Abstract
Background
While patient-derived xenografts (PDXs) offer a powerful modality for translational cancer research, a precise evaluation of how accurately patient responses correlate with matching PDXs in a large, heterogeneous population is needed for assessing the utility of this platform for preclinical drug-testing and personalized patient cancer treatment.
Patients and methods
Tumors obtained from surgical or biopsy procedures from 237 cancer patients with a variety of solid tumors were implanted into immunodeficient mice and whole-exome sequencing was carried out. For 92 patients, responses to anticancer therapies were compared with that of their corresponding PDX models.
Results
We compared whole-exome sequencing of 237 PDX models with equivalent information in The Cancer Genome Atlas database, demonstrating that tumorgrafts faithfully conserve genetic patterns of the primary tumors. We next screened PDXs established for 92 patients with various solid cancers against the same 129 treatments that were administered clinically and correlated patient outcomes with the responses in corresponding models. Our analysis demonstrates that PDXs accurately replicate patients’ clinical outcomes, even as patients undergo several additional cycles of therapy over time, indicating the capacity of these models to correctly guide an oncologist to treatments that are most likely to be of clinical benefit.
Conclusions
Integration of PDX models as a preclinical platform for assessment of drug efficacy may allow a higher success-rate in critical end points of clinical benefit.
Keywords: patient-derived xenograft, PDX, tumorgraft, chemotherapy, translational model
Introduction
The large percentage of investigational cancer drugs fail to demonstrate clinical activity due to an inability to identify patients that are most likely to respond [1, 2]. With the understanding that each individual cancer is characterized by patient-specific molecular events, it is becoming apparent that cell-line xenografts diverge substantially from the tumors they were derived, and cannot recapitulate patients’ responses to therapy. Furthermore, genetically engineered models develop lesions triggered by the same initiating oncogenic hit(s), lack genetic diversity, and do not represent the wide heterogeneity that occurs on a population basis [3, 4]. Hence, new strategies that more closely mimic individual tumors are required to maximize drug development success.
Patient-derived-xenografts (PDXs), established from tumor tissue directly engrafted into immunodeficient mice, are one possible solution. Increasing evidence indicate that PDXs faithfully conserve biological features of the parental malignancies (such as histologic architecture, gene-expression, mutational status and metastatic potential), as well as the complex interplay between cancer cells and tumor microenvironment [5–10], establishing PDXs as a more suitable model of human cancer.
Given the potential application of PDXs to clinical decision-making and drug-development, extensive research has been devoted to assessing the response of these models to anticancer treatment. Several retrospective studies comparing patients’ responses to conventional anticancer therapies with that of corresponding PDXs reported comparable treatment outcomes in various solid tumor types [11–17]. Furthermore, a remarkable similarity in response rates was observed between PDXs and respective clinical trials [13, 18–22]. Consequently, a number of very small prospective studies have been conducted focusing on the integration of PDXs into clinical-care paradigms [5–8, 23, 24]. While these studies suggest that PDXs may predict individual drug responses [5, 8], they have focused on specific tumor types or drug(s) combinations and have limited value in predicting patient responses at a population level due to the small number of models used. Although a recent large-scale in vivo drug screen that profiled over 60 treatment regimens against an extensive collection of PDXs (1075 models across 15 cancer-types) has shown that tumorgrafts were able to reproduce treatment responses from previous clinical trials [25], a comprehensive analysis of the clinical accuracy of these models across a larger, more heterogeneous patient cohort and matched PDX counterparts has not been previously reported.
Here we carried out whole-exome next-generation sequencing (WES) on 237 early passage PDXs (including 4 tumorgrafts with matched primary tumor), and compared PDX sequencing data with equivalent information in The Cancer Genome Atlas (TCGA) database, demonstrating that tumorgrafts faithfully conserved genetic patterns of the primary tumors. To underscore the promise of this technology as a tool for guiding therapeutic responses, we have screened PDXs established for 92 patients with various solid cancers against the same 129 treatments that were administered clinically (some patients having undergone multiple treatments). Our analysis revealed that tumor growth regression in PDXs accurately parallels clinical responses in patients, indicating that these models have the potential to correctly guide an oncologist to treatments that are most likely to be of clinical benefit. Furthermore, PDXs retained the capacity to identify viable treatments for recurrent disease arising months after the initial presentation, suggesting the potential to guide therapeutic decisions as multiple cycles of disease progression and therapy occur.
Methods
Patient inclusion
Patients were not selected by any specific criteria other than the presence of a drug correlation. All patients whose tumor models were tested and received therapy in the clinical setting were included.
Model generation
All animal procedures were carried out at Champions Oncology following Institutional Animal Care and Use Committee protocols. Methods concerning in vivo experiments are provided in supplementary experimental procedures, available at Annals of Oncology online.
Characterization of patient tumor and PDX histology
Details on patient tumor and PDX histology are provided in supplementary experimental procedures, available at Annals of Oncology online.
Mutation profiling and CNV analysis
Details on sequencing, mutation profiling and CNV analysis are provided in supplementary experimental procedures, available at Annals of Oncology online.
Tumor growth regression and RECIST
Upon study completion, percent tumor regression (%TR) values were calculated using initial (i) and final (f) tumor measurements for the treatment (T) group by the formula: %TR = [1 − (Tf/Ti)] × 100. Responses to therapy were then converted to clinical outcomes (based on RECIST) from changes in tumor volume over the course of treatment (details available at Annals of Oncology online).
Statistical analysis
Differences in tumor volumes between treated and control animals during drug screening were analyzed using Student’s t-test, where P < 0.05 was considered significant. Associations between patient clinical outcome and the response in PDXs were examined using Fisher's exact test, with P < 0.05 set as the threshold for significance. All statistical analyses were two-sided.
