Summary
Bacteria control major nutrient cycles and directly influence plant, animal, and human health. However, we know relatively little about the forces shaping their large-scale ecological ranges. Here, we reveal patterns in the distribution of individual bacterial taxa at multiple levels of phylogenetic resolution within and between Earth’s major habitat types. Our analyses suggest that while macro-scale habitats structure bacterial distribution to some degree, abundant bacteria (i.e., detectable using 16S rRNA gene sequencing methods) are confined to single assemblages. Additionally, we show that the most cosmopolitan taxa are also the most abundant in individual assemblages. These results add to the growing body of data that support that the diversity of the overall bacterial metagenome is tremendous. The mechanisms governing microbial distribution remain poorly understood, but our analyses provide a framework with which to test the importance of macro-ecological environmental gradients, relative abundance, neutral processes and the ecological strategies of individual taxa in structuring microbial communities.
Introduction
Forces shaping the biogeography of macroorganisms - including dispersal limitations, habitat differentiation, competition, and adaptive radiation – have been a central focus of ecology for more than a century (Brown 1998). Yet, while microorganisms are the most abundant and diverse organisms on Earth (Whitman et al. 1998), relatively little is known about the patterns of, or controls over, microbial distribution within and between the planet’s major habitat types. One common theory holds that the tremendous dispersal potential of microbes will lead to everything being everywhere (i.e., no dispersal limitations), with environmental selection determining which species are abundant (Martiny et al. 2006). However, until recently, methodological limitations have prevented large-scale tests of ideas about where certain microorganisms exist, and why (Hugenholtz et al. 1998; Prosser et al. 2007).
Over the last decades, however, molecular phylogenetic approaches have revolutionized microbiology, expanding our view of microbial diversity and our appreciation of the complexity of microbial communities (Hugenholtz et al. 1998). While these techniques do not provide an exhaustive sampling of any but the simplest microbial assemblages, they do provide information on the dominant members of the community, allowing ecologically meaningful questions to be addressed about the distribution of these lineages. These methods have been used to reveal that some microorganisms exhibit distinct biogeographical patterns (Horner-Devine et al. 2004; Green & Bohannan 2006; Martiny et al. 2006), which appear to be controlled by differences in environmental variables in some cases (Horner-Devin et al. 2004), and geographical distance in others (Cho & Tiedje 2000; Whitaker et al. 2003). Other work investigating overall community composition supports the role of environmental gradients in structuring both lake and soil bacterial assemblages (Fierer & Jackson 2006; Van der Gucht et al. 2007). Biotic interactions may also be important in determining microbial community composition; a recent study showed that microbial communities exhibit more segregation of taxa than would be predicted by chance, suggesting that competitive interactions and/or niche specialization may be important in structuring bacterial biogeography (Horner-Devine et al. 2007).
To date, however, most studies of microbial biogeography have focused on a single habitat type or on a phylogenetically restricted set of taxa; thus, broader patterns in the distribution of microorganisms among Earth’s major ecosystems remain poorly understood. Recently, Lozupone and Knight (2007) demonstrated that salinity is the primary driver of community-level phylogenetic differentiation among bacterial assemblages sampled from different habitat types. Yet, we know that many bacterial phyla are widely distributed across multiple habitats (Madigan et al. 2000). Thus, to further investigate the role of macro-scale habitats in structuring the biogeographic patterns exhibited by individual bacterial taxa, we examined the distribution of taxa at multiple levels of phylogenetic resolution (98, 95, 92 and 89% 16S rRNA gene sequence identity), both within and across different habitat types. Here, we show that there is minimal taxon overlap between assemblages both within and between habitat types, and that the most abundant taxa are also the most widely distributed.
Results
We examined the distribution of 16S rRNA genes in a dataset of clone libraries assembled from a variety of habitat types (Lozupone & Knight 2007) and expanded upon (Table S1). Operational taxonomic units (OTUs) were selected at four different levels of sequence identity: 98%, 95%, 92% and 89% and the collection of OTUs present in a given sample was considered an individual assemblage. Although there is controversy about the amount of sequence differentiation that constitutes a particular taxonomic ranking, there is some consensus that these levels of divergence are less than those required to differentiate phyla (Hugenholtz et al. 1998; Dojka et al. 2000) and have been used to correspond roughly to species, genus, family, and order, respectively (Stackebrandt & Goebel 1994; Schloss & Handelsman 2004).
