ABSTRACT
Characterisation of the Arabidopsis RNA polymerase II (RNAPII) elongation complex revealed an assembly of a conserved set of transcript elongation factors associated with chromatin remodellers, histone modifiers as well as with various pre-mRNA splicing and polyadenylation factors. Therefore, transcribing RNAPII streamlines the processes of mRNA synthesis and processing in plants.
Keywords: chromatin, polyadenylation, RNA polymerase II, splicing, transcript elongation
Introduction
In addition to controlling the initiation step, transcription by RNA polymerase II (RNAPII) is dynamically regulated during the elongation phase of transcript synthesis by various mechanisms. Accordingly, a range of so-called transcript elongation factors (TEFs) were identified that modulate the transcription process after initiation. Consequently, TEFs typically are detected along the transcribed part of active genes. The heterogeneous TEFs can be divided into functional groups including factors that (i) modulate the catalytic activity of RNAPII, (ii) facilitate progression of the enzyme through repressive chromatin or (iii) covalently modify histones of nucleosomes situated in the transcribed region of genes.1,2 The requirement for TEF activities very much depends on the context and likely reflects the variety of challenges that elongating RNAPII encounters. In many cases in the absence of a certain TEF only a relatively small subset of genes is differentially expressed, while the majority of genes is ordinarily transcribed. In line with that, mutations in genes encoding TEFs commonly cause rather distinct developmental phenotypes.3,4
Isolation of various affinity-tagged TEFs from Saccharomyces cerevisiae cells demonstrated that several TEFs co-purified with each other and with RNAPII.5,6 This set of TEFs comprised SPT4/SPT5 and TFIIS which can modulate RNAPII activity, the histone chaperones FACT and SPT6, the chromatin remodeller CHD1, PAF1C which can promote transcription-related histone modifications, and the SPT6-interactor IWS1. Therefore, these factors may associate with the twelve-subunit RNAPII to form the yeast elongation complex. Importantly, the heptapeptide repeats of the carboxy-terminal domain (CTD) of the largest subunit (NRPB1) of RNAPII are differentially modified during the transcription cycle, for instance, with Ser5 and Ser2 phosphorylation marking early and later transcript elongation stages, respectively.7,8 Moreover, the RNAPII-CTD modifications are critical for the coordination of ongoing transcription and co-transcriptional pre-mRNA processing events.
The Arabidopsis RNAPII transcript elongation complex (TEC)
Relatively little is known about the RNAPII elongation complex in plants. Compared to the yeast model, the situation in plants is complicated by the fact that several genes encoding TEFs are duplicated. The Arabidopsis genome, for instance, encodes alternative versions of SPT4, SPT5 and SPT6 that may be differentially expressed and/or could serve (partially) distinct functions.9,10 A recent approach expressing tagged versions of various TEFs in Arabidopsis cells followed by affinity purification and mass spectrometry identified a network of interactions between different TEFs and RNAPII.11 As exemplified by TFIIS and the PAF1C subunit ELF7 (orthologue of yeast PAF1), the Ser2 phosphorylated form of elongating RNAPII is enriched in the TEF eluates relative to the non-phosphorylated RNAPII. Like in yeast,5,6 FACT, SPT6, TFIIS, SPT4/SPT5 and the six-subunit PAF1C co-purified with each other and with RNAPII, whereas P-TEFb was not among the interactors. Therefore, the core plant RNAPII elongation complex appears to resemble that of yeast cells (Fig. 1). In addition, various ATP-dependent chromatin remodelling factors,12 NAP1 histone chaperones13 and several enzymes involved in histone acetylation including Elongator14 repeatedly co-purified with the Arabidopsis TEFs.11 Remodelling factors may support elongation by facilitating the passage of RNAPII through nucleosomes.15 NAP1 occurs in four versions in Arabidopsis13 and three of them repeatedly co-eluted with the TEFs.11 Therefore, perhaps in collaboration with the FACT and SPT6 histone chaperones NAP1 proteins could promote chromatin transcription.13 Histone acetyltransferases (HATs, i.e. Elongator, SWR1/NuA4) and histone deacetylases (HDACs) co-purified repeatedly with Arabidopsis TEFs, while enzymes involved in histone methylation and ubiquitination were hardly detected. Whereas Elongator and different HDACs co-eluted with various TEFs, SWR1/NuA4 rather specifically was detected in the affinity purification of the P-TEFb subunit CDKC;2.11 The HATs and HDACs may control dynamic histone acetylation within gene bodies to modulate the efficiency of transcript elongation, but generally HDACs (in collaboration with chromatin remodelling enzymes) maintain a low acetylation level to counteract deleterious, cryptic transcription within transcribed regions.16,17 Interestingly, in contrast to studies in yeast,5,6 the SPT6-interactor IWS1 was not found associated with Arabidopsis TEFs, although both SPT6 and SPT6L were robustly identified in the affinity purifications of various TEFs.11 IWS1 interacts directly with the N-terminal region of SPT6,18 and it was reported to regulate transcription in Arabidopsis.19,20 Therefore, it remains unclear whether IWS1 is a component of the plant RNAPII elongation complex.