Results
Establishment of PDX models
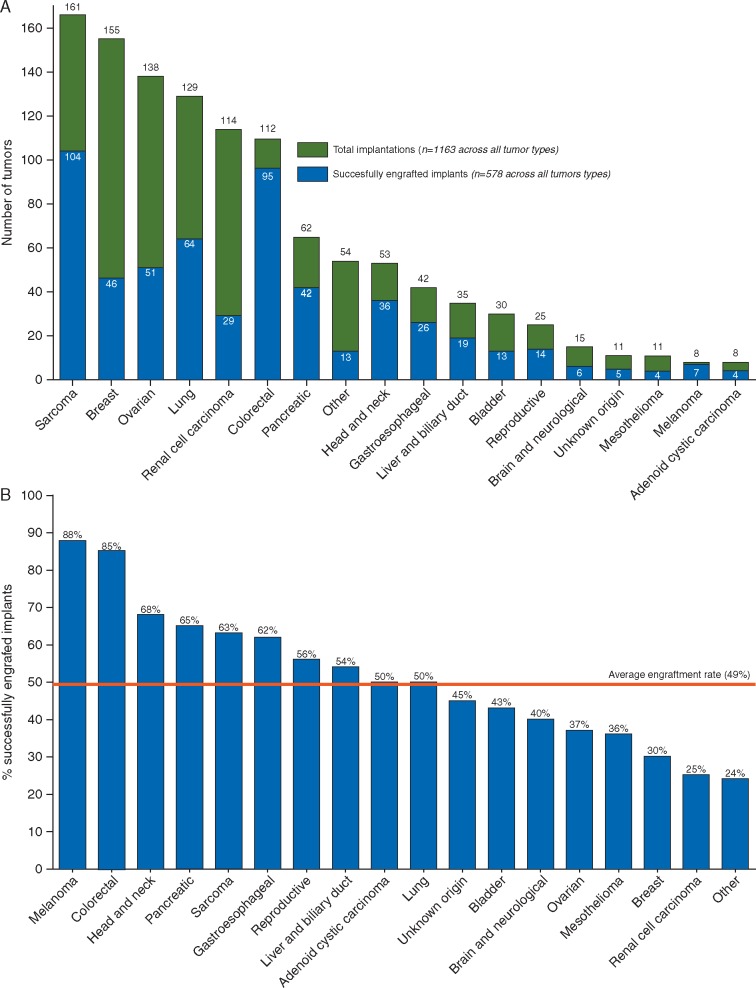
Tumor samples from 1163 patients with variety of advanced solid cancers were implanted into immunodeficient mice. The tumors ranged from common subtypes such as colorectal, lung, and breast, to rarer tumors including adenoid cystic carcinoma and cholangiocarcinoma (Figure 1A). Of these samples, 578 successfully engrafted to generate a PDX model, at an overall engraftment rate of 49% (Figure 1B). Engraftment rates varied within each tumor type, ranging from 85% for colorectal tumors (n = 112) to 30% for breast (n = 155) (Figure 1B). The average time from engraftment to drug exposure across all implanted models was 16 weeks, with substantial variability within each tumor type (supplementary Table S1, available at Annals of Oncology online).
Figure 1.
Engraftment rate by tumor type. (A) Graph depicting different tumor types implanted into immunodeficient mice to establish PDX models. Green bars represent the total number of implantations carried out for a particular tumor type, whereas blue bars depict a number of successfully engrafted implants (generating at least one model in immunodeficient mice). (B) Graph depicts the percent of engraftment for PDX models generated from different tumor types. Red line shows the average engraftment rate across all tumors.
PDXs preserve histopathology and genetic landscape of the parental tumor
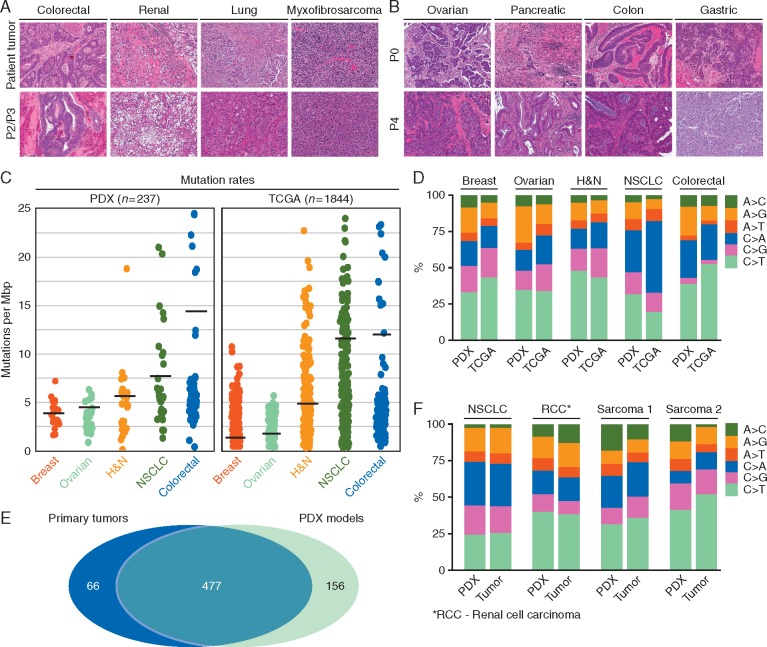
The histology of parental tumors and corresponding PDXs used for the subsequent correlation analysis was compared by an independent pathologist. Primary lesions and matching PDXs from 4 different tumor types are shown in Figure 2A. PDX models faithfully conserved histopathological features of the parental lesions (Figure 2A), and maintained histopathological fidelity over multiple passages (Figure 2B). Out of 578 tumorgrafts, WES data was available for 237 models (109 colorectal, 30 ovarian, 38 lung, 30 head and neck, and 30 breast cancers). This data was compared with equivalent information in the TCGA database (224 colorectal, 316 ovarian, 227 lung, 306 head and neck and 772 breast cancers). Our analysis revealed that the background mutation frequencies in PDXs and primary TCGA tumors were highly comparable (Figure 2C). The slightly higher frequencies for PDX models in some cancers are likely due to the lack of matched normal for analysis. Nonetheless, mutation frequencies of mostly mutated genes in PDXs of different tumor types highly resembled mutation frequencies reported for these genes in TCGA (supplementary Figure S1, available at Annals of Oncology online). Furthermore, the frequencies of specific base-pair substitutions in PDXs and TCGA database showed high proximity, although differences were observed in smoking-related C>A transversions in NSCLC and A>G transitions in general (Figure 2D). Additionally, we compared somatic single nucleotide variants (SNVs) and copy number variations (CNVs) in 4 primary tumors [non-small-cell lung cancer adenocarcinoma (NSCLC), renal cell carcinoma, synovial sarcoma and dedifferentiated liposarcoma] and matched early passage PDX counterparts. We detected 543 and 634 aberrations across 4 primary tumors and 4 PDX models, respectively (supplementary Table S2, available at Annals of Oncology online), with 88% of mutations identified in parental tumors present in the corresponding PDXs (Figure 2E; supplementary Figure S2A and Table S3, available at Annals of Oncology online). Except for A>C transversions, the majority of specific mutation types in tumors and matched PDX models showed tight correlation (Figure 2F). There were no well-characterized drivers among genes mutated in PDXs only; however, mutations in cancer genes were detected in each PDX-primary tumor pair (supplementary Table S4, available at Annals of Oncology online). Notably, while the allelic frequencies of mutations detected