Abstract
Background
Cancer initiation and development are driven by key mutations in driver genes. Applying high-throughput sequencing technologies and bioinformatic analyses, The Cancer Genome Atlas (TCGA) project has identified panels of somatic mutations that contributed to the etiology of various cancers. However, there are few studies investigating the germline genetic variations in these significantly mutated genes (SMGs) and lung cancer susceptibility.
Patients and methods
We comprehensively evaluated 1655 tagged single nucleotide polymorphisms (SNPs) located in 127 SMGs identified by TCGA, and test their association with lung cancer risk in large-scale case–control study. Functional effect of the validated SNPs, gene mutation frequency and pathways were analyzed.
Results
We found 11 SNPs in 8 genes showed consistent association (P < 0.1) and 8 SNPs significantly associated with lung cancer risk (P < 0.05) in both discovery and validation phases. The most significant association was rs10412613 in PPP2R1A, with the minor G allele associated with a decreased risk of lung cancer [odds ratio = 0.91, 95% confidence interval (CI): 0.87–0.96, P = 2.3 × 10−4]. Cumulative analysis of risk score built as a weight sum of the 11 SNPs showed consistently elevated risk with increasing risk score (P for trend = 9.5 × 10−9). In stratified analyses, the association of PPP2R1A:rs10412613 and lung cancer risk appeared stronger among population of younger age at diagnosis and never smokers. The expression quantitative trait loci analysis indicated that rs10412613, rs10804682, rs635469 and rs6742399 genotypes significantly correlated with the expression of PPP2R1A, ATR, SETBP1 and ERBB4, respectively. From TCGA data, expression of the identified genes was significantly different in lung tumors compared with normal tissues, and the genes’ highest mutation frequency was found in lung cancers. Integrative pathway analysis indicated the identified genes were mainly involved in AKT/NF-κB regulatory pathway suggesting the underlying biological processes.
Conclusion
This study revealed novel genetic variants in SMGs associated with lung cancer risk, which might contribute to elucidating the biological network involved in lung cancer development.
Keywords: significantly mutated genes, TCGA, tagSNP, eQTL, pathway analysis, lung cancer
Introduction
Lung cancer is one of the most common cancers worldwide and is a leading cause of cancer deaths [1]. While smoking is the primary risk factor for lung cancer, only ∼15% of lifelong smokers develop the deadly disease [2]. In recent years, studies found that genetics may play a role in the development of lung cancer [3, 4].
Identifying key genes whose alterations drive cancer initiation and development is an important area of cancer research. Recent advances in high-throughput DNA sequencing technologies have enabled the comprehensive characterization of somatic mutations in cancers [5]. Utilizing novel technology and bioinformatics analysis, The Cancer Genome Atlas (TCGA) project previously has identified panels of genetic mutations that contributed to or were associated with the etiology of a variety of cancers [6]. In lung cancer, considerable progress in personalized treatment has been made by targeting significantly mutated genes (SMGs) in tumor, such as mutations in epidermal growth factor receptor (EGFR) and anaplastic lymphoma receptor tyrosine kinase (ALK) [7, 8]. Given the significance of somatic mutations in SMGs in tumor development and growth, the germline risk factors involving SMGs have not been systematically investigated. Previous genome-wide association studies (GWAS) have identified several susceptibility loci in a region of 15q25 contributing to smoking behavior, nicotine addiction and lung cancer risk [9, 10]. However, to our knowledge, no study has systematically investigated the common single nucleotide polymorphisms (SNPs) within SMGs in lung cancer. Since SMGs may represent key drivers for tumor development and progression [6], identifying the SNPs in SMGs associated with lung cancer may uncover new susceptibility loci that contribute to disease etiology with biological plausibility.