Figure 1.
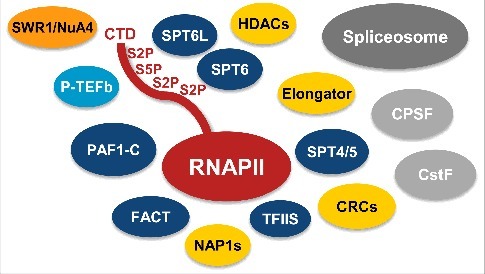
Scheme depicting the Arabidopsis RNAPII elongation complex based on affinity purification of TEFs in combination with mass spectrometry analyses.11 The TEFs FACT, TFIIS, SPT4/SPT5, SPT6, SPT6L and PAF1-C (dark blue) robustly co-purified with each other and with RNAPII (red), while P-TEFb (light blue) was not enriched in these experiments. Moreover, further chromatin factors (yellow) also repeatedly co-purified with the TEFs, except for SWR1/NuA4 (orange), which was primarily isolated along with P-TEFb. In addition to the transcription related proteins, many spliceosomal components (dark grey) and polyadenylation factors (light grey) co-purified with the TEFs.
Mutant plants defective in TEFs are phenotypically affected to very different extents. Thus, plants lacking TFIIS or the PAF1C subunit CDC73 have essentially wild type appearance,21-23 while plants lacking the PAF1C subunits ELF7, ELF8 are more severely affected24,25 and the loss of SPT5-2, SPT6L or of the FACT subunit SSRP1 is lethal.9,10,26 The analysis of double-mutant plants defective in different combinations of TEFs revealed various genetic interactions between genes encoding TEFs.11 In line with the physical interaction of TFIIS and PAF1C in yeast and mammals27,28 and synthetic growth defects of yeast cells lacking TFIIS and PAF1,29 plants deficient in TFIIS and ELF7 exhibit (relative to the parental lines) severe synergistic defects, for instance, regarding plant size (Fig. 2) and the leaf vein patterning.11 Both FACT and TFIIS can facilitate RNAPII transcription through nucleosomes,30,31 and the analysis of double-mutant plants lacking TFIIS in combination with expressing reduced levels of the FACT subunits SSRP1 or SPT16 indicated that the genes encoding the FACT subunits are epistatic to TFIIS regarding bolting time and seed set.11 Therefore, Arabidopsis double-mutants may prove a valuable tool for further studies addressing the functional interplay of TEFs in a higher eukaryote model, supplementing biochemical interaction studies.
Figure 2.

tfIIs elf7 double-mutant Arabidopsis plants in comparison to the respective single-mutants and the Col-0 wild type. While tfIIs has essentially wild type appearance and elf7 is relatively mildly affected, the growth of tfIIs elf7 double-mutant plants is severely reduced, which is obvious from the rosette diameter. Plants at 28 days after stratification grown under long-day conditions are shown.
Cooperation of RNAPII-TEFs with other RNA polymerases?
