ABSTRACT
Autophagy is an essential cellular process that degrades cytoplasmic organelles and components. Precise control of autophagic activity is achieved by context-dependent signaling pathways. Recent studies have highlighted the involvement of transcriptional programs during autophagic responses to various signals. Here, we summarize the current understanding of the transcriptional regulation of autophagy.
KEYWORDS: autophagy, BRD4, histone, lysosome, modification, TFEB, transcriptional regulation of autophagy
Introduction
Macroautophagy (hereafter referred to as autophagy) is a conserved catabolic process responsible for the turnover of intracellular organelles and constituents.1 Autophagy is essential to maintain cellular homeostasis and dysregulation of this process is implicated in various diseases such as cancer, neuronal degeneration, and immune diseases.1 This process initiates with the formation of a lipid membrane structure called a phagophore (also known as isolation membrane) that engulfs cytoplasmic materials.2 As the phagophore expands and the edges of the membrane fuse, it forms a sphere-like double membrane structure called an autophagosome. Autophagosomes then fuse with lysosomes to become autolysosomes where the materials inside are degraded by lysosomal hydrolases.
Great advances have been made in the identification of genes involved in these steps, which has established the basic mechanisms of autophagy.2,3 The core molecular machinery includes initiation by the Unc-51-Like Kinase (ULK) complex and the generation of phosphatidylinositol 3-phosphate (PI3P)-enriched membrane compartments by a class III Phosphoinositide 3-Kinase (PI3K) complex followed by the recruitment of the ATG12-5-16L1 complex that mediates LC3 lipidation and subsequent autophagosome formation.2,3 In addition, various signaling pathways that control autophagy in response to cellular stresses have been described.3,4 For example, during the cellular response to nutrient deprivation, autophagy is induced via signaling cascades that result from the inhibition of the nutrient-sensing kinase complex, mechanistic Target Of Rapamycin Complex 1 (mTORC1) and activation of AMP-activated protein Kinase (AMPK).3,4
Interestingly, recent studies have uncovered that transcriptional regulation of autophagy genes plays an important role in autophagic responses to specific stimuli.5–7 Several transcription factors and histone modifications that regulate autophagy gene expression have been identified. Alteration of post-translational modifications of these transcription factors and histones in response to cellular stresses affects their function and autophagy gene expression. These findings add another layer of complexity to the control of autophagy. In this Point-of-View article, we focus on the emerging role of transcriptional programs in the regulation of autophagy.
TFEB is a master transcriptional regulator of autophagy
Transcriptional regulation of autophagy was first recognized about 10 years ago, when two transcription factors, p53 and Forkhead box O3 (FOXO3), were shown to induce autophagy.8,9 Since then, a number of studies have characterized transcription factors that activate or repress the expression of core autophagy genes and/or their upstream regulators.5 Among them, Transcription Factor EB (TFEB) is considered a master transcriptional regulator of autophagy as it upregulates a subset of autophagy and lysosome genes and activates the whole autophagy-lysosome pathway.10 TFEB binds to DNA sequences, termed Coordinated Lysosomal Expression And Regulation (CLEAR) elements, that are present in numerous autophagy and lysosome genes and induces their expression.11,12 This increases autophagosome formation, promotes the fusion of autophagosomes with lysosomes, and enhances lysosome function and biogenesis.
The activity of TFEB is tightly controlled by environmental conditions via post-translational modifications and is usually suppressed under stress-free conditions. It is highly phosphorylated by various kinases including mTORC1, Extracellular signal-Regulated Kinase 2 (ERK2), and AKT and sequestered in the cytoplasm under nutrient rich conditions (Fig. 1).12-14 Following nutrient deprivation and subsequent mTORC1 inactivation, TFEB is dephosphorylated and translocates to the nucleus. In addition to mTORC1 inactivation, calcium ion release from lysosomes also contributes to TFEB nuclear translocation through calcineurin-mediated dephosphorylation of TFEB (Fig. 1).15
Figure 1.