in PDXs and tumors were largely comparable (supplementary Figure S2B, available at Annals of Oncology online), a number of subclonal events in lung and renal patients displayed clonal expansion during PDX generation, supporting the notion that engraftment and passaging of primary tumors in the murine host induces dynamic changes in clonal subpopulations [9, 10, 26]. Nevertheless, the overall concordance in the mutational landscape and CNVs (supplementary Figure S2A–C, available at Annals of Oncology online) suggests that clonal composition in the PDX models parallels, to an extent, the genetic heterogeneity seen in the primary lesions.
Figure 2.
PDX models preserve the histopathology and genetic landscape of the parental tumor. (A) Histology (H&E) of PDX models at early passage (2 or 3) and the parental tumor from which they were established. Four primary lesions and matching PDX models from four different tumor types are shown. ×10 magnification. (B) Histology (H&E) of four PDX models at passage 0 (first transplantation into immunodeficient mice) and passage 4. Four PDX models from 4 different tumor types are shown. ×10 magnification. (C) Background mutation rates from WES analysis of 237 early passage PDX models of different tumor types (109 colorectal, 30 ovarian, 38 lung, 30 head and neck and 30 breast cancers) and samples from the TCGA database (224 colorectal, 316 ovarian, 227 lung, 306 head and neck and 772 breast cancers). (D) Spectrum of specific base-pair substitutions in PDX models and TCGA samples across different cancer types analyzed. (E) Venn diagram summarizes single-nucleotide mutations concurrently detected by WES in four primary tumors and their matched early passage PDX counterparts. (F) Spectrum of specific base-pair substitutions in 4 PDX models and parental tumor from which they were established.
Positive clinical outcomes can be reproduced by PDX models
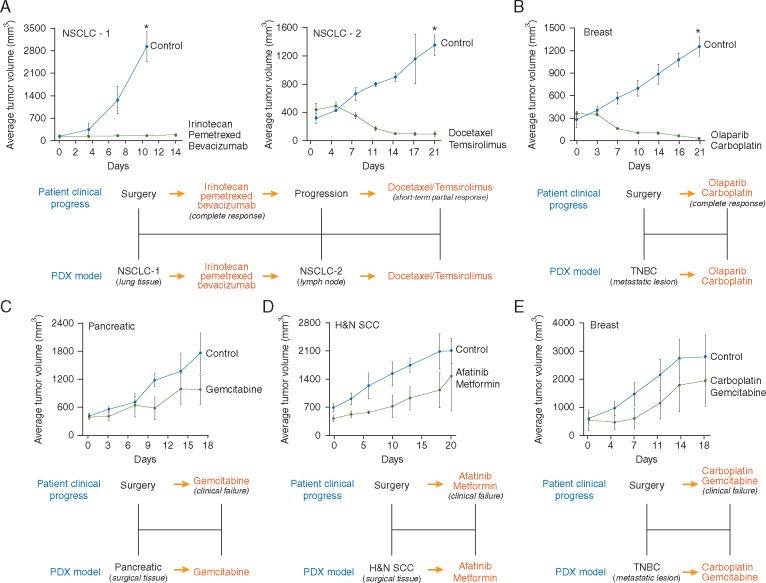
We first tested a number of PDXs against therapies that yielded a positive outcome in the corresponding patient (Figure 3A and B). These and other examples that follow are derived from our clinical correlation dataset (see below). Patient responses to therapy were determined by the treating oncologists using radiological RECIST criteria. PDXs were screened against the same therapy received by the patient, and responses assessed using changes in tumor volume as described in the methods section. A PDX response was designated positive and correlative to the patient response if it showed the RECIST equivalent of complete response (CR), partial response (PR), or stable disease (SD), all of which are generally considered therapeutically beneficial in a clinical setting. As an example, a PDX was established after a lung bilobectomy for a 45-year-old male with a poorly differentiated, mucin-producing NSCLC that had invaded through the interlobular pleura. The patient was treated with irinotecan, pemetrexed, and bevacizumab, showing a durable CR lasting 14 months. In keeping with the patient response, PDX growth was profoundly inhibited by this drug combination (Figure 3A, left panel). This patient eventually progressed and a new PDX was generated from a lymph node metastasis. The patient began treatment with a combination of docetaxel and temsirolimus, demonstrating a short-lasting PR. The new PDX model was tested against the aforementioned treatment and similarly to the patient, responded to the docetaxel/temsirolimus combination (Figure 3A, right panel). Figure 3B shows the PDX results for a 48-year-old female with stage III triple-negative breast cancer (TNBC). A PDX model was developed from a metastatic lesion in the supraclavicular fossa. Based on the presence of a BRCA1 mutation, the patient was treated with carboplatin and olaparib, which resulted in a near CR. After treatment with the same regimen, almost total regression of the engrafted tumor was observed.