TEFs were originally characterised as factors that regulate RNAPII-mediated transcription after the initiation stage.1,2 However, several studies (primarily in yeast) suggested that RNAPII-related TEFs cooperate also with RNAPI (and RNAPIII).32 The affinity purification of various TEFs from Arabidopsis cells did not yield conclusive evidence for the association of the TEFs with RNA polymerases other than RNAPII,11 which could be due to structural differences between RNA polymerases as well as the decoration with polymerase-specific assistant factors.33 Moreover, according to immunofluorescence analyses several TEFs including SPT5, SPT6 and ELF7 are excluded from the nucleoli of Arabidopsis nuclei,9,11 arguing against an involvement in RNAPI transcription. In line with that in various Arabidopsis mutants defective in TEFs the levels of the 35S pre-rRNA were unaffected and chromatin immunoprecipitation experiments revealed that the analysed TEFs associated with RNAPII-transcribed genes, but not with sites transcribed by RNAPI and RNAPIII.11 These findings suggest that in Arabidopsis the RNAPII-TEFs are not involved in RNAPI/III-mediated transcription. Interestingly, the plant-specific protein SPT5L and AGO4 that are components of the RNAPV-mediated RNA-directed DNA methylation (RdDM)34,35 robustly co-purified with SPT4, but not with the other analysed TEFs.9,11 SPT4 directly binds SPT5L9 and it can modulate SPT5L-specific RdDM.36 Therefore, Arabidopsis SPT4 occurs in SPT4/SPT5 (which is conserved among eukaryotes) regulating RNAPII transcript elongation and in SPT4/SPT5L modulating RNAPV-mediated transcriptional silencing.
Association of mRNA processing factors with the RNAPII TEC
It is well established in yeast and metazoa that the RNAPII TEC is pivotal in coordinating synthesis and co-transcriptional processing of pre-mRNAs by 5´ capping, splicing and 3´ end processing.37 In plants, relatively little is known about the functional coupling of ongoing transcription and pre-mRNA processing. Accordingly, it was of great interest that many spliceosomal components38 were identified that co-purified with TEFs (i.e. SPT4, SPT16, ELF7) from Arabidopsis cells. These splicing factors included multiple constituents of U1, U2, U5, Sm and NTC sub-complexes,11 suggesting that spliceosomes of various assembly stages associate with the TEC. In addition, several components of the CPSF polyadenylation complex39 co-eluted with SPT4 and ELF7.11 In a reverse experiment, various polyadenylation and splicing factors co-purified with the CstF77 subunit of the CstF polyadenylation complex,11 illustrating the interplay of polyadenylation and splicing factors that recently was also reported in plants.40 Interestingly, several Arabidopsis PAF1C subunits co-purified with the CstF complex.11 In line with that human PAF1C can recruit CstF to transcribed genes41 and PAF1C modulates mRNA 3´end processing.42 Transcript elongation can modulate the exact outcome of alternative splicing and polyadenylation events. Thereby the RNAPII elongation rate may influence the selection of splice/polyadenylation sites and/or the recruitment of splicing/polyadenylation factors to the transcription machinery couples mRNA synthesis and processing.43,44 According to a recent pilot study in Arabidopsis, similar mechanisms appear to be relevant also for plant gene expression,45 but future work will show to which extent plant transcriptome diversity is influenced by transcript elongation, which may depend on genome complexity. Surprisingly, the targeted proteomics approaches with several Arabidopsis TEFs11 did not provide evidence for an interaction of the RNAPII TEC with the 5´ mRNA capping machinery and the THO/TREX complex that plays a central role in linking ongoing transcription with the different mRNA processing events and mRNA export. Consistently, a study addressing protein interactions of the Arabidopsis THO/TREX complex also did not reveal a clear association with the RNAPII TEC.46 In other systems, both the capping enzymes47 and THO/TREX48 associate co-transcriptionally with the TEC. However, in order to favour the detection of protein interactions rather than (indirect) interactions mediated by nucleic acids, the Arabidopsis protein extracts used for the isolation of TEC and THO/TREX were extensively treated with the nuclease benzonase.11,46 Since efficient association of the capping complex49 and of THO/TREX48 with the TEC in addition to protein interactions requires RNA, it is possible that degradation of RNAs resulted in dissociation of these complexes from the TEC, which needs further clarification. In conclusion, the biochemical characterisation of the Arabidopsis RNAPII TEC in combination with the initial analysis of double-mutant plants defective in TEFs has demonstrated that transcribing RNAPII serves not only as a platform for interactions between TEFs, but also as a centre coordinating transcript synthesis and processing. Studying the Arabidopsis model in this context has the potential to extend the findings obtained with yeast and metazoa particularly regarding developmental aspects, as well as to discover plant peculiarities of which to date presumably only few have been unveiled.
Funding Statement
KDG acknowledges funding by the German Research Foundation (DFG) through grants SFB960/A6 and Gr1159/14-1.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Alex Pfab and Wojtek Antosz for the image documenting Arabidopsis mutant phenotypes and for critically reading the manuscript.