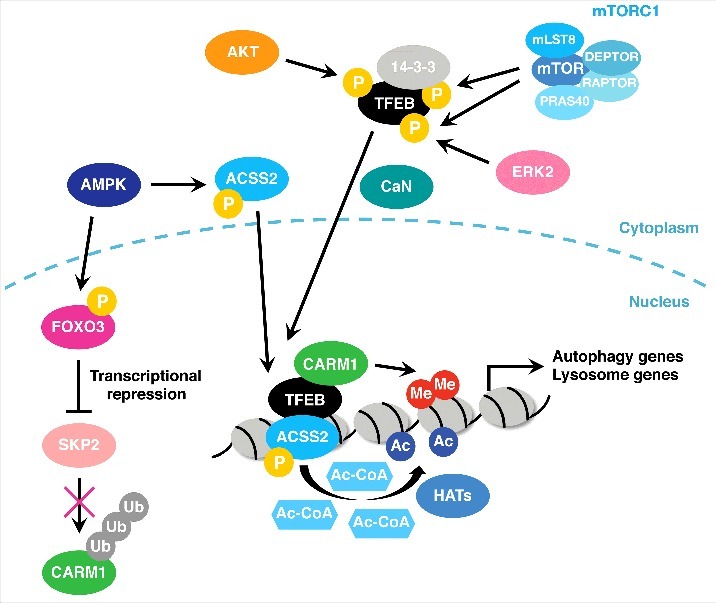
Regulation of autophagy gene transcription by TFEB. TFEB is phosphorylated by mTORC1, ERK2, and AKT and is sequestered in the cytoplasm via interaction with 14-3-3 under nutrient rich conditions. Upon nutrient deprivation, TFEB is dephosphorylated by calcineurin (CaN) and translocates to the nucleus where it activates autophagy and lysosome gene transcription. FOXO3 is phosphorylated by AMPK and represses SKP2 expression, which leads to CARM1 stabilization. CARM1 then binds to TFEB and increases H3R16 dimethylation at autophagy and lysosome gene promoters. AMPK also phosphorylates ACSS2 and facilitates its nuclear translocation. ACSS2 interacts with TFEB and increases local acetyl-CoA and histone acetylation. HAT: histone acetyltransferase.
Interestingly, recent studies have shown that transcriptional activation of autophagy and lysosome genes by TFEB also involves AMPK-dependent changes in histone modifications.16,17 These mechanisms mainly operate under glucose-starved conditions. Upon glucose deprivation, the AMP/ATP ratio increases and AMPK becomes active, leading to stabilization of Coactivator-Associated arginine (R) Methyltransferase 1 (CARM1) by suppressing the expression of ubiquitin ligase S-phase Kinase-associated Protein 2 (SKP2) via FOXO3. CARM1 then interacts with TFEB and co-activates TFEB-mediated transcription by depositing dimethylation at H3R17 (Fig. 1).16 AMPK also phosphorylates Acetyl-CoA synthetase 2 (ACSS2) and facilitates its nuclear translocation and interaction with TFEB. This leads to local production of acetyl donor acetyl-CoA and an increase in histone H3 acetylation at TFEB target gene promoters for transcriptional activation (Fig. 1).17 These two mechanisms seem to be responsible for transcriptional activation of autophagy genes in response to glucose deprivation. Taking these reports together, following nutrient deprivation, mTORC1 inactivation and AMPK activation cooperatively induce autophagy and lysosome genes by altering TFEB localization and the chromatin environment of its target genes.