In this study, leveraging an ongoing case–control study of lung cancer patients treated at The University of Texas MD Anderson Cancer Center, we systematically evaluated 1655 tagged SNPs (tagSNPs) located in cancer SMGs identified by the TCGA studies, and assessed the association of these SNPs with risk of lung cancer. The results were then validated by an independent population from dbGAP database. The cumulative effect of the risk SNPs and their association stratified by age, gender and smoking status were also analyzed. Functional significance of the identified SNPs was investigated using bioinformatics tools and public databases. The significant SNPs and genes identified from the study were further assessed for gene expression, mutation frequency in major cancers, and involvement of putative intracellular pathways. To our knowledge, this is the first large scale investigation of germline polymorphisms of SMGs in modulating the risk of lung cancer, which further supports the role of the SMGs in lung cancer etiology.
Methods
Detailed description of study methods can be found in supplementary methods, available at Annals of Oncology online.
Study population and data collection
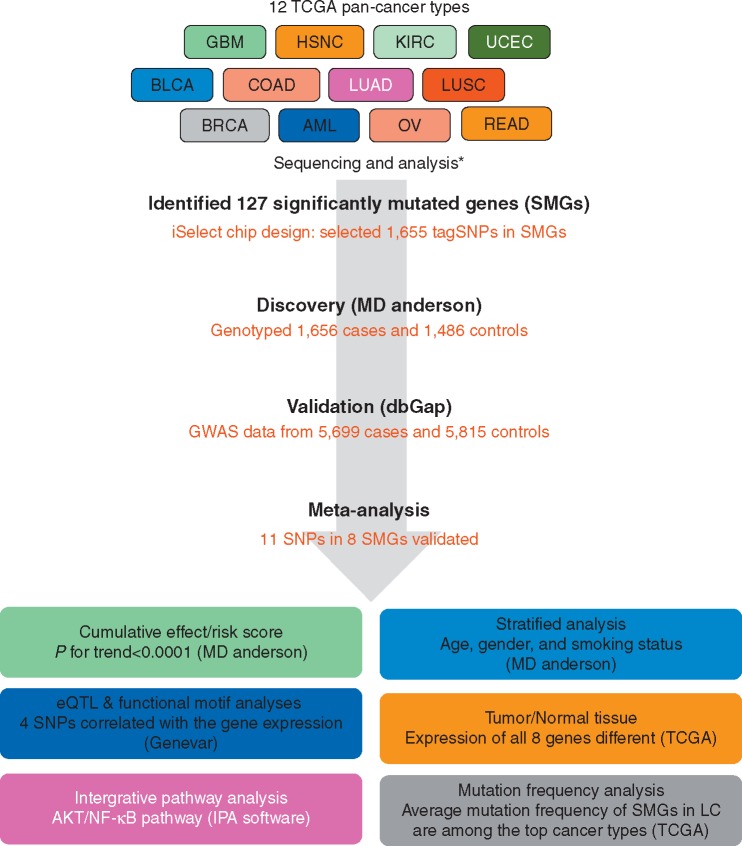
A schematic diagram of the study design is displayed in Figure 1. The study was approved by the Institutional Review Board of UT MD Anderson Cancer Center, and written informed consent to participate in this study was obtained from all participants before biospecimens, and data were collected.
Figure 1.
Schematic of study design. BRCA, breast adenocarcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; UCEC, uterine corpus endometrial carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; COAD, READ; colon and rectal carcinoma; BLCA, bladder urothelial carcinoma; KIRC, kidney renal clear cell carcinoma; OV, ovarian serous carcinoma; AML, acute myeloid leukemia; LC; lung cancer. *Refer to reference 6.
SNP selection and genotyping
SMGs were identified by TCGA Pan-Cancer effort in 12 major cancer types (http://cancergenome.nih.gov/ (May 2015, date last accessed)) [6]. All identified genes have a mutation rate of at least 1% in at least one cancer type. TagSNPs were selected in 127 SMGs using Tagger program (NCBI B36 assembly, dbSNP b126) from HapMap (www.hapmap.org (May 2015, date last accessed)). Genomic DNA isolation and genotyping were carried out as previously described [11].