References
- [1].Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16:167-77. http://doi.org/10.1038/nrm3953. PMID:25693130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sims RJ, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and the long of it. Genes Dev. 2004;18:2437-68. http://doi.org/10.1101/gad.1235904. PMID:15489290 [DOI] [PubMed] [Google Scholar]
- [3].Smith E, Shilatifard A. Transcriptional elongation checkpoint control in development and disease. Genes Dev. 2013;27:1079-88. http://doi.org/10.1101/gad.215137.113. PMID:23699407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Van Lijsebettens M, Grasser KD. Transcript elongation factors: shaping transcriptomes after transcript initiation. Trends Plant Sci. 2014;19:717-26. http://doi.org/10.1016/j.tplants.2014.07.002. PMID:25131948 [DOI] [PubMed] [Google Scholar]
- [5].Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol. 2002;22:6979-92. http://doi.org/10.1128/MCB.22.20.6979-6992.2002. PMID:12242279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lindstrom DL, Squazzo SL, Muster N, Burckin TA, Wachter KC, Emigh CA, McCleery JA, Yates JR III, Hartzog GA. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol Cell Biol. 2003;23:1368-78. http://doi.org/10.1128/MCB.23.4.1368-1378.2003. PMID:12556496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hajheidari M, Koncz C, Eick D. Emerging roles for RNA polymerase II CTD in Arabidopsis. Trends Plant Sci. 2013;18:633-43. http://doi.org/10.1016/j.tplants.2013.07.001. PMID:23910452 [DOI] [PubMed] [Google Scholar]
- [8].Jeronimo C, Collin P, Robert F. The RNA polymerase II CTD: the increasing complexity of a low-complexity protein domain. J Mol Biol. 2016;428:2607-22. http://doi.org/10.1016/j.jmb.2016.02.006. PMID:26876604 [DOI] [PubMed] [Google Scholar]
- [9].Dürr J, Lolas IB, Sørensen BB, Schubert V, Houben A, Melzer M, Deutzmann R, Grasser M, Grasser KD. The transcript elongation factor SPT4/SPT5 is involved in auxin-related gene expression in Arabidopsis. Nucleic Acids Res. 2014;42:4332-47. http://doi.org/10.1093/nar/gku096. PMID:24497194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gu XL, Wang H, Huang H, Cui XF. SPT6L encoding a putative WG/GW-repeat protein regulates apical-basal polarity of embryo in Arabidopsis. Mol Plant. 2012;5:249-59. http://doi.org/10.1093/mp/ssr073. PMID:21948524 [DOI] [PubMed] [Google Scholar]
- [11].Antosz W, Pfab A, Ehrnsberger HF, Holzinger P, Köllen K, Mortensen SA, Bruckmann A, Schubert T, Längst G, Griesenbeck J, et al.. Composition of the Arabidopsis RNA polymerase II transcript elongation complex reveals the interplay between elongation and mRNA processsing factors. Plant Cell. 2017;29:854-70. http://doi.org/10.1105/tpc.16.00735. PMID:28351991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gentry M, Hennig L. Remodelling chromatin to shape development of plants. Exp Cell Res. 2014;321:40-46. http://doi.org/10.1016/j.yexcr.2013.11.010. PMID:24270012 [DOI] [PubMed] [Google Scholar]
- [13].Zhou W, Zhu Y, Dong A, Shen W-H. Histone H2A/H2B chaperones: from molecules to chromatin-based functions in plant growth and development. Plant J. 2015;83:78-95. http://doi.org/10.1111/tpj.12830. PMID:25781491 [DOI] [PubMed] [Google Scholar]
- [14].Woloszynska M, Le Gall S, Van Lijsebettens M. Plant Elongator-mediated transcriptional control in a chromatin and epigenetic context. Biochim Biophys Acta. 2016;1859:1015-33. [DOI] [PubMed] [Google Scholar]
- [15].Subtil-Rodríguez A, Reyes JC. To cross or not to cross the nucleosome, that is the elongation question… RNA Biol. 2011;8:389-93. http://doi.org/10.4161/rna.8.3.14334. PMID:21445002 [DOI] [PubMed] [Google Scholar]