Transcriptional repression of autophagy
Autophagy gene expression is suppressed under nutrient replete conditions. Several transcription factors such as Zinc finger with KRAB and SCAN domains 3 (ZKSCAN3) and Forkhead box K (FOXK) engage in this transcriptional repression (Fig. 2).18,19 ZKSCAN3 functions as a transcriptional repressor of the autophagy-lysosome pathway.18 It is recruited to promoter regions of various autophagy and lysosome genes and represses gene expression under nutrient rich conditions. Knockdown of ZKSCAN3 upregulates a subset of autophagy and lysosome genes and enhances autophagic flux and lysosome function and biogenesis. Nutrient deprivation leads to the nuclear export of ZKSCAN3 and de-repression of autophagy gene expression. However, the mechanisms by which ZKSCAN3 represses autophagy gene expression and how its localization is regulated remain unknown. A recent report has shown that c-Jun N-terminal Kinase 2 (JNK2) and p38 Mitogen-Activated Protein Kinase (MAPK), that are activated by Protein Kinase Cδ (PKCδ), phosphorylate ZKSCAN3 and facilitate its nuclear export.20 It would therefore be interesting to know whether this phosphorylation contributes to ZKSCAN3 nuclear export during starvation.
Figure 2.
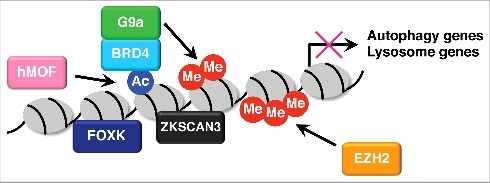
Transcriptional repression of autophagy genes. Under nutrient rich conditions, ZKSCAN3 and FOXK transcription factors bind to promoter regions of autophagy genes and repress transcription. BRD4 is recruited to autophagy and lysosome gene promoters via H4K16 acetylation catalyzed by hMOF. BRD4 then interacts with G9a which deposits dimethylation at H3K9 and represses gene expression. H3K27 trimethylation catalyzed by EZH2 suppresses autophagy by repressing negative regulators of mTORC1 pathway.
Similarly, FOXK transcriptionally suppresses the early stages of autophagy.19 It binds to promoter regions of early-stage autophagy genes including components of the ULK and class III PI3K complexes and recruits the SIN3A- Histone deacetylase (HDAC) repressor complex to these regions under nutrient rich conditions.19 During starvation, it translocates from the nucleus to the cytoplasm in an mTOR-dependent manner. FOXO3 transcription factor, in turn, is recruited to these promoter regions and activates transcription of these genes. These transcriptional repressors may therefore function to prevent over-activation of autophagy by suppressing gene transcription.
Autophagy gene regulation by histone modifications
In addition to the transcription factors described above, the post-translational modification status on histones is also linked to autophagy gene regulation.7 To date, several histone modifications, including histone H4K16 acetylation, H3K9 dimethylation, and H3K27 trimethylation have been reported to influence autophagic activity.7 H4K16 acetyltransferase human Males absent On the First (hMOF) has been described as both a positive and negative regulator of autophagy.21,22 In the course of prolonged autophagy activation by mTOR inhibition caused by rapamycin treatment or starvation, H4K16 acetylation and autophagy gene expression decline via hMOF degradation and/or Sirtuin1 (SIRT1)-dependent histone deacetylation.21 This is considered as a negative feedback mechanism that restrains over-activation of autophagy and subsequent cell death.21 In contrast, another report has shown that knockdown of hMOF enhances autophagic flux and promotes the formation of autolysosomes.22
Histone lysine methyltransferase G9a acts as a suppressor of autophagy by dimethylating H3K9 and repressing autophagy gene expression.23 Nutrient starvation causes G9a dissociation from promoters and subsequent H3K9 demethylation, which leads to transcriptional activation of autophagy genes.
Different from H4K16 acetylation and H3K9 dimethylation which regulate the expression of core autophagy genes, H3K27 trimethylation catalyzed by Enhancer of Zeste Homolog 2 (EZH2) represses the expression of negative regulators of the mTORC1 signaling components and leads to mTORC1 activation and autophagy inhibition (Fig. 2).24 Collectively, histone modification status is also an important determinant of transcriptional responses to autophagic stimuli.