Figure 3.
PDX models accurately replicate both positive and negative patient responses. (A, B) PDX models were screened against the corresponding therapies received by the patient. Graphs show the average tumor volume for three to nine animals ± SD. *Treated groups significantly different from untreated controls at the end point of the experiment (Student's t-test; P < 0.05). (C–E) As per (A) and (B). Graphs show the average tumor volume for three to four animals ± SD. No treated group was significantly different from the corresponding untreated control group at the end point of the experiment (Student's t-test; P < 0.05).
Negative clinical outcomes can be reproduced by PDX models
We next screened a number of models developed from tumors failing to respond to treatment against the same therapeutic regimens (Figure 3C and E). A 71-year-old male diagnosed with stage-IV pancreatic ductal adenocarcinoma was treated with gemcitabine and failed to respond, with continued disease progression observed clinically. Gemcitabine was also unable to reduce tumor growth in the PDX established for this patient (Figure 3C). A PDX was established for a 61-year-old female with stage III, heavily pre-treated head and neck squamous cell carcinoma while the patient was placed on a new regimen of the off-label, pan-ERBB inhibitor, afatinib (due to a somatic mutation in the ERBB4 gene), along with metformin. This treatment could not prevent further disease progression and consistent with this outcome, also failed to reduce growth of the implanted tumor (Figure 3D). The final example is a 34-year-old female presenting with stage-IV TNBC. A model was developed from a pulmonary metastasis. During the establishment of the model, the patient was treated with carboplatin/gemcitabine combination, which failed to limit continued disease progression. Similarly, these drugs did not inhibit growth in the PDX (Figure 3E).
PDXs retain therapeutic accuracy over time
Among the challenges facing the implementation of PDXs for pre-clinical evaluation of anticancer agents, is variable engraftment rates and the long time required to establish the model and test for drug sensitivity. Although new models can be generated as cycles of disease progression and therapy occurs, it would be more practical to use models established from early surgical resections for screening drug sensitivity over time. To test whether PDXs developed from early resections retain the ability to replicate treatment outcomes observed for recurrent disease, we screened such models against all therapies employed clinically from disease presentation to subsequent progression. Three examples are presented in Figure 4.
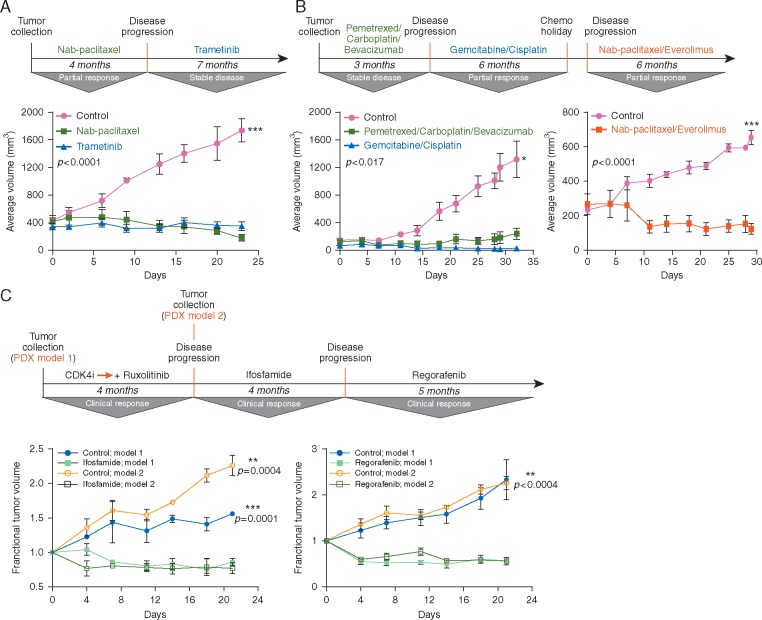
Figure 4.
PDX models retain the capacity to replicate multiple lines of therapy. Fragments of each patient tumor [ovarian for (A), lung for (B), and liposarcoma for (C)] were implanted into immunodeficient mice to establish a PDX model. These models were screened against the therapies that had been used to treat each patient as they cycled through disease stabilization/regression and disease recurrence. Graphs depict the average tumor volume (mm3) for the different treatment groups at each measurement point, with standard deviations plotted (n = 3–6 mice/group). Fractional tumor volume was calculated as the ratio of the tumor volume at time t to the tumor volume at time 0. Treated groups significantly different from untreated controls at the end point of the experiment (Student's t-test; P < 0.05).
Example 1 . A PDX model was established from a hepatic lesion resected from a 71-year-old female with stage-IV endometrioid ovarian cancer with multiple nodules in the liver and lungs (Figure 4A). A number of treatments were subsequently clinically employed, including nab-paclitaxel, yielding a 4-month PR, followed by trametinib (an MEK inhibitor) upon progression, which led to disease stabilization for 7 months. Importantly, each treatment regimen strongly inhibited tumor growth in the PDX model originally generated from the single hepatic lesion upon disease presentation.