- [16].Swygert SG, Peterson CL. Chromatin dynamics: interplay between remodeling enzymes and histone modifications. Biochim Biophys Acta. 2014;1839:728-36. http://doi.org/10.1016/j.bbagrm.2014.02.013. PMID:24583555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tanny JC. Chromatin modification by the RNA Polymerase II elongation complex. Transcription. 2014;5:e988093.https://doi.org/ 10.4161/21541264.2014.988093. PMID:25494544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Diebold ML, Koch M, Loeliger E, Cura V, Winston F, Cavarelli J, Romier C. The structure of an Iws1/Spt6 complex reveals an interaction domain conserved in TFIIS, Elongin A and Med26. EMBO J. 2010;29:3979-91. http://doi.org/10.1038/emboj.2010.272. PMID:21057455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li L, Ye H, Guo H, Yin Y. Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression. Proc Natl Acad Sci USA. 2010; 107: 3918-23. http://doi.org/10.1073/pnas.0909198107. PMID:20139304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Widiez T, El Kafafi S, Girin T, Berr A, Ruffel S, Krouk G, Vayssières A, Shen W-H, Coruzzi GM, Gojon A, et al.. High nitrogen insensitive 9 (HNI9)-mediated systemic repression of root NO3- uptake is associated with changes in histone methylation. Proc Natl Acad Sci USA. 2011;108:13329-334. http://doi.org/10.1073/pnas.1017863108. PMID:21788519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Grasser M, Kane CM, Merkle T, Melzer M, Emmersen J, Grasser KD. Transcript elongation factor TFIIS is involved in Arabidopsis seed dormancy. J Mol Biol. 2009;386:598-611. http://doi.org/10.1016/j.jmb.2008.12.066. PMID:19150360 [DOI] [PubMed] [Google Scholar]
- [22].Park S, Oh S, Ek-Ramos J, van Nocker S. PLANT HOMOLOGOUS TO PARAFIBROMIN is a component of the PAF1 complex and assists in regulating expression of genes within H3K27ME3-enriched chromatin. Plant Physiol. 2010;153:821-31. http://doi.org/10.1104/pp.110.155838. PMID:20363855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yu X, Michaels SD. The Arabidopsis Paf1c complex component CDC73 participates in the modification of FLOWERING LOCUS C chromatin. Plant Physiol. 2010;153:1074-84. http://doi.org/10.1104/pp.110.158386. PMID:20463090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].He Y, Doyle MR, Amasino RM. PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 2004;18:2774-84. http://doi.org/10.1101/gad.1244504. PMID:15520273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Oh S, Zhang H, Ludwig P, van Nocker S. A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. Plant Cell. 2004;16:2940-53. http://doi.org/10.1105/tpc.104.026062. PMID:15472079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lolas IB, Himanen K, Grønlund JT, Lynggaard C, Houben A, Melzer M, Van Lijsebettens M, Grasser KD. The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2. Plant J. 2010;61:686-97. http://doi.org/10.1111/j.1365-313X.2009.04096.x. PMID:19947984 [DOI] [PubMed] [Google Scholar]
- [27].Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell. 2010;140:491-503. http://doi.org/10.1016/j.cell.2009.12.050. PMID:20178742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Xu Y, Bernecky C, Lee CT, Maier KC, Schwalb B, Tegunov D, Plitzko JM, Urlaub H, Cramer P. Architecture of the RNA polymerase II-Paf1C-TFIIS transcription elongation complex. Nat Commun. 2017;8:15741.https://doi.org/ 10.1038/ncomms15741. PMID:28585565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Squazzo SL, Costa PJ, Lindstrom DL, Kumer KE, Simic R, Jennings JL, Link AJ, Arndt KM, Hartzog GA. The Paf complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 2002;21:1764-1174. http://doi.org/10.1093/emboj/21.7.1764. PMID:11927560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090-93. http://doi.org/10.1126/science.1085703. PMID:12934006 [DOI] [PubMed] [Google Scholar]
- [31].Bondarenko VA, Steele LM, Ujvári A, Gaykalova DA, Kulaeva OI, Polikanov YS, Luse DS, Studitsky VM. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol Cell. 2006;24:469-79. http://doi.org/10.1016/j.molcel.2006.09.009. PMID:17081995 [DOI] [PubMed] [Google Scholar]
- [32].Zhang Y, Najmi SM, Schneider DA. Transcription factors that influence RNA polymerases I and II: To what extent is mechanism of action conserved? Biochim Biophys Acta. 2017;1860:246-55. http://doi.org/10.1016/j.bbagrm.2016.10.010. PMID:27989933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Vannini A, Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell. 2012;45:439-46. http://doi.org/10.1016/j.molcel.2012.01.023. PMID:22365827 [DOI] [PubMed] [Google Scholar]
