies of amplification detection in ctDNA from BC [7, 13]. The estimated ctDNA fraction was significantly higher for cases with an amplification detected compared with cases without (supplementary Figure S2B, available at Annals of Oncology online).
Genomic profiles of matched blood and tissue samples collected within 60 days of each other were available for 14 cases. We compared GAs assessed in both ctDNA and tissue (Figure 1D; supplementary Table S3, available at Annals of Oncology online). For short variant mutations, 89% (17/19) that were detected in tissue were also detected in ctDNA. Six mutations were detected in ctDNA only and two mutations were in tissue only. One ctDNA only ESR1 mutation (patient-5) was found in a second tissue sample from a distinct metastatic site, collected 408 days before blood sampling. One case (patient-10) harbored one shared and one tissue only ESR1 mutation; the allele frequency (AF) for the shared mutation (AF = 34%) was 10-fold higher compared with the tissue only mutation (AF = 3%). Two cases harbored both shared and ctDNA only mutations for the same gene (patient-8: ESR1; patient-12: PIK3CA): the shared mutation had a higher AF than the ctDNA only mutations in both cases (ESR1: twofold; PIK3CA: threefold), suggesting that ctDNA only mutations occur in less represented clones that may not be detected in a single tumor biopsy, consistent with clonal heterogeneity. For amplifications, 27% (3/11) that were detected in tissue were also detected in ctDNA; no amplifications were detected in ctDNA only. The estimated ctDNA fraction was higher for two cases where at least one amplification was detected in both tissue and ctDNA than for cases where amplification was detected in tissue only.
Landscape of ESR1 alterations in ctDNA
A total of 131 ESR1 GA were observed in 80 ER+ cases (supplementary Figure S3A, available at Annals of Oncology online), including both ER+/HER2− and ER+/HER2+ cases (Figure 1A and B); whereas, only 1 ESR1 GA (amplification) was observed in the ctDNA of 74 ER- cases (P = 0.0001, Fisher’s exact test, two-tailed, supplementary Figure S3B, available at Annals of Oncology online). For the 130 ER+ patients with available clinical information regarding AI treatment, 35% (40/115) of all AI-treated patients had an ESR1 GA, and consistent with previous studies [9], ESR1 GAs were more frequent in patients treated with AI in the metastatic setting (40%, 36/91) versus patients treated with adjuvant AI only (11%, 2/19); all patients (40/40) with ESR1 GA had received prior AI (supplementary Table S2, available at Annals of Oncology online).
The most frequent ESR1 GAs were D538G, Y537S, Y537N, and E380Q. All observed ESR1 mutations are activating or occur at the L536 position where multiple activating mutations have been characterized (Figure 2A). Of the 80 ESR1-altered cases, 40% had >1 ESR1 GA (range 2–4) (Figure 2B). In comparison, 24% (19/79) of PIK3CA-altered cases harbored >1 PIK3CA GA (range 2–8) and 23% (22/96) of TP53-altered cases harbored >1 TP53 GA (range 2–11). In cases with >1 ESR1 mutation, no one ESR1 mutation had a consistently greater AF than co-occurring ESR1 mutations, suggesting that diverse ESR1 mutations could contribute to AI resistance (supplementary Figure S4, available at Annals of Oncology online).
Figure 2.
Landscape of ESR1 alterations in ctDNA. (A) Graph represents the number of cases with each ESR1 GA. The percent indicate the frequency of each ESR1 GA relative to all 131 ESR1 GA identified. AMP, amplification; RE, rearrangement. (B) Percent of cases with 1, 2, 3, or 4 ESR1 GAs. (C) Compound mutations in ESR1: in case-1 all instances of L536F and D538G were observed on the same read in cis. In case-2 ESR1 c.1609T>A (Y537N) was observed alone in the majority of reads, and ESR1 c.1609T>A and c.1610A>G were seen together as compound mutations in a portion of reads to generate ESR1 Y537S. (D) ESR1 variants of unknown significance identified in this study. (E) ESR1 rearrangements identified in this study. Numbered boxes represent exons and numbers above indicate amino acid position. Rearrangement break points in ESR1 were in exon 4 or exon 5. (F) Assessment of GAs that co-occur with ESR1 GA: frequency of ESR1-altered cases (N = 80) with alterations in the genes indicated. ‘Multiple’ represents cases harboring multiple classes of genomic alteration.