Example 2 . A model was developed from a metastatic lesion in the tibia for a 58-year-old male with stage-IV, poorly differentiated NSCLC (Figure 4B). The patient was initially treated with pemetrexed and carboplatin combination, resulting in disease stabilization, followed by slow progression. At the time of progression, the patient started treatment with gemcitabine and cisplatin, leading to a PR lasting 6 months. He was subsequently treated with a combination of nab-paclitaxel and the mTORC1 inhibitor, everolimus, with another PR observed clinically (for 6 months). Similarly, the PDX model first established from the metastatic site in the tibia remained sensitive to all of these agents (Figure 4B).
Example 3 . Since evolutionary clonal dynamics imposed by the selective pressure of anticancer therapies may impact the capacity of PDXs developed early in the disease course to continue replicating patient outcomes as tumors clinically progress, we screened two individual PDX models developed from a 55-year-old male with liposarcoma. The first model was developed before treatment and the second model after therapy for progressive disease (Figure 4C). After the first model was developed, the patient was treated with a CDK4 inhibitor (P1446A-05), which was later supplemented with the JAK2 inhibitor, ruxolitinib. This initially led to a decrease in tumor growth before new lesions appeared. A new PDX model was established from one of these lesions before the patient being treated with ifosfamide, which yielded a PR clinically. At progression, the patient was treated with the multi-kinase inhibitor, regorafenib, with additional tumor regression observed. Most significantly, when each model was screened against ifosfamide and regorafenib, both showed substantial tumor growth inhibition in response to these agents, in keeping with the positive clinical response seen in the patient (Figure 4C). Collectively, these results suggest that PDXs established early in the disease course may retain the capacity to reproduce patient outcomes to therapies used months later, despite probable tumor evolution during treatment.
PDXs are accurate and clinically relevant tumor models
Although our observations further support the potential of this technology for guiding therapeutic decisions [8, 14, 16], to the best of our knowledge, no studies have attempted to calculate specific performance metrics that define how applicable PDXs are to clinical decision-making. We obtained 129 clinical correlations across 92 patients with advanced solid tumors for whom a PDX was successfully developed and drug testing completed (supplementary Table S5, available at Annals of Oncology online). Out of 129 correlations described in this manuscript 94 are new and have not been previously reported. Parallel responses in patients and mice for each one of the 129 correlations shown in supplementary Table S6, available at Annals of Oncology online. Patient treatment response was provided by the consulting oncologist. To assess drug responses in mice, we calculated changes in tumor volume over the course of treatment by using percent tumor regression (%TR), and then converted %TR values to a clinical outcome equivalent RECIST criteria [27]. Drug dosing and schedules employed for PDX screening of patient therapies are listed in supplementary Table S7, available at Annals of Oncology online. For a number of reasons highlighted in supplementary Table S8, available at Annals of Oncology online, we were unable to obtain clinical correlations for the remaining 486 patients who had a tumor successfully implanted.
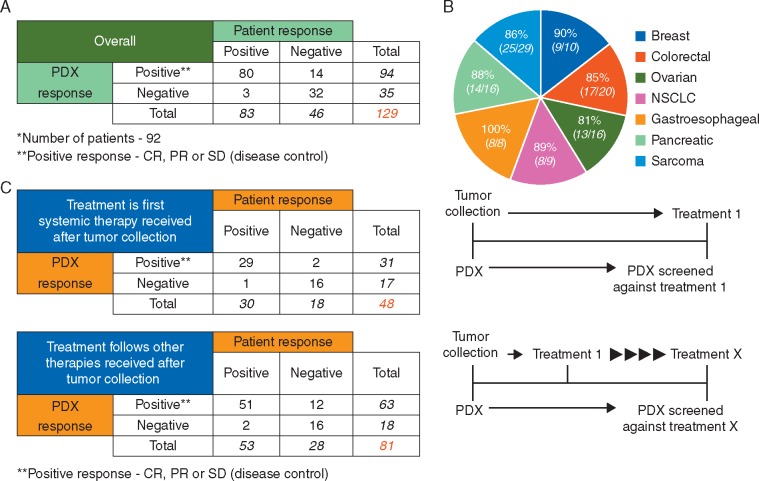
PDXs that demonstrated RECIST values equivalent to CR, PR, or SD were designated as positive for drug response. A significant association was observed between drug responses in patient and corresponding PDXs in 87% (112/129) of the therapeutic outcomes (Figure 5A, Fisher's exact test; P = 5.06× 10−16, supplementary Table S6, available at Annals of Oncology online). Our analysis revealed a sensitivity for the PDX drug screens of 96%, with a lower 95% confidence interval (CI) of 89% (which meets the acceptance criteria of 80%), and specificity of 70%, with a lower 95% CI of 54% (Table 1, top panel). This resulted in positive and negative predictive values (PPVs and NPVs) of 85% and 91%, respectively. Notably, the percentage of correlative therapeutic responses did not vary significantly among different cancer types (Figure 5B). If the response was considered to correlate only if the RECIST equivalent in xenografts was exactly concordant with RECIST-defined clinical responses in patients, a significant association was still observed in 91 of 129 (71%) therapeutic tests (supplementary Figure S3 and Table S6, available at Annals of Oncology online). To exclude the possibility that PDXs are generally sensitive/insensitive to treatment, we have collected data from a large number of PDX models treated with several standard-of-care regimens (supplementary Table S9, available at Annals of Oncology online). A wide range of responses to each therapy suggests that drug sensitivity is not a general feature, but is intrinsic and specific to the individual model. A range of responses to different therapies within individual models (supplementary Table S10, available at Annals of Oncology online), further supports this suggestion.
Figure 5.