- [34].Bies-Etheve N, Pontier D, Lahmy S, Picart C, Vega D, Cooke R, Lagrange T. RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep. 2009;10:649-54. http://doi.org/10.1038/embor.2009.31. PMID:19343051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].He XJ, Hsu YF, Zhu S, Wierzbicki AT, Pontes O, Pikaard CS, Liu HL, Wang C-S, Jin H, Zhu JK. An effector of RNA-directed DNA methylation in Arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell. 2009;137:498-508. http://doi.org/10.1016/j.cell.2009.04.028. PMID:19410546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Köllen K, Dietz L, Bies-Etheve N, Lagrange T, Grasser M, Grasser KD. The zinc-finger protein SPT4 interacts with SPT5L/KTF1 and modulates transcriptional silencing in Arabidopsis. FEBS Lett. 2015;589:3254-57. http://doi.org/10.1016/j.febslet.2015.09.017. PMID:26424658 [DOI] [PubMed] [Google Scholar]
- [37].Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688-700. http://doi.org/10.1016/j.cell.2009.02.001. PMID:19239889 [DOI] [PubMed] [Google Scholar]
- [38].Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 2014;15:108-21. http://doi.org/10.1038/nrm3742. PMID:24452469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hunt AG, Xing D, Li QQ. Plant polyadenylation factors: conservation and variety in the polyadenylation complex in plants. BMC Genomics. 2012;13:641.https://doi.org/ 10.1186/1471-2164-13-641. PMID:23167306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Li QQ, Liu Z, Lu W, Liu M. Interplay between alternative splicing and alternative polyadenylation defines the expression outcome of the plant unique OXIDATIVE TOLERANT-6 gene. Sci Rep. 2017;7:2052.https://doi.org/ 10.1038/s41598-017-02215-z. PMID:28515442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Rozenblatt-Rosen O, Nagaike T, Francis JM, Kaneko S, Glatt KA, Hughes CM, LaFramboise T, Manley JL, Meyerson M. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc Natl Acad Sci USA. 2009;106:755-60. http://doi.org/10.1073/pnas.0812023106. PMID:19136632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Nagaike T, Logan C, Hotta I, Rozenblatt-Rosen O, Meyerson M, Manley JL. Transcriptional activators enhance polyadenylation of mRNA precursors. Mol Cell. 2011;41:409-18. http://doi.org/10.1016/j.molcel.2011.01.022. PMID:21329879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Moreira A. Integrating transcription kinetics with alternative polyadenylation and cell cycle control. Nucleus. 2011;2:556-61. http://doi.org/10.4161/nucl.2.6.18064. PMID:22127258 [DOI] [PubMed] [Google Scholar]
- [44].Naftelberg S, Schor IE, Ast G, Kornblihtt AR. Regulation of alternative splicing through coupling with transcription and chromatin structure. Ann Rev Biochem. 2015;84:165-98. http://doi.org/10.1146/annurev-biochem-060614-034242. PMID:26034889 [DOI] [PubMed] [Google Scholar]
- [45].Dolata J, Guo Y, Kolowerzo A, Smolinski D, Brzyzek G, Jarmolowski A, Swiezewski S. NTR1 is required for transcription elongation checkpoints at alternative exons in Arabidopsis. EMBO J. 2015;34:544-58. http://doi.org/10.15252/embj.201489478. PMID:25568310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sørensen BB, Ehrnsberger HF, Esposito S, Pfab A, Bruckmann A, Hauptmann J, Meister G, Merkl R, Schubert T, Längst G, et al.. The Arabidopsis THO/TREX component TEX1 functionally interacts with MOS11 and modulates mRNA export and alternative splicing events. Plant Mol Biol. 2017;93:283-98. http://doi.org/10.1007/s11103-016-0561-9. PMID:28004241 [DOI] [PubMed] [Google Scholar]
- [47].Ghosh A, Lima CD. Enzymology of RNA cap synthesis. WIREs RNA. 2010;1:152-72. PMID:21956912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Heath CG, Viphakone N, Wilson SA. The role of TREX in gene expression and disease. Biochem J. 2016;473:2911-35. http://doi.org/10.1042/BCJ20160010. PMID:27679854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Martinez-Rucobo FW, Kohler R, van de Waterbeemd M, Heck AJ, Hemann M, Herzog F, Stark H, Cramer P. Molecular basis of transcription-coupled pre-mRNA capping. Mol Cell. 2017;58:1079-89. http://doi.org/10.1016/j.molcel.2015.04.004 [DOI] [PubMed] [Google Scholar]