We first examined the distribution of OTUs across all 238 assemblages examined. At the species level of sequence identity, more than 85% of all OTUs were not detected in more than one assemblage and no single OTU was observed in more than 12% of assemblages (Fig. 1A). At higher levels of sequence divergence, more OTUs were widespread; for example, at the order level of identity, 35% of OTUs were found in two or more assemblages. However, at all levels of phylogenetic resolution, distribution patterns featured a similar pattern with the majority of OTUs found in no more than one assemblage and small numbers of OTUs that were more highly distributed. All OTUs that were detected in greater than 20% of assemblages belonged to the Proteobacteria, specifically the α–, β– and γ– proteobacteria. Although ‘unclassifiable’ OTUs comprised 10% of the original dataset (Fig. 1B), no single unclassifiable OTU was observed in more than 6% of all assemblages for any OTU definition.
Fig. 1.
A. The number of OTUs that were found in different proportions of assemblages within our clone library dataset (Table S1), which contains 28,115 sequences and 238 assemblages. At all OTU definitions, the vast majority of lineages were observed in only a single assemblage. B. The relative abundance of different phyla within the clone library dataset. Phyla that represent at least 2% of all sequences are labeled.
To examine how much of this limited distribution was driven by environment type, we explored patterns of occurrence across the fourteen different habitats. At the species level, 97% of OTUs were found in no more than one habitat type and no single OTU was detected in more than 6 habitats (Fig. 2). Although less pronounced, these patterns were also discernable at lower levels of phylogenetic resolution, with 92%, 88% and 84% of genus, family and order-level OTUs, respectively, detected in no more than a single habitat type. OTUs detected in more than five habitat types were related to the Comamonadaceae, Pseudomonadaceae, Aeromonas, Staphylococcus, and Propionibacterium (Table 1). We performed PERMANOVAs (Anderson 2001) to test the hypothesis that habitat types structure the distribution of bacteria. This is an analysis of variance test that uses permutation to examine the significance of factors (in this case, habitat types) in partitioning variation within multivariate datasets (in this case, an assemblage by OTU presence-absence matrix). These analyses revealed that, while most variation in assemblage composition was accounted for within habitat types (83–95%), there was a significant amount (5–17%) of variation between different habitat types (P < 0.001).
Fig. 2.
Rank distribution plots displaying the presence of OTUs in different numbers of habitat types. At all OTU definitions, the vast majority of lineages were observed in only a single habitat type.
Table 1.
Phylogenetic identities of ecologically ‘widely distributed’ OTUs 529 at the 98% minimum sequence identity cutoff. Table shows the OTU identifier (OTU #), the number of habitat types it was detected in, and its classification. Note that our classification system did not allow all OTUs to be identified at the same level of phylogenetic resolution; some were resolved to the genus level while others were resolved to the family level.
| OTU # | Number of Habitat types |
Classification |
|---|---|---|
| 467 | 6 | Comamonadaceae |
| 178 | 5 | Pseudomonadaceae |
| 243 | 5 | Propionibacterium |
| 469 | 5 | Pseudomonas |
| 63 | 5 | Staphylococcus |
| 107 | 5 | Aeromonas |
| 163 | 6 | Pseudomonas |
| 144 | 6 | Comamonadaceae |
The distribution of OTUs across assemblages was then examined within the six habitat types for which we had the most samples: soil, lakewater, freshwater sediment, seawater, marine sediment, and insect-associated assemblages. Again, distribution was assayed at four different OTU cutoffs. At all levels of phylogenetic resolution, all habitat types revealed a distribution pattern featuring a few widely dispersed lineages and many more confined lineages (Fig. 3).
Fig. 3.
The rank in distribution plotted against the percent of assemblages each OTU was found in for (A) soils (n=49), (B) lakewater (n=21), (C) freshwater sediments (n=15), (D) seawater (n=40), (E) saline sediments (n=36), (F) and insect associated samples (n=15) for the clone library data. Those OTUs that were most widely dispersed within habitat types are indicated. Within habitat types, some OTUs were widely distributed among assemblages while the majority were limited to only a few assemblages.