PDX drug responses correlate with patient outcomes. (A) Contingency table for correlations between 129 drug responses across 92 PDX models and clinical outcomes seen in patient from whom they were established. **SD, PR and CR were all considered positive test results (disease control) whereas PD was considered a negative test result. Positive and negative PDX responses were defined from measurements of changes in tumor volume and based on RECIST criteria. Positive and negative patient response information was provided by the consulting oncologist. (B) Associations between drug responses in patients and the corresponding PDXs for seven different types of cancer where more than eight patients were available. (C) Contingency tables for correlations between drug responses in PDXs and matched patients when results were stratified for screens against treatments used immediately following surgery (top) or if additional treatments had been employed before the one against which the PDX was screened (bottom). **SD, PR and CR were all considered positive test results (disease control) whereas PD was considered a negative test result. PDX responses were defined from measurements of changes in tumor volume and based on RECIST criteria. Patient responses were provided by the consulting oncologist.
Table 1.
Values and confidence intervals for parameters of analytical and clinical accuracy for PDX model drug screening
| Measurer | Result | 95% CI | |
|---|---|---|---|
| Overall | Sensitivity | 96% (80/83) | 89%–99% |
| Specificity | 70% (32/46) | 54%–82 | |
| PPV | 85% (80/94) | 76%–91% | |
| NPV | 91% (32/35) | 76%–98% | |
| Treatment is first therapy received after tumor collection | Sensitivity | 97% (29/30) | 81%–99% |
| Specificity | 89% (16/18) | 64%–98% | |
| PPV | 94% (29/31) | 77%–99% | |
| NPV | 94% (16/17) | 69%–99% | |
| Treatment follows other therapies received after tumor collection | Sensitivity | 96% (51/53) | 86%–99% |
| Specificity | 57% (16/28) | 37%–75 | |
| PPV | 81% (51/63) | 69%–89% | |
| NPV | 89% (16/18) | 64%–98% |
Values for sensitivity, specificity, and positive and negative predictive values were calculated using the online tool: http://vassarstats.net/. The top table shows parameters calculated from all 129 correlations in our test population, whilst the middle and bottom tables show the parameter values when correlations were stratified for screens against treatments used immediately following surgery (middle) or if additional treatments had been employed before the one against which the PDX was screened (bottom).
CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value.
To evaluate if the diagnostic performance of the models changes as patients progress through multiple lines of therapy, we stratified our correlation data according to whether or not the patient treatment was the first received after resection of the tumor to develop the PDX. There was essentially no change in sensitivity and a modest decrease in the PPV (from 94% to 81%) and NPV (from 94% to 89%) if the tested therapy was not the first received after resection (Figure 5C, Table 1, middle and bottom panels). Notably, the decrease in specificity and the PPV was primarily due to twelve patients whose PDX models showed tumor regression in response to treatment yet who failed to respond to that therapy clinically. Nevertheless, these results highlight that most drug responses in PDXs accurately replicate patients’ clinical outcomes, even as patients continue to undergo several cycles of therapy overtime.
Discussion
The poor success in oncology drug development is due in part to the inability to tailor therapy to a group of patients that are more likely to benefit from the treatment. PDXs maintain the histopathological and molecular features of human tumors [5–8, 23], and offer a potential solution. Cross-analysis of mice harboring xenografts from the same tumor enables evaluation of numerous therapies simultaneously, including those that might not otherwise be indicated for a particular tumor type, and may also help avoid exposure to toxicities associated with therapies that provide no benefit.
A number of retrospective studies have reported a correlation between drug responses in PDXs and patients’ clinical outcomes [11–17, 20]. Furthermore, remarkable similarity was seen between response rates to standard-of-care drugs in PDXs and respective clinical trials [13, 18–22]. Lately, small prospective studies have demonstrated that PDXs may replicate patient outcomes across multiple solid tumors and different treatments [5–8, 23, 24]. A notable example that PDXs may offer a preclinical tool for drug-development is a recent pilot clinical study in which refractory advanced cancers were propagated in immunodeficient mice and treated with 63 drugs across 232 treatment regimens. Eleven of 13 patients benefited from PDX-directed therapies, including patients placed on treatments not typically recommended in clinical practice [5]. Although promising, these studies have focused on specific tumor types or drugs/drug combinations and the ability to conduct statistically appropriate correlation analyses was limited by the small number of cases evaluated. A recent invivo drug screen expanded such observations to a greater number of human cancers and demonstrated that PDXs with a diverse set of driver mutations may reproduce treatment responses known from previous clinical trials [25]. However, a precise evaluation of how accurately patient responses correlate with matching PDXs in a large, heterogeneous population and real treatment settings is important for assessing the utility of this platform.
In order for PDXs to be considered a reliable preclinical tool for drug testing, it is critical that tumorgrafts retain the genetic changes found in primary cancers. Here we demonstrate that genomic alterations identified via WES analyses of PDX models and matched parental tumors were nearly identical. Moreover, sequencing of 237 early-passage PDXs established from patients representing five major solid tumor types showed remarkable mutational fidelity to TCGA database. Our dataset represents one of the largest genomic landscape analysis of PDX models to date, and further supports the notion that PDXs retain driver mutations and molecular characteristics of primary tumors [25], even across serial passaging [9, 10].
Advances in high-throughput genomic technologies allow characterization of the cancer genome in a time frame compatible with treatment decisions. The PDX platform can be used to test different empirical treatment strategies potentially targeting the genomic aberrations that drive tumor behavior, offering the unique opportunity to increase clinical benefit [6]. However, predicting treatment response to known oncogenic pathways is still not straightforward, and future studies are warranted to determine the feasibility of this approach.