Analysis of expression quantitative trait loci, functional motif of SNPs, mutation frequency of genes, and integrative network analysis (IPA)
The potential effect of validated SNPs on gene expression and functional motif of SNPs were evaluated by expression quantitative trait loci (eQTL) association using the Genevar database [12] and HaploReg v4.1 from Broad Institute (http://www.broadinstitute.org/mammals/haploreg/haploreg.php (May 2015, date last accessed)), respectively. Genes corresponding to the identified SNPs were analyzed in TCGA Pan-Cancer database [6], and their mutation frequency was ascertained for 12 major cancer types. Genes of the identified SNPs were further analyzed for cell signaling network by IPA.
Expression of identified genes in lung cancer tumor and normal lung tissues
Gene expression derived from TCGA data was obtained from Broad GDAC Firehose pipeline (http://gdac.broadinstitute.org (March 2016, date last accessed), dbGaP Study Accession: phs000178.v1.p1). Expression data were generated by Illumina HiSeq RNA sequencing of 108 normal lung tissues and lung cancer tumor tissues from patients with lung adenocarcinoma (n = 488) and squamous cell carcinoma (n = 482).
Statistical analysis
For demographic variables, Pearson χ2 or Fisher exact test was used to analyze the difference in distribution of categorical variables and Wilcoxon rank sum test or Student’s t-test was used to analyze continuous variables between cases and controls. The association of each SNP with the risk of lung cancer was estimated using the odds ratio (OR) and 95% confidence interval (CI). In the discovery population, multivariate unconditional logistic regression was used adjusting for age, gender and smoking status. In the validation population, multivariate unconditional logistic regression was used adjusting for age, gender, study (four studies listed in NCI dbGAP GWAS: EAGLE, ATBC, PLCO and CPS-II), and significant principal components, following the procedure as described previously [13]. Meta-analysis was based on logistic regression combining estimates in discovery and validation sets under fixed-effect model. Cumulative effects for the candidate SNPs were assessed by summing risk alleles showing significant association with lung cancer risk (P < 0.05). Risk scores were a weighed sum of top association and were then categorized into quartiles.
Results
Host characteristics
In discovery phase, a total of 1656 patients and 1486 controls were enrolled in this study. supplementary Table S1, available at Annals of Oncology online summarized the host characteristics of the discovery population. The mean (±SD) age of the subjects was 62.7 ± 10.7 for cases and 62.3 ± 10.7 years for controls. We observed significant differences between cases and controls with regard to gender (P = 0.002) and smoking status (P < 0.001) (supplementary Table S1, available at Annals of Oncology online). For the validation group, host characteristic data were obtained from dbGaP (supplementary Table S1, available at Annals of Oncology online). In the validation group, gender but not age distribution was significantly different between cases and controls (P < 0.001), and there was greater proportion of males (81%) versus females compared with the discovery group (54%).
Genetic variants associated with risk of lung cancer
Among 1655 selected SNPs in 127 SMGs, 168 SNPs were found significantly associated with lung cancer risk (supplementary Table S2, available at Annals of Oncology online). Of these, 11 SNPs consistently associated with risk of lung cancer (P < 0.1) in both discovery and validation populations and 8 SNPs were significant (P < 0.05, Table 1). The most significant SNP was rs10412613 (OR = 0.84, 95% CI: 0.75–0.94, P = 0.002) in PPP2R1A, which is also significant in validation set (OR = 0.93, 95% CI: 0.88–0.98, P = 0.01). Combining results from the two populations, we carried out a meta-analysis for the 11 identified SNPs, and all variants were significantly associated with lung cancer risk, among which rs10412613 in PPP2R1A being the most significant (OR = 0.91, 95% Cl: 0.87–0.96, P = 2.3 × 10−4, Table 1).
Table 1.