Transcriptional regulation of autophagy by a “reader” of histone modification: Bromodomain protein BRD4 transcriptionally represses autophagy and lysosome programs
Post-translational modifications on histones are recognized by so-called “readers” such as bromodomain, chromodomain, and tudor domain proteins that mediate chromatin-based processes including transcription.25 We recently identified the Bromodomain and Extra-Terminal (BET) family protein Bromodomain containing 4 (BRD4) as a novel autophagy repressor that links histone modifications and autophagy gene transcription.26 BRD4 is recruited to autophagy and lysosome gene promoters through hMOF-mediated H4K16 acetylation. It then interacts with histone methyltransferase G9a, which facilitates H3K9 dimethylation and represses autophagy gene transcription (Fig. 2). During nutrient starvation, AMPK disrupts the interaction between histone deacetylase SIRT1 and its inhibitory molecule Deleted in Breast Cancer protein 1 (DBC1), leading to SIRT1 activation. It then facilitates H4K16 deacetylation and BRD4 dissociation from the promoters, driving transcription of autophagy and lysosome genes. BRD4 knockdown induces autophagy and lysosome gene expression and activates a series of autophagic processes including the formation of phagophores/autophagosomes, the fusion between autophagosomes and lysosomes, and lysosome function and biogenesis. Therefore, these results suggest that BRD4 functions to restrain autophagic activity under nutrient rich conditions and its de-repression contributes to sustained autophagy during prolonged starvation.
Concluding remarks and perspectives
In the course of nutrient starvation, autophagy is regulated by multiple mechanisms. Nutrient sensing kinases, such as mTORC1 and AMPK, rapidly respond to a change in cellular energy status and initiate autophagy programs by directly altering the post-translational modification status of core autophagy molecules such as ULK1 and Beclin 1.2,3 It is becoming clear that transcriptional upregulation of autophagy genes also contributes to starvation-induced autophagy.5–7 In contrast to the direct regulation of core autophagy proteins by post-translational modifications that leads to rapid activation of autophagy, transcriptional regulation of autophagy genes usually occurs over a longer time period. Hence, these transcriptional programs seem to contribute to sustained autophagy and/or function to adjust the autophagic activity during prolonged starvation. Of note, many of the transcriptional regulators that modulate autophagy gene expression are controlled by common upstream kinases such as mTORC1 and AMPK. This allows coordinated activation of transcriptional activators (i.e. TFEB and FOXO3) and inhibition of transcriptional repressors (i.e. FOXK and BRD4), enabling induction of a broad range of autophagy molecules that are sufficient to sustain autophagy during long periods of nutrient shortage. Many of the studies on the transcriptional regulation of autophagy to date mainly focus on how these mechanisms respond to nutrient starvation, such as amino acid and glucose starvation. It thus would be interesting to explore whether other autophagic stimuli also affect the autophagy gene expression and, if so, how much it contributes to autophagy activation.
Given that dysregulation of autophagy is implicated in various diseases,1 it would be intriguing to explore how these transcriptional programs are regulated or dysregulated in disease contexts. A recent report has shown that TFEB and its family members Transcription Factor E3 (TFE3) and MIcrophthalmia-associated Transcription Factor (MITF) interact with Importin 8 (IPO8) and are constitutively localized in the nucleus in pancreatic ductal adenocarcinoma (PDAC).27 This leads to autophagy activation and provides nutrient sources to support PDAC growth. In another instance, we found that the chromosomal translocation involving NUclear protein in Testis (NUT) gene and BRD4, a driver of NUT midline carcinoma (NMC),28 strongly suppresses autophagy in NMC.26 However, whether autophagy suppression contributes to NMC development is still an open question. Similarly, chromosomal translocation of TFEB and TFE3 genes are also found in renal cell carcinoma,10 though their effects on autophagy are currently unknown. In addition, histone modifications that affect the outcomes of autophagy are also implicated in carcinogenesis.29 These findings suggest that dysregulation of these transcriptional programs may contribute to disease onset and/or progression.