We next screened PDXs established for 92 patients with various solid tumors against the same treatments that were administered clinically and correlated patient outcomes with the responses in corresponding models. Percent tumor growth inhibition (%TGI) (which is commonly used to assess therapeutic efficacy in PDXs) is calculated relative to the untreated control group. This approach is quite different from the criteria used to evaluate response rates in the clinical setting, where control is not available, and may therefore lead to an overestimation in some cases. To more adequately assess antitumor drug efficacy in mice, we have calculated control-independent %TR rate for each regimen tested, and then converted %TR values to clinical outcomes equivalent according to RECIST criteria [27].
Positive and negative predictive values provide a quantitative way to assess whether a specific outcome in PDXs is likely to yield the same outcome in the patient. Based on our data, positive and negative predictive values calculated for PDX screenings were of a magnitude rarely observed with any other type of in vitro testing. Our study highlights the utility of this platform for drug screening in co-clinical trials, where drug studies in PDXs and patients are evaluated in parallel [20, 28, 29], allowing rapid integration of information from PDX experiments to human trials. These types of studies will be particularly important for identifying biomarkers allowing response prediction in patient subpopulations using the established PDXs. Supporting this rationale, a growing number of studies have demonstrated the value of PDXs for discovering prospective biomarkers that warrant clinical validation [8, 13, 18, 24, 25]. Furthermore, PDXs or PDX-derived short term exvivo cultures [10] can continue to predict responses in pre-clinical drug screening long after a trial is completed, even if the patient has succumbed to disease.
Although most of the PDX drug responses reliably correlated with patient outcomes, in three cases, the patient’s tumor responded to treatment while the PDX was resistant (false negatives). False negative responses may occur due to the differences in absorption, distribution and pharmacokinetics of antitumor agents between mice and humans [30, 31]. Mice may eliminate certain compounds faster, causing underestimation of their clinical efficacy. Future trials designed to determine the differences in pharmacology of cancer therapeutics between mouse and human may help to better forecast whether drug exposures needed for assessment of preclinical efficacy could be achieved in mice. Additionally, we observed fourteen cases where the patient’s tumor did not respond to treatment, while the corresponding PDX was sensitive (false positives). This typically occurred when the duration between PDX generation and drug testing was long and patients received additional therapy before the treatment against which the model was screened. Although the basis of drug-induced clonal dynamics and the speed with which it occurs is not yet understood, it is possible that administration of other drugs allowed the outgrowth of tumor cell sub-populations that were intrinsically more resistant to the new treatment, whereas the model established from the original tumor remained sensitive. Nevertheless, although PDXs may be unable to provide treatment-guidance for systemic therapy administered immediately following surgery, we showed only a modest decrease in predictive value and retention of high accuracy even if the patient underwent subsequent treatments after resection. Moreover, we have shown that PDXs retain the capacity to identify viable treatments for recurrent disease appearing months after the initial presentation, making it interesting to speculate that these models may be useful in helping manage cancer as a chronic disease in patients with long or recurring responses.
Although our data support PDXs as a valuable modality for translational research, there are a number of challenges limiting their broad application. One of the concerns is that establishing PDXs requires large quantities of fresh tumor tissue, which sometimes may not be available. To address this challenge, we have conducted a number of engraftments using tissue originating from biopsies (supplementary Figure S4, available at Annals of Oncology online). While our data suggests that biopsies have less variance in growth rates, which may be attributed to lower heterogeneity, the engraftment rate, biological features and clinical parameters of models established from biopsies were comparable to surgical explants. However, this observation may reflect the more aggressive, late-stage tumors examined in this study, and it remains to be determined whether small amounts of tissue from early-stage patients engraft and grow as readily. As molecular analysis of tumors becomes standard, the amount of tumor material available for implantation decreases. While a number of promising approaches [32] have enhanced the engraftment rate in mice, the ability of tumors to engraft is not universal and PDX cannot be generated for every patient [33].
It is also apparent that use of PDXs as a standard modality for modeling human cancer has additional limitations, such as loss of tumor microenvironment and immune-response [32, 34], selection for clonal subpopulations different from the original tumor [9, 26], differences in drug metabolism [30, 31] and cost-effectiveness [33]. Although at the present time, tumorgrafts are predominantly pharmaceutical research tools, with the dismal response rates observed with repeated lines of traditional chemotherapy, the value of this tool in addressing the continuing challenges in clinical oncology will likely increase over time.
Supplementary Material
Acknowledgements
We are grateful to all the patients and oncologists who participated in this study.
Funding
National Institutes of Health (P50DE019032 and R01CA163594).
Disclosure
DS is the chairman of the board of directors of Champions Oncology. The terms of this arrangement are being managed by Johns Hopkins University in accordance with its conflict of interest policies. KP, AK, DV, DC, IS, IBZ, AD and RM are current or former employees of Champions Oncology. MH is a founder and stock share holder of Champions Oncology. All remaining authors have declared no conflicts of interest.