Significant association of SNPs in cancer-related significantly mutated genes and risk of lung cancer (MD Anderson, n = 3142; dbGAP, n = 11 514)
| Minor allele | Discovery |
Validation |
Meta-analysis |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Gene | Modela | ORb(95% CI) | P | ORc(95% CI) | P | OR (95% CI) | P | Pheterogeneity | |||
| rs10412613 | PPP2R1A | G | ADD | 0.84 (0.75–0.94) | 2.3 × 10−3 | 0.93 (0.88–0.98) | 9.7 × 10−3 | 0.91 (0.87–0.96) | 2.3 × 10−4 | 0.11 | ||
| rs4802905 | PPP2R1A | G | DOM | 1.23 (1.02–1.47) | 0.027 | 1.10 (1.00–1.21) | 0.041 | 1.13 (1.04–1.23) | 5.7 × 10−3 | 0.29 | ||
| rs10804682 | ATR | A | DOM | 1.21 (1.03–1.41) | 0.021 | 1.11 (1.03–1.20) | 9.5 × 10−3 | 1.13 (1.05–1.21) | 5.6 × 10−4 | 0.33 | ||
| rs2819365 | ELF3 | A | DOM | 0.86 (0.73–1.00) | 0.05 | 0.91 (0.84–0.98) | 9.1 × 10−3 | 0.90 (0.84–0.96) | 2.9 × 10−3 | 0.53 | ||
| rs6747637 | ERBB4 | C | REC | 0.81 (0.67–0.98) | 0.033 | 0.89 (0.81–0.98) | 0.017 | 0.87 (0.80–0.95) | 1.8 × 10−3 | 0.39 | ||
| rs6742399 | ERBB4 | A | ADD | 0.88 (0.79–0.98) | 0.021 | 0.95 (0.90–1.00) | 0.063 | 0.94 (0.89–0.98) | 6.2 × 10−3 | 0.22 | ||
| rs6740117 | ERBB4 | G | REC | 0.82 (0.67–1.00) | 0.046 | 0.90 (0.81–0.99) | 0.031 | 0.88 (0.81–0.97) | 6.7 × 10−3 | 0.42 | ||
| rs434645 | SFTA1P | A | DOM | 1.21 (1.02–1.43) | 0.029 | 1.09 (1.00–1.18) | 0.054 | 1.11 (1.03–1.20) | 5.0 × 10−3 | 0.28 | ||
| rs635469 | SETBP1 | G | DOM | 0.83 (0.69–1.00) | 0.049 | 0.92 (0.85–1.00) | 0.059 | 0.90 (0.84–0.97) | 8.5 × 10−3 | 0.32 | ||
| rs1052669 | ARHGAP35 | A | REC | 0.73 (0.54–0.98) | 0.035 | 0.87 (0.75–1.00) | 0.05 | 0.84 (0.74–0.96) | 9.1 × 10−3 | 0.30 | ||
| rs11175985 | LRRK2 | A | DOM | 1.24 (1.04–1.48) | 0.016 | 1.07 (0.98–1.17) | 0.01 | 1.10 (1.02–1.19) | 0.016 | 0.14 | ||
Model of inheritance selected for the most significant model: dom, dominant; rec, recessive; add, additive.
Adjusted for age, gender and smoking status.
Adjusted for age, gender, study, and significant principal component.
In the stratified analysis, the association of PPP2R1A:rs10412613 and the risk of lung cancer appeared stronger among population of younger age at diagnosis (OR = 0.82, 95% CI 0.70–0.95, P = 0.008) and never smokers (OR = 0.84, 95% CI 0.74–0.96, P = 0.009). The association of ATR: rs10804682 and risk of lung cancer was stronger among population of younger age at diagnosis (OR = 1.35, 95% CI 1.06–1.68, P = 0.006) and males (OR = 1.35, 95% CI 1.08–1.68, P = 0.009). As for ARHGAP35:rs1052669, the association was only stronger in population of younger age at diagnosis (OR = 0.59, 95% CI 0.39–0.88, P = 0.009) but not in other stratified groups (supplementary Table S3, available at Annals of Oncology online). No significant interactions were observed between all identified SNPs and age, gender or smoking status.