We also examined OTU distribution across two pyrosequencing-based soils datasets including an inter-continental analysis of 88 samples (Lauber et al. 2009) and a study of 27 samples from within a single hectare of tropical rainforest. The advantage of examining both of these datasets separately is that we can look for similarities and differences in patterns of distribution that exist over both large (inter-continental) and small (intra-hectare) scales. Although we did not examine multiple OTU definitions for the short sequences generated via pyrosequencing because of known inconsistencies (Elshahed et al. 2008), both of these datasets contained more than 1000 16S rRNA gene sequences per soil, and thus are much better sampled than the clone library data. Another advantage of the pyrosequencing studies is that it is possible to consider the relationship between relative abundance and distribution patterns, which is impossible for the compiled dataset because of irregularities in analyzing and reporting abundance between studies (Lozupone & Knight 2007).
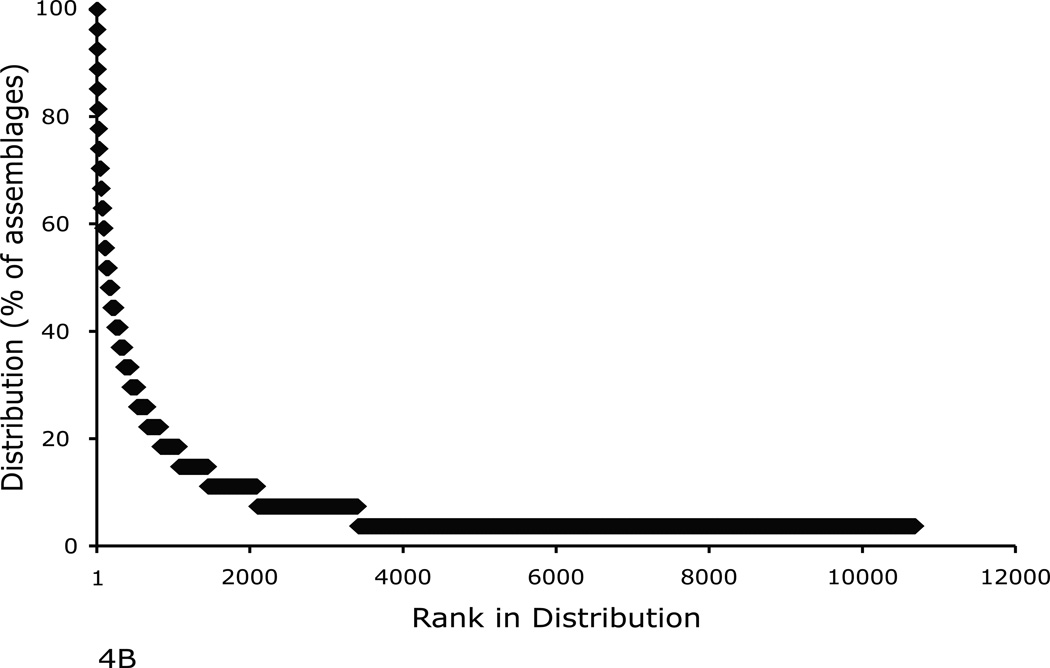
The distribution of OTUs within the pyrosequencing datasets shows the same basic pattern as was seen in the clone library data (Fig. 4). For the large-scale dataset, 75% of OTUs were not found in more than one soil (Lauber et al. 2009) while 68% from the smaller-scale tropical forest site were detected in a single soil sample. The top four most widely distributed OTUs from the large-scale dataset were detected in between 70 and 88% of soils, and were all related to the Bradyrhizobiales. Three OTUs related to the α-proteobacteria and one OTU related to the δ-proteobacteria were detected in all of the tropical forest soils.
Fig. 4.
The rank in distribution plotted against the percent of assemblages each OTU was found in for (A) the inter-continental soils dataset (n=88) and (B) the tropical forest (intra-hectare) soils dataset (n=27).