Multiple ESR1 GAs in the same sample are thought to be polyclonal in origin [9, 10]. We carried out a pairwise assessment of all co-occurring ESR1 mutations to determine whether any mutation pairs existed as compound mutations on the same allele (supplementary Table S4, available at Annals of Oncology online). Out of 67 mutation pairs, 49 were close enough to be evaluated on the same sequencing read; compound mutations were observed in 2/49 mutation pairs (Figure 2C). In case-1, ESR1 L536F/D538G were observed as compound mutations on all reads. In case-2, ESR1 Y537N occurred as a single mutation in most reads, but a subset of reads harbored a compound mutation at the Y537 codon that resulted in conversion of Y537N to Y537S; the existence of two subsets of reads suggests sequential mutational events.
In addition to the GAs described above, 21 ESR1 variant of unknown significance (VUS) mutations were identified (Figure 2D) in 15 ER+ cases including 5 cases with no ESR1 GAs (supplementary Figure S3A, available at Annals of Oncology online); no ESR1 VUS was observed in 74 ER- cases. To evaluate compound mutations, we analyzed 51 co-occurring ESR1 mutation pairs that involved an ESR1 VUS, and 20 could be evaluated on the same sequencing read. Compound mutations were observed in 4/20 mutation pairs and 2 mutation pairs existed as compound mutations in only a subset of sequencing reads (supplementary Figure S5, available at Annals of Oncology online).
ESR1 rearrangements were observed in three cases: two ESR1 rearrangements had potential 3’ fusion partners (AKAP12, NKAIN2) and one ESR1 rearrangement was fused to an intergenic region (Figure 2E). ESR1-AKAP12 is recurrent in BC and all three ESR1 rearrangements resulted in loss of the ligand-binding domain (LBD), which likely results in constitutive ER activation [14, 15]. Each ESR1-rearranged case harbored concurrent ESR1 mutation, suggesting prior AI exposure: we confirmed prior adjuvant AI and fulvestrant treatment of the patient with ESR1 fused to intergenic space.
Co-occurring GAs with ESR1
To inform therapeutic strategies for AI refractory patients, we evaluated co-occurring alterations with ESR1 GAs and identified concurrent GAs that have been associated with responses to targeted therapy in BC [4] including PIK3CA (35%), FGFR1 (16%), ERBB2 (8%), BRCA1/2 (5%), and AKT1 (4%) (Figure 2F).
For cases with concurrent PIK3CA/ESR1 mutation, the PIK3CA:ESR1 AF ratio was ≥1 for 75% (21/28) of cases, consistent with PIK3CA being a truncal driver and ESR1 arising following AI (supplementary Figure S6, available at Annals of Oncology online). Concurrent ESR1/ERBB2 mutation was more frequent in ctDNA than tissue: in ctDNA, 4% (3/79) of ESR1-mutated cases had concurrent ERBB2 mutation; whereas, in the FM database, 0.6% (6/969) of ESR1-mutated BC tissue samples had concurrent ERBB2 mutation.
Discussion
Genomic profiling of ctDNA has the potential to capture GAs that drive recurrent disease or therapeutic resistance and may provide an alternative when tissue biopsy is challenging. However, genomic profiles of ctDNA from ER+ mBC have not been extensively studied. We describe GAs identified in ctDNA from the blood of 254 patients with ER+ mBC.
Eighty-four percent of samples had evidence of ctDNA in the blood, consistent with a study of ctDNA release in mBC [8]. For cases with no evidence of ctDNA in the blood, lack of detectable somatic alterations is, in part, likely associated with insufficient ctDNA release into the blood at the time point of sampling that can be affected by clinical parameters including disease stage, tumor size, number of metastatic sites, albumin level, and number of lines of treatment [8, 16]; these parameters were variable in this study of unselected cases (supplementary Table S2, available at Annals of Oncology online).
Alterations were identified in genes that have been associated with responses to targeted therapy in ER+ BC (PIK3CA, ESR1, ERBB2, FGFR1, BRCA1/2, AKT1) [4]. Compared with genomic studies of ER+ BC tissue biopsies, we identified similar mutation frequencies in ctDNA [1–3]. Tumor burden can be monitored by longitudinal assessment of variant AFs in ctDNA [8]. However, genomic profiling of large numbers of genes is best-suited for guiding therapy selection, but may be cost-prohibitive for serial testing. Instead, genomic profiling could establish GAs present in ctDNA at baseline for a patient, and guide design of personalized serial monitoring assays. GAs reported here could inform prioritization of genes to include for limited sequencing panels for longitudinal disease monitoring of ER+ mBC.
For a smaller subset of patients with temporally matched ctDNA and tissue samples, 89% of short variant mutations that were detected in tissue were also detected in ctDNA. Additional ESR1, TP53, and PIK3CA mutations were identified only in ctDNA; other studies have similarly observed additional mutations for each of these genes in ctDNA compared with matched tissue [5, 10, 17]. Additional mutations in ctDNA could reflect the utility of liquid biopsy to capture heterogeneity of metastatic sites in ER+ mBC [6]. Consistent with this idea, for paired cases with both shared and ctDNA only mutations in one gene, the shared mutation AF was two to threefold higher than the ctDNA only mutation AF. This hypothesis warrants confirmation in prospective trials and may be more relevant in clinical settings where targeted therapies are routinely employed.