Cumulative effect of risk alleles
To examine the cumulative effect of 11 replicated SNPs, the risk score built as a weight sum of the SNPs estimated in case and control subjects, and the effect of lung cancer risk was analyzed. Compared with lowest-risk score (≤0.90), subjects with 0.91–1.16, 1.17–1.35, and >1.35, had 1.4-fold (OR = 1.40, 95% CI 1.12–1.76, P = 2.8 × 10−3), 1.6-fold (OR = 1.58, 95% CI 1.26–1.98, P = 6.3 × 10−5) and 1.9-fold (OR = 1.90, 95% CI 1.53–2.37, P = 8.4 × 10−9) increased risk of lung cancer, respectively, and these risks showed increasing trend with increasing risk scores (P for trend = 9.5 × 10−9, Table 2). Similar results were also observed in cumulative unfavorable genotype analysis (supplementary Table S4, available at Annals of Oncology online).
Table 2.
Genetic risk score of discovery set for SNPs of significantly mutated genes in lung cancer (MD Anderson, n = 3142)
| Risk score | Cases, n (%) | Controls, n (%) | ORa(95% CI) | P |
|---|---|---|---|---|
| ≤0.90 | 301 (44.79) | 371 (55.21) | 1 (Reference) | |
| 0.91–1.16 | 414 (51.56) | 389 (48.44) | 1.40 (1.12–1.76) | 2.8 × 10−3 |
| 1.17–1.35 | 419 (54.13) | 355 (45.87) | 1.58 (1.26–1.98) | 6.3 × 10−5 |
| >1.35 | 521 (58.61) | 368 (41.39) | 1.90 (1.53–2.37) | 8.4 × 10−9 |
| P for trend | 9.5 × 10−9 |
Adjusted for age, gender and smoking status.
SNPs function
To understand the functional impact of the identified SNPs from cancer-related SMGs, the SNP associated gene expression was tested by eQTL analysis in Genevar public database (supplementary Figure S1, available at Annals of Oncology online). Seven SNPs were available in Genevar database and 4 of them were significantly associated with gene expression in normal tissues: rs10412613 and PPP2R1A expression in skin cells (P = 0.04), rs10804682 and ATR expression in fibroblasts (P = 0.03), rs635469 and SETBP1 expression in lymphoblastoid cells (P = 0.04) and rs6742399 and ERBB4 expression in CEU population (P = 0.003).
Bioinformatics analysis in ENCODE database revealed that 8 of 11 SNPs that were associated with risk of lung cancer might alter the intracellular regulatory motifs (P < 0.05) (supplementary Table S5, available at Annals of Oncology online). In TCGA database, gene expression of lung cancer tumors (n = 970) and normal tissues (n = 108) were available for all 8 genes of the identified SMG SNPs: PPP2R1A, ATR, ELF3, ERBB4, SFTA1P, LRRK2, SETBP1, and GATA3. The expression of these genes was significantly altered in tumors compared with normal lung tissues (P < 0.0001, supplementary Table S6, available at Annals of Oncology online). The mutation frequency of the 8 genes across 12 major cancer sites was analyzed in TCGA Pan-Cancer database. Interestingly, the highest average mutation frequency for the identified genes was found in lung cancers (adenocarcinoma: 5.1%; squamous cell carcinoma: 4.9%) compared with other cancer types (supplementary Table S7, available at Annals of Oncology online). Among these genes, the highest mutation frequency was found for SETBP1, whose expression could be affected by rs635469 genotypes and whose mutations appeared in 12.7% of lung adenocarcinomas.
Integrative network analysis
We also carried out integrative network analysis to examine the underlying biological processes that might be associated with risk of lung cancer development. The eight candidate genes were analyzed by IPA analysis, which revealed an interaction network consisting of these genes and AKT/NF-κB regulatory pathway (supplementary Figure S2, available at Annals of Oncology online). Several known genes that have been implicated in lung cancer development (e.g. CHRNA3 and BCL2L2) were found in the network.