References
- 1. Denison TA, Bae YH.. Tumor heterogeneity and its implication for drug delivery. J Control Release 2012; 164: 187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Simon R. Clinical trial designs for evaluating the medical utility of prognostic and predictive biomarkers in oncology. Per Med 2010; 7: 33–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jonkers J, Berns A.. Conditional mouse models of sporadic cancer. Nat Rev Cancer 2002; 2: 251–265. [DOI] [PubMed] [Google Scholar]
- 4. Huse JT, Holland EC.. Genetically engineered mouse models of brain cancer and the promise of preclinical testing. Brain Pathol 2009; 19: 132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hidalgo M, Bruckheimer E, Rajeshkumar NV. et al. A pilot clinical study of treatment guided by personalized tumorgrafts in patients with advanced cancer. Mol Cancer Ther 2011; 10: 1311–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garralda E, Paz K, Lopez-Casas PP. et al. Integrated next generation sequencing and avatar mouse models for personalized cancer treatment. Clin Cancer Res 2014; 9: 2476–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Villarroel MC, Rajeshkumar NV, Garrido-Laguna I. et al. Personalizing cancer treatment in the age of global genomic analyses: PALB2 gene mutations and the response to DNA damaging agents in pancreatic cancer. Mol Cancer Ther 2011; 10: 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stebbing J, Paz K, Schwartz GK. et al. Patient-derived xenografts for individualized care in advanced sarcoma. Cancer 2014; 120: 2006–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eirew P, Steif A, Khattra J. et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 2015; 518: 422–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bruna A, Rueda OM, Greenwood W. et al. A biobank of breast cancer explants with preserved intra-tumor heterogeneity to screen anticancer compounds. Cell 2016; 167: 260–274.e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weroha SJ, Becker MA, Enderica-Gonzalez S. et al. Tumorgrafts as in vivo surrogates for women with ovarian cancer. Clin Cancer Res 2014; 5: 1288–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fichtner I, Slisow W, Gill J. et al. Anticancer drug response and expression of molecular markers in early-passage xenotransplanted colon carcinomas. Eur J Cancer 2004; 40: 298–307. [DOI] [PubMed] [Google Scholar]
- 13. Garrido-Laguna I, Uson M, Rajeshkumar NV. et al. Tumor engraftment in nude mice and enrichment in stroma- related gene pathways predict poor survival and resistance to gemcitabine in patients with pancreatic cancer. Clin Cancer Res 2011; 17: 5793–5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dong X, Guan J, English JC. et al. Patient-derived first generation xenografts of non-small cell lung cancers: promising tools for predicting drug responses for personalized chemotherapy. Clin Cancer Res 2010; 16: 1442–1451. [DOI] [PubMed] [Google Scholar]
- 15. Stewart EL, Mascaux C, Pham NA. et al. Clinical utility of patient-derived xenografts to determine biomarkers of prognosis and Map resistance pathways in EGFR-mutant lung adenocarcinoma. J Cln Oncol 2015; 33: 2472–2480. [DOI] [PubMed] [Google Scholar]
- 16. Zhang X, Claerhout S, Prat A. et al. A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer Res 2013; 73: 4885–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marangoni E, Vincent-Salomon A, Auger N. et al. A new model of patient tumor-derived breast cancer xenografts for preclinical assays. Clin Cancer Res 2007; 13: 3989–3998. [DOI] [PubMed] [Google Scholar]
- 18. Bertotti A, Migliardi G, Galimi F. et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov 2011; 1: 508–523. [DOI] [PubMed] [Google Scholar]
- 19. Julien S, Merino-Trigo A, Lacroix L. et al. Characterization of a large panel of patient-derived tumor xenografts representing the clinical heterogeneity of human colorectal cancer. Clin Cancer Res 2012; 18: 5314–5328. [DOI] [PubMed] [Google Scholar]
- 20. Hidalgo M, Amant F, Biankin AV. et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov 2014; 4: 998–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Migliardi G, Sassi F, Torti D. et al. Inhibition of MEK and PI3K/mTOR suppresses tumor growth but does not cause tumor regression in patient-derived xenografts of RAS-mutant colorectal carcinomas. Clin Cancer Res 2012; 18: 2515–2525. [DOI] [PubMed] [Google Scholar]
- 22. Keysar SB, Astling DP, Anderson RT. et al. A patient tumor transplant model of squamous cell cancer identifies PI3K inhibitors as candidate therapeutics in defined molecular bins. Mol Oncol 2013; 7: 776–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morelli MP, Calvo E, Ordonez E. et al. Prioritizing phase I treatment options through preclinical testing on personalized tumorgraft. J Clin Oncol 2012; 30: e45–e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jimeno A, Tan AC, Coffa J. et al. Coordinated epidermal growth factor receptor pathway gene overexpression predicts epidermal growth factor receptor inhibitor sensitivity in pancreatic cancer. Cancer Res 2008; 68: 2841–2849. [DOI] [PubMed] [Google Scholar]
- 25. Gao H, Korn JM, Ferretti S. et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med 2015; 21: 1318–1325. [DOI] [PubMed] [Google Scholar]
- 26. Kreso A, O'Brien CA, van Galen P. et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science 2013; 339: 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Eisenhauer EA, Therasse P, Bogaerts J. et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009; 45: 228–247. [DOI] [PubMed] [Google Scholar]
- 28. Garrido-Laguna I, Tan AC, Uson M. et al. Integrated preclinical and clinical development of mTOR inhibitors in pancreatic cancer. Br J Cancer 2010; 103: 649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Von Hoff DD, Ramanathan RK, Borad MJ. et al. Gemcitabine plus nab-paclitaxel is an active regimen in patients with advanced pancreatic cancer: a phase I/II trial. J Clin Oncol 2011; 29: 4548–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peterson JK, Houghton PJ.. Integrating pharmacology and in vivo cancer models in preclinical and clinical drug development. Eur J Cancer 2004; 40: 837–844. [DOI] [PubMed] [Google Scholar]
- 31. Thompson J, Stewart CF, Houghton PJ.. Animal models for studying the action of topoisomerase I targeted drugs. Biochim Biophys Acta 1998; 1400: 301–319. [DOI] [PubMed] [Google Scholar]
- 32. Izumchenko E, Meir J, Bedi A. et al. Patient derived xenografts as tools in pharmaceutical development. Clin Pharmacol Ther 2016; 6: 612–621. [DOI] [PubMed] [Google Scholar]
- 33. Aparicio S, Hidalgo M, Kung AL.. Examining the utility of patient-derived xenograft mouse models. Nat Rev Cancer 2015; 15: 311–316. [DOI] [PubMed] [Google Scholar]
- 34. Martinez-Garcia R, Juan D, Rausell A. et al. Transcriptional dissection of pancreatic tumors engrafted in mice. Genome Med 2014; 6: 27.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.