In this study, genomic profiling was carried out as part of routine clinical care for unselected patients, including patients with low tumor burden; therefore, many samples had a low estimated ctDNA fraction (supplementary Table S2, available at Annals of Oncology online). The sensitivity for amplification detection in ctDNA was 27% for the 14 matched ctDNA-tissue pairs. Amplifications (including CCND1, FGFR1, ERBB2) were detected in ctDNA at lower frequencies than tissue; specifically ERBB2 amplification was identified in 29% of HER2+ cases. Detection of amplifications was associated with higher estimated ctDNA fraction. Our findings are consistent with NGS studies [using NGS panels or alternative approaches for amplification detection such as low coverage whole genome sequencing (plasma-Seq)] that highlighted the limitations for robust detection of amplifications in the context of low ctDNA fractions [18, 19]. Other studies identified similarly low frequencies of ERBB2 amplification (21%–32%) in ctDNA from HER2+ BC and detected other common BC amplifications (including CCND1, FGFR1) at significantly lower frequencies in ctDNA compared with matched tissue [7, 13]. Therefore, although amplifications may be detected in a subset of cases with sufficient ctDNA fraction, tissue-based genomic testing may be a more reliable method of detection. In BC, ERBB2 amplification remains the only established clinically utilized copy number biomarker, but amplifications including FGFR1 and 11q13 are being evaluated as biomarkers in trials [4]; tissue-based testing may be the preferable method for treatment selection based on copy number biomarkers.
We observed a high frequency of ESR1 GAs that are associated with AI resistance, as expected for this patient population of mostly mBC with prior AI treatment [9]. The ESR1 mutation frequency reported here is consistent with studies of AI-treated, ER+ mBC that used ddPCR to assess selected ESR1 mutations in ctDNA, including frequencies reported in several phase 3 trials [4, 9–11].
We observed a similar distribution of ESR1 mutations compared with a study of common ESR1 mutations in ctDNA using ddPCR [10]. Consistent with other studies, we frequently observed cases harboring >1 ESR1 GA [9, 10]. Multiple ESR1 mutations are thought to reflect convergent evolution of distinct clones during AI resistance [6, 10]; for the few cases evaluated using dual mutation-specific ddPCR probes, different ESR1 mutations existed on separate alleles [17, 20]. We confirmed that most ESR1 mutation pairs occur on distinct sequencing reads, likely reflecting polyclonal origin; however, we also identified cases with ESR1 compound mutations on the same allele. Studies of ESR1 have focused on single mutations; ESR1 compound mutations in cis warrant further study, and such mutations might display differential therapeutic sensitivities compared with characterized single mutations.
Diverse ESR1 alterations were observed, including rearrangements with break points resulting in loss of the LBD. Similar ESR1 rearrangements with variable 3’ fusion partners have been described and are activating [14, 15]. ESR1 rearrangements demonstrate preclinical resistance to AI and SERDs, therefore, detection of ESR1 rearrangements may be important for therapy selection [15]. ESR1 VUSs reported here could represent novel functional mutations that warrant characterization.
We identify co-occurring alterations with ESR1, which could represent alternative targets or rational targets for combination therapy with SERDs [4]. Some of these GAs have been successfully targeted in the context of co-occurring ESR1 GA: responses have been observed for patients with concurrent ESR1/AKT1 mutation (to AZD5363) [21], ESR1/PIK3CA mutation (to alpelisib) [22], and for a patient from this study with ESR1/BRCA2 mutation (to olaparib; Dr S. Blau, personal communication).
Concurrent ESR1/ERBB2 activating mutations occurred in ER+/HER2− BC and were more frequently observed in ctDNA compared with tissue, suggesting that ERBB2 and ESR1 mutations may commonly reside on distinct clones that may not be detected in a single tissue biopsy; ESR1 mutations were also observed in 25% of ER+/HER2+ BC. Combinations of SERDs with HER2-targeted therapy could be relevant for such cases.
Here, we demonstrate the clinical implementation of genomic profiling of ctDNA from patients with ER+ BC and identify clinically relevant GAs. Blood-based testing may provide an alternative or complementary approach to tissue-based genomic testing for patients with mBC.
Supplementary Material
Acknowledgement
The authors would like to recognize the important contributions from other researchers whose work could not be cited due to space constraints.
Funding
None declared.