Discussion
It is well-known that mutations in key cancer-related genes drive cancer initiation and tumor cell proliferation while inhibiting apoptosis. The somatic profile of SMGs may help to identify these driver genes, which has drawn significant attention in cancer research. However, the influence of germline genetic variation of these genes on lung cancer risk has not been extensively explored. To our knowledge, this study is the first large-scale, integrative association analysis of SMG genetic variants with susceptibility to lung cancer. We comprehensively incorporated genetic polymorphism data, gene expression data, mutation frequency data, SNP functional analysis and bioinformatic analysis. We identified 11 SNPs, including 4 potentially functional SNPs, in 8 genes as novel lung cancer susceptibility loci. The functional characterization using eQTL analysis suggested the correlation of functional SNP genotypes with gene expression. Indeed, the expression of these genes was significantly different in tumors compared with normal tissues. The fact that the cancer types bearing the highest mutation frequency for the identified genes were lung cancers also supported the notion of these SMGs playing potential role as cancer drivers. Furthermore, IPA analysis suggested that the identified genes were mainly involved in cellular proliferation and developmental signaling of the AKT-NF-κB regulatory pathway.
The most significant SNP (rs10412613) associated with risk of lung cancer is located in PPP2R1A gene, which encodes a regulatory subunit of protein phosphatase 2 (PP2A). PP2A is one of the major cellular Ser/Thr protein phosphatases which play key roles in regulating cell growth, apoptosis, and transformation by dephosphorylating target proteins in several signaling pathways of both normal and cancer cells [14, 15]. Some studies reported that PP2A in cancer development was regulated by activation of ATM and Rad3-related (ATR) to initiate DNA damage response [16]. Of interest, in our study, we also found that genetic variant, rs10804682, in ATR was also associated with lung cancer risk. In lung cancer, Yang et al. [17] and Zienolddiny et al. [18] recently identified the association of rs11453459 in PPP2R1A and rs10804682 in ATR with susceptibility to lung cancer. Our study is in line with these studies; we identified three distinct genetic signals: rs10412613 and rs4802905 in PPP2R1A and rs10804682 in ATR being associated with risk of lung cancer with two of the three SNPs, rs10412613 and rs10804682, having potential functional effect as determined by bioinformatic analyses.
The genotypes of rs10412613 in PPP2R1A and rs10804682 in ATR both correlated with gene expressions in tissues or cell lines in eQTL analysis. As predicted from the ENCODE dataset, the polymorphism of rs10804682 perturbs a regulatory binding site, which may potentially affect the interaction with NKX2-5 resulting in alternative splicing with multiple transcript variants that may further modify lung cancer risk. Therefore, rs10412613 and rs10804682 may affect RNA stability and its subsequent production. Further functional characterization may be necessary to dissect the mechanistic basis for the variants’ association with lung cancer risk.
E74-like factor 3 (ELF3) is the prototypic member of a novel subset of the ETS transcription factor family [19] and has been reported to involve in a variety of pathophysiologic processes, including cancer and inflammatory disorders. Liu et al. [20] found that loss of PTEN and SMAD4 resulted in ELF3 and ErbB2 pathway activation and suggested the importance of the ErbB2/Akt/ELF3 signaling pathway as both lung cancer biomarker and therapeutic drug target for tumor cells. Other recent studies also reported similar findings in lung cancer [21, 22]. Our result showing genetic variant of ELF3 associated with lung cancer susceptibility supports this notion. Further functional analysis using the ENCODE dataset also showed that rs2819365 might impact the interaction with TLX1 and NF1C, which are nuclear transcription factors that are involved in development and transcriptional machinery, respectively.
The presence of activating driver mutations of ERBB4 has been demonstrated in lung cancer [23]. Although ERBB4 is a member of the EGFR subfamily of receptor tyrosine kinases and 5.4% of non-small cell lung cancers harbor ERBB4 missense mutations [24, 25], to our knowledge, ERBB4 polymorphisms have not been studied in lung cancer, while studies in breast and prostate cancer have been investigated [26]. Rokavec et al. [27] demonstrated that SNP rs62626348 in ERBB4 conferred highest risk of breast cancer in a large case–control study. In the present study, the significant association of three SNPs in ERBB4 (rs6747637, rs6740117 and rs6742399) with lung cancer risk suggests that germline genetic variations of this gene may play an important role similar to the effect of somatic mutations to initiate or support cancer development in the lung.