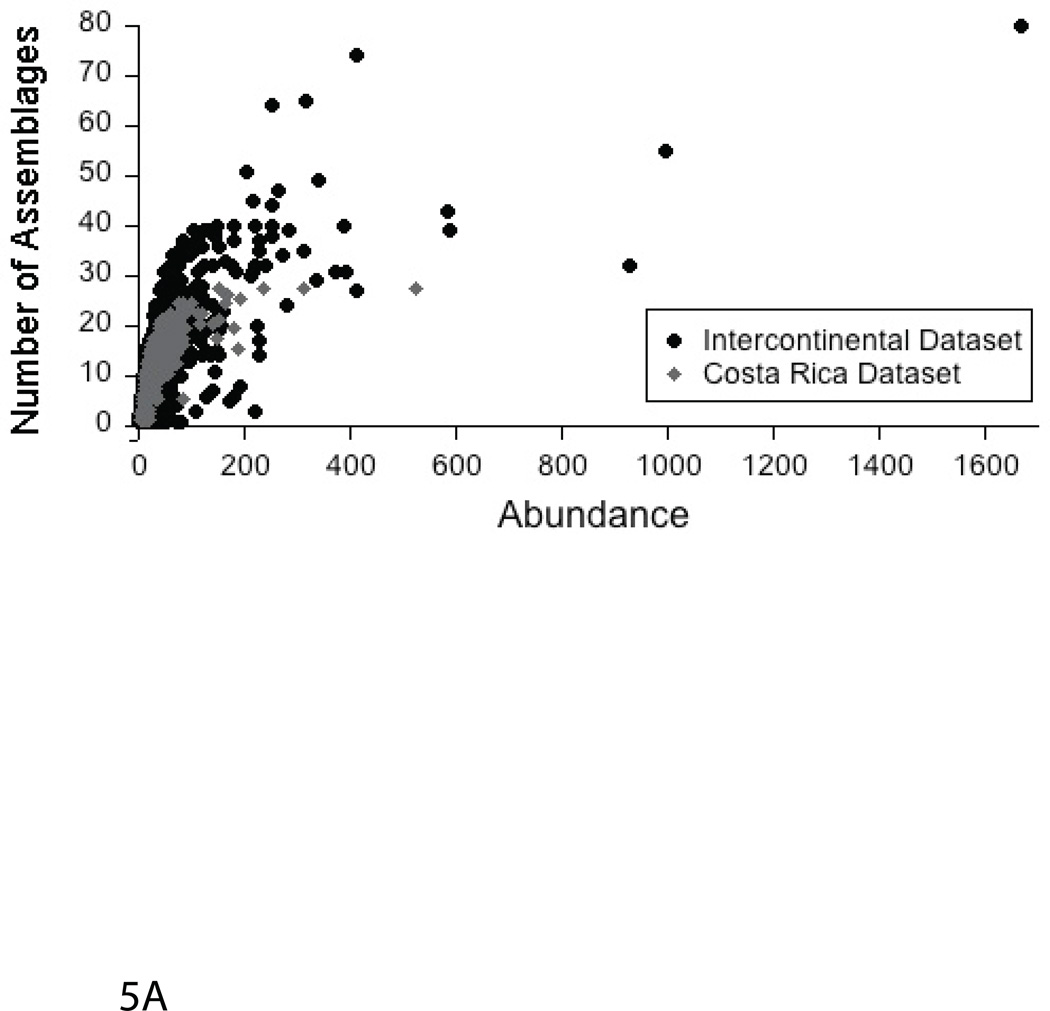
For each OTU, we plotted its total abundance across all assemblages against the number of assemblages in which it was detected, revealing a significant, positive relationship (Fig. 5A). We also plotted the average of the relative abundance of each OTU across all assemblages against its distribution, which did not change the shape of the function (data not shown). The top ten most abundant OTUs from each sample were found in an average of 28% of assemblages (vs. 2% for all OTUs) for the large-scale dataset. For the tropical forest dataset, the top ten most abundant OTUs from each sample were found in an average of 69% of soils (vs. 8% for all OTUs). Interestingly, the high abundance values for the overall top ten OTUs in each dataset were not driven by a few assemblages with high proportions of these sequences. Rather, they reflect moderate abundances (relative to the total abundance of that OTU within the dataset) across many samples (Fig. 5B).