Disclosure
JHC, DP, RH, LY, ABS, LMG, GMF, PJS, DL, TIM, VAM, JSR, and SMA are employees and hold equity stake in Foundation Medicine, Inc. BHP is a paid consultant for Foundation Medicine, Inc. and has research contracts with Foundation Medicine, Inc. All remaining authors have declared no conflicts of interest.
References
- 1. Ciriello G, Gatza ML, Beck AH. et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 2015; 163(2): 506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fumagalli D, Wilson TR, Salgado R. et al. Somatic mutation, copy number and transcriptomic profiles of primary and matched metastatic estrogen receptor-positive breast cancers. Ann Oncol 2016; 27(10): 1860–1866. [DOI] [PubMed] [Google Scholar]
- 3. Lefebvre C, Bachelot T, Filleron T. et al. Mutational profile of metastatic breast cancers: a retrospective analysis. PLOS Med 2016; 13(12): e1002201.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Turner NC, Neven P, Loibl S, Andre F.. Advances in the treatment of advanced oestrogen-receptor-positive breast cancer. Lancet 2016. doi:10.1016/S0140-6736(16)32419-9. [DOI] [PubMed] [Google Scholar]
- 5. Parsons HA, Beaver JA, Cimino-Mathews A. et al. Individualized molecular analyses guide efforts (IMAGE): a prospective study of molecular profiling of tissue and blood in metastatic triple-negative breast cancer. Clin Cancer Res 2017; 23(2): 379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Savas P, Teo ZL, Lefevre C. et al. The subclonal architecture of metastatic breast cancer: results from a prospective community-based rapid autopsy program “CASCADE”. PLOS Med 2016; 13(12): e1002204.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chae YK, Davis AA, Jain S. et al. Concordance of genomic alterations by next-generation sequencing (NGS) in tumor tissue versus circulating tumor DNA in breast cancer. Mol Cancer Ther 2017; molcanther.0061.2017. [DOI] [PubMed] [Google Scholar]
- 8. Siravegna G, Marsoni S, Siena S, Bardelli A.. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017. doi:10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 9. Schiavon G, Hrebien S, Garcia-Murillas I. et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med 2015; 7(313): 313ra182–313ra182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Spoerke JM, Gendreau S, Walter K. et al. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat Commun 2016; 7: 11579.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fribbens C, O’Leary B, Kilburn L. et al. Plasma ESR1 mutations and the treatment of estrogen receptor-positive advanced breast cancer. J Clin Oncol 2016; 34(25): 2961–2968. [DOI] [PubMed] [Google Scholar]
- 12. Stephens PJ, Clark T, Kennedy M. et al. Analytic validation of a clinical circulating tumor DNA assay for patients with solid tumors. Ann Oncol 2016. doi:10.1093/annonc/mdw380.01. [Google Scholar]
- 13. Liang DH, Ensor JE, Liu Z. et al. Cell-free DNA as a molecular tool for monitoring disease progression and response to therapy in breast cancer patients. Breast Cancer Res Treat 2016; 155(1): 139–149. [DOI] [PubMed] [Google Scholar]
- 14. Veeraraghavan J, Ma J, Hu Y, Wang X-S.. Recurrent and pathological gene fusions in breast cancer: current advances in genomic discovery and clinical implications. Breast Cancer Res Treat 2016; 158(2): 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li S, Shen D, Shao J. et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep 2013; 4(6): 1116–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jovelet C, Ileana E, Le Deley M-C. et al. Circulating cell-free tumor DNA analysis of 50 genes by next-generation sequencing in the prospective MOSCATO trial. Clin Cancer Res 2016; 22(12): 2960–2968. [DOI] [PubMed] [Google Scholar]
- 17. Chu D, Paoletti C, Gersch C. et al. ESR1 mutations in circulating plasma tumor DNA from metastatic breast cancer patients. Clin Cancer Res 2016; 22(4): 993–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lanman RB, Mortimer SA, Zill OA. et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS One 2015; 10(10): e0140712.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ulz P, Belic J, Graf R. et al. Whole-genome plasma sequencing reveals focal amplifications as a driving force in metastatic prostate cancer. Nat Commun 2016; 7: 12008.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang P, Bahreini A, Gyanchandani R. et al. Sensitive detection of mono- and polyclonal ESR1 mutations in primary tumors, metastatic lesions, and cell-free DNA of breast cancer patients. Clin Cancer Res 2016; 22(5): 1130–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hyman DM, Smyth LM, Donoghue MTA. et al. AKT inhibition in solid tumors with AKT1 mutations. J Clin Oncol 2017; JCO.2017.73.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mayer IA, Abramson VG, Formisano L. et al. A phase Ib study of alpelisib (BYL719), a PI3Kα-specific inhibitor, with letrozole in ER+/HER2− metastatic breast cancer. Clin Cancer Res 2017; 23(1): 26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.