The Rho GTPase-activating protein 35 (ARHGAP35) is an important Rho family GTPase-activating protein, and was identified as a crucial regulator for cell proliferation, spreading and migration whose alteration may determine cancer cell types. Campbell et al. [28] suggested that lung adenocarcinoma lacking receptor tyrosine kinase-Ras–Raf pathway alterations have mutations in SOS1, VAV1, RASA1 and ARHGAP35. The association between genetic variants in ARHGAP35 and lung cancer risk is previously unknown. However, polymorphism of ARHGAP35 has been demonstrated to associate with osteosarcoma risk and prognosis [29]. In this study, rs1052669 in ARHGAP35 was associated with lung cancer risk, and genotypes of this SNP significantly correlated with expression of ARHGAP35 which suggested functional relevance. Finally, using bioinformatics approach, we found the identified genes were mainly associated with AKT/NF-κB pathway genes and network. The genes in this network, including AKT, NF-κB, CHRNA3 and BCL2L2, have been demonstrated to associate with lung cancer [9, 30].
There are several strengths in this study. First, the relatively large sample size with complete host characteristics and clinical information of patients in MD Anderson Cancer Center allowed us to have sufficient power to test the cancer risk association while fully adjusting possible confounders and stratifying by important risk variables. Second, the design of two independent phases for discovery and validation enabled us to minimize potential false positive findings. Third, broad selection of genetic variants from 127 SMGs ensure adequate coverage of important genes involved in the pathogenesis of major cancer types, although some functional SNPs might be missed due to the tagSNP selection strategy.
In conclusion, the results of this large, integrated multi-phase case–control study suggest for the first time that genetic variations of SMGs may contribute to lung cancer susceptibility. Future independent studies are warranted to confirm the results. Distinct genetic markers could provide translational application in risk stratification and uncover the novel mechanistic basis of lung carcinogenesis.
Funding
This work was supported in part by grants from the Cancer Prevention and Research Institute of Texas (RP1300502); and National Cancer Institute (P50 CA070907 and R01 CA176568). Additional funding was provided by MD Anderson institutional support for the Center for Translational and Public Health Genomics and Duncan Family Institute for Cancer Prevention and Risk Assessment (no grant numbers apply).
Disclosure
The authors have declared no conflicts of interest.
Key Message
Alterations in key driver genes initiate cancer development. This integrated multi-phase study indicates for the first time that genetic variations of significantly mutated genes could contribute to lung cancer susceptibility. Distinct genetic markers may provide translational application in risk stratification and uncover novel mechanistic basis of lung carcinogenesis.
Supplementary Material
References
- 1. Siegel RL, Miller KD, Jemal A.. Cancer statistics, 2015. CA Cancer J Clin 2015; 65: 5–29. [DOI] [PubMed] [Google Scholar]
- 2. Ruano-Ravina A, Figueiras A, Barros-Dios JM.. Lung cancer and related risk factors: an update of the literature. Public Health 2003; 117: 149–156. [DOI] [PubMed] [Google Scholar]
- 3. Sato M, Shames DS, Gazdar AF, Minna JD.. A translational view of the molecular pathogenesis of lung cancer. J Thorac Oncol 2007; 2: 327–343. [DOI] [PubMed] [Google Scholar]
- 4. Meyerson M, Franklin WA, Kelley MJ.. Molecular classification and molecular genetics of human lung cancers. Semin Oncol 2004; 31: 4–19. [DOI] [PubMed] [Google Scholar]
- 5. Watson IR, Takahashi K, Futreal PA, Chin L.. Emerging patterns of somatic mutations in cancer. Nat Rev Genet 2013; 14: 703–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kandoth C, McLellan MD, Vandin F. et al. Mutational landscape and significance across 12 major cancer types. Nature 2013; 502: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tan CS, Gilligan D, Pacey S.. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol 2015; 16: e447–4e459. [DOI] [PubMed] [Google Scholar]