With regard to the relation between autophagy and human disease, the growing evidence suggests that modulation of autophagic activity has a beneficial effect on diseases including neurodegeneration and cancer and modulating autophagy gene programs is sufficient to ameliorate these diseases.10,30 For example, TFEB overexpression enhances autophagic clearance of protein aggregates and ameliorates pathology in mouse models of neurodegeneration, and conversely knockdown of TFEB suppresses PDAC growth.10,27 In addition, our recent work has shown that autophagy activation by BRD4 knockdown promotes autophagic degradation of pathogenic protein aggregates that cause neurodegeneration. Importantly, some of these transcriptional programs can be pharmacologically targeted by small molecules such as BET and EZH2 inhibitors. In light of this, modulating the autophagy gene programs by these small molecules may be an attractive strategy for the treatment of these diseases. Further studies will therefore advance our understanding of the role of transcriptional regulation of autophagy, its relevance to human diseases, and its therapeutic potential.
Funding Statement
Work in the Ryan laboratory is supported by Cancer Research UK (C596/A17196) and Worldwide Cancer Research (16-1194).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank members of the Ryan laboratory for helpful discussions and apologize to the authors whose work could not be included due to space limitations.
References
- 1.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728–41. doi: 10.1016/j.cell.2011.10.026. PMID:22078875 [DOI] [PubMed] [Google Scholar]
- 2.Ktistakis NT, Tooze SA. Digesting the Expanding Mechanisms of Autophagy. Trends Cell Biol. 2016;26(8):624–35. doi: 10.1016/j.tcb.2016.03.006. PMID:27050762 [DOI] [PubMed] [Google Scholar]
- 3.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14(12):759–74. doi: 10.1038/nrm3696. PMID:24201109 [DOI] [PubMed] [Google Scholar]
- 4.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40(2):280–93. doi: 10.1016/j.molcel.2010.09.023. PMID:20965422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fullgrabe J, Ghislat G, Cho DH, Rubinsztein DC. Transcriptional regulation of mammalian autophagy at a glance. J Cell Sci. 2016;129(16):3059–66. doi: 10.1242/jcs.188920. PMID:27528206 [DOI] [PubMed] [Google Scholar]
- 6.Fullgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol. 2014;15(1):65–74. doi: 10.1038/nrm3716. PMID:24326622 [DOI] [PubMed] [Google Scholar]
- 7.Baek SH, Kim KI. Epigenetic Control of Autophagy: Nuclear Events Gain More Attention. Mol Cell. 2017;65(5):781–5. doi: 10.1016/j.molcel.2016.12.027. PMID:28257699 [DOI] [PubMed] [Google Scholar]
- 8.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6(6):472–83. doi: 10.1016/j.cmet.2007.11.004. PMID:18054316 [DOI] [PubMed] [Google Scholar]
- 9.Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126(1):121–34. doi: 10.1016/j.cell.2006.05.034. PMID:16839881 [DOI] [PubMed] [Google Scholar]
- 10.Napolitano G, Ballabio A. TFEB at a glance. J Cell Sci. 2016;129(13):2475–81. doi: 10.1242/jcs.146365. PMID:27252382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. . A gene network regulating lysosomal biogenesis and function. Science. 2009;325(5939):473–7. doi: 10.1126/science.1174447. PMID:19556463 [DOI] [PubMed] [Google Scholar]
- 12.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. . TFEB links autophagy to lysosomal biogenesis. Science. 2011;332(6036):1429–33. doi: 10.1126/science.1204592. PMID:21617040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmieri M, Pal R, Nelvagal HR, Lotfi P, Stinnett GR, Seymour ML, Chaudhury A, Bajaj L, Bondar VV, Bremner L, et al. . mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat Commun. 2017;8:14338. doi: 10.1038/ncomms14338. PMID:28165011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et al. . A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31(5):1095–108. doi: 10.1038/emboj.2012.32. PMID:22343943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, et al. . Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol. 2015;17(3):288–99. doi: 10.1038/ncb3114. PMID:25720963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin HJ, Kim H, Oh S, Lee JG, Kee M, Ko HJ, Kweon MN, Won KJ, Baek SH. AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature. 2016;534(7608):553–7. doi: 10.1038/nature18014. PMID:27309807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Yu W, Qian X, Xia Y, Zheng Y, Lee JH, Li W, Lyu J, Rao G, Zhang X, et al. . Nucleus-Translocated ACSS2 Promotes Gene Transcription for Lysosomal Biogenesis and Autophagy. Mol Cell. 2017;66(5):684-97 e9. doi: 10.1016/j.molcel.2017.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chauhan S, Goodwin JG, Chauhan S, Manyam G, Wang J, Kamat AM, Boyd DD. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell. 2013;50(1):16–28. doi: 10.1016/j.molcel.2013.01.024. PMID:23434374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bowman CJ, Ayer DE, Dynlacht BD. Foxk proteins repress the initiation of starvation-induced atrophy and autophagy programs. Nat Cell Biol. 2014;16(12):1202–14. doi: 10.1038/ncb3062. PMID:25402684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Xu M, Ding X, Yan C, Song Z, Chen L, Huang X, Wang X, Jian Y, Tang G, et al. . Protein kinase C controls lysosome biogenesis independently of mTORC1. Nat Cell Biol. 2016;18(10):1065–77. doi: 10.1038/ncb3407. PMID:27617930 [DOI] [PubMed] [Google Scholar]
- 21.Fullgrabe J, Lynch-Day MA, Heldring N, Li W, Struijk RB, Ma Q, Hermanson O, Rosenfeld MG, Klionsky DJ, Joseph B. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 2013;500(7463):468–71. doi: 10.1038/nature12313. PMID:23863932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hale CM, Cheng Q, Ortuno D, Huang M, Nojima D, Kassner PD, Wang S, Ollmann MM, Carlisle HJ. Identification of modulators of autophagic flux in an image-based high content siRNA screen. Autophagy. 2016;12(4):713–26. doi: 10.1080/15548627.2016.1147669. PMID:27050463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Artal-Martinez de Narvajas A, Gomez TS, Zhang JS, Mann AO, Taoda Y, Gorman JA, Herreros-Villanueva M, Gress TM, Ellenrieder V, Bujanda L, et al. . Epigenetic regulation of autophagy by the methyltransferase G9a. Mol Cell Biol. 2013;33(20):3983–93. doi: 10.1128/MCB.00813-13. PMID:23918802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei FZ, Cao Z, Wang X, Wang H, Cai MY, Li T, Hattori N, Wang D, Du Y, Song B, et al. . Epigenetic regulation of autophagy by the methyltransferase EZH2 through an MTOR-dependent pathway. Autophagy. 2015;11(12):2309–22. doi: 10.1080/15548627.2015.1117734. PMID:26735435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Musselman CA, Lalonde ME, Cote J, Kutateladze TG. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol. 2012;19(12):1218–27. doi: 10.1038/nsmb.2436. PMID:23211769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakamaki JI, Wilkinson S, Hahn M, Tasdemir N, O'Prey J, Clark W, Hedley A, Nixon C, Long JS, New M, et al. . Bromodomain Protein BRD4 Is a Transcriptional Repressor of Autophagy and Lysosomal Function. Mol Cell. 2017;66(4):517–32 e9. doi: 10.1016/j.molcel.2017.04.027. PMID:28525743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perera RM, Stoykova S, Nicolay BN, Ross KN, Fitamant J, Boukhali M, Lengrand J, Deshpande V, Selig MK, Ferrone CR, et al. . Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015;524(7565):361–5. doi: 10.1038/nature14587. PMID:26168401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.French CA. NUT midline carcinoma. Cancer Genet Cytogenet. 2010;203(1):16–20. doi: 10.1016/j.cancergencyto.2010.06.007. PMID:20951314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fullgrabe J, Heldring N, Hermanson O, Joseph B. Cracking the survival code: autophagy-related histone modifications. Autophagy. 2014;10(4):556–61. doi: 10.4161/auto.27280. PMID:24429873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galluzzi L, Bravo-San Pedro JM, Levine B, Green DR, Kroemer G. Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat Rev Drug Discov. 2017;6(7):487-511. doi: 10.1038/nrd.2017.22. PMID:28529316. [DOI] [PMC free article] [PubMed] [Google Scholar]