- 8. Peifer M, Fernandez-Cuesta L, Sos ML. et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012; 44: 1104–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Amos CI, Wu X, Broderick P. et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet 2008; 40: 616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hung RJ, McKay JD, Gaborieau V. et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature 2008; 452: 633–637. [DOI] [PubMed] [Google Scholar]
- 11. Liang D, Meyer L, Chang DW. et al. Genetic variants in MicroRNA biosynthesis pathways and binding sites modify ovarian cancer risk, survival, and treatment response. Cancer Res 2010; 70: 9765–9776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang TP, Beazley C, Montgomery SB. et al. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics 2010; 26: 2474–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Landi MT, Chatterjee N, Yu K. et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet 2009; 85: 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Soprano KJ, Purev E, Vuocolo S, Soprano DR.. Rb2/p130 and protein phosphatase 2A: key mediators of ovarian carcinoma cell growth suppression by all-trans retinoic acid. Oncogene 2006; 25: 5315–5325. [DOI] [PubMed] [Google Scholar]
- 15. Westermarck J, Hahn WC.. Multiple pathways regulated by the tumor suppressor PP2A in transformation. Trends Mol Med 2008; 14: 152–160. [DOI] [PubMed] [Google Scholar]
- 16. Cimprich KA, Cortez D.. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 2008; 9: 616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang R, Yang L, Qiu F. et al. Functional genetic polymorphisms in PP2A subunit genes confer increased risks of lung cancer in southern and eastern Chinese. PLoS One 2013; 8: e77285.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zienolddiny S, Campa D, Lind H. et al. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis 2006; 27: 560–567. [DOI] [PubMed] [Google Scholar]
- 19. Wang H, Fang R, Cho JY. et al. Positive and negative modulation of the transcriptional activity of the ETS factor ESE-1 through interaction with p300, CREB-binding protein, and Ku 70/86. J Biol Chem 2004; 279: 25241–25250. [DOI] [PubMed] [Google Scholar]
- 20. Liu J, Cho SN, Akkanti B. et al. ErbB2 pathway activation upon Smad4 loss promotes lung tumor growth and metastasis. Cell Rep 2015; S2211–1247(15)00142–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ali SA, Justilien V, Jamieson L. et al. Protein kinase Ciota drives a NOTCH3-dependent stem-like phenotype in mutant KRAS Lung Adenocarcinoma. Cancer Cell 2016; 29: 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Erdogan E, Klee EW, Thompson EA, Fields AP.. Meta-analysis of oncogenic protein kinase Ciota signaling in lung adenocarcinoma. Clin Cancer Res 2009; 15: 1527–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kurppa KJ, Denessiouk K, Johnson MS, Elenius K.. Activating ERBB4 mutations in non-small cell lung cancer. Oncogene 2016; 35: 1283–1291. [DOI] [PubMed] [Google Scholar]
- 24. Forbes SA, Bhamra G, Bamford S. et al. The catalogue of somatic mutations in cancer (COSMIC). Curr Protoc Hum Genet 2008; Chapter 10; Unit 10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ding L, Getz G, Wheeler DA. et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008; 455: 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murabito JM, Rosenberg CL, Finger D. et al. A genome-wide association study of breast and prostate cancer in the NHLBI's Framingham Heart Study. BMC Med Genet 2007; 8(Suppl 1): S6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rokavec M, Justenhoven C, Schroth W. et al. A novel polymorphism in the promoter region of ERBB4 is associated with breast and colorectal cancer risk. Clin Cancer Res 2007; 13: 7506–7514. [DOI] [PubMed] [Google Scholar]
- 28. Campbell JD, Alexandrov A, Kim J. et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016; 48: 607–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhao J, Xu H, He M.. Rho GTPase-activating protein 35 rs1052667 polymorphism and osteosarcoma risk and prognosis. Biomed Res Int 2014; 2014: 396947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shishodia S, Koul D, Aggarwal BB.. Cyclooxygenase (COX)-2 inhibitor celecoxib abrogates TNF-induced NF-kappa B activation through inhibition of activation of I kappa B alpha kinase and Akt in human non-small cell lung carcinoma: correlation with suppression of COX-2 synthesis. J Immunol 2004; 173: 2011–2022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.