Fig. 5.
The relationship between abundance and distribution of OTUs. A. The grey diamonds represent OTUs from the tropical forest dataset; the black circles represent OTUs from the trans-continental (Lauber et al. 2009) dataset. Here, we plotted the total abundance (within the entire dataset) of each OTU against its distribution. However, we also examined the relationship between the average of the relative abundance of each OTU within all assemblages against its distribution, which yielded similar results (data not shown). B. Heatmaps of the top ten most abundant OTUs for each study (tropical forest soils = top, intercontinental soils= bottom) showing the abundance of each OTU in each assemblage relative to its total abundance across the dataset. Each column represents a different assemblage; each row represents a different OTU; the color of the cells represents the relative abundance of that OTU within specific assemblages.
Discussion
Our results support that, for the most abundant organisms from these assemblages, macro-scale habitats structure bacterial distribution (Fig. 2). Indeed, as has been shown in other work (Tanner et al. 1998) close relatives of the eight OTUs that were detected in five or more habitat types (Table 1) are among the most abundant organisms found on human skin (e.g., Staphylococcus, Propionibacterium) or have been found in low-organic matter water supplies (e.g., Comamonadaceae, Pseudomonadaceae, Aeromonas) (Burtscher et al. 2009; Costello et al. 2009), suggesting that these ‘widely distributed’ bacteria actually may be contaminants introduced during sample processing or PCR amplification.
Other studies have shown that habitat types harbor different overall communities of bacteria (Lozupone & Knight 2007) and archaea (Auguet 2010), but have not determined if habitat type also shapes the distribution of individual microbial taxa. For example, one possible explanation for the difference in community composition between habitat types may be that bacteria are widespread across multiple habitat types, but that different habitat types support different combinations of organisms. Our data support that, for the most part, abundant (i.e., detectable using 16S rRNA gene sequencing methods) bacteria are confined to specific environments and that a significant fraction of the variation in the distribution of bacteria is related to habitat type.
Although macro-scale habitats do structure bacterial distribution, our results also suggest that, within a habitat type, most bacterial taxa are still restricted to a relatively small number of assemblages (Figs. 1, 3 and 4) and that there is a positive relationship between the relative abundance of an organism and its distribution across assemblages (Fig. 5). We discuss the implications of these observations below.
Most bacteria are confined to one assemblage
We found that between 65 and 85% of OTUs at all levels of sequence identity examined were present in only a single assemblage (Fig. 1). This pattern of limited distribution has also been observed between assemblages within individual habitat types (e.g., Figs. 4 and 5), including coastal waters (Pommier et al. 2007), and soils (Noguez et al. 2005; Fulthorpe et al. 2008), but has not been documented across habitats. For example, Fulthorpe et al. (2008) examined four soils from different sites in North and South America using pyrosequencing of SSU rRNA genes (Roesch et al. 2007). They generated between ~26,000 and 53,000 gene sequences per soil and showed that, at the 97% identity cutoff, 74% of OTUs were confined to a single assemblage. Likewise, in the Lauber et al. (2009) study 75% of sequences at the 97% OTU cutoff were detected in only a single sample (Fig. 5).
Other studies have used more sensitive methods to support endemism among particular groups of microorganisms. For example, Cho and Teidje (2000) isolated fluorescent pseudomonads from ten sites on four continents. Using a method for genomic fingerprinting, they revealed no overlap in genotypes between sites or between continents. Likewise, Wawrik et al. (2007) used tRFLPs to examine 16S rRNA and type II polyketide synthase genes of actinomycetes from soils collected in New Jersey and Asia and showed that fewer than 1% of phylotypes were found in more than 50% of soils that they examined. Geographical isolation has also been demonstrated for archaea in hot springs (Whitaker et al. 2003) and β-proteobacteria in sediments (Horner-Devine et al. 2004). Thus, several studies using a variety of methods support microbial endemism over a range of environments.
Although sampling limitations constrain our ability to conclude that the distribution patterns that we observed reflect microbial endemism, we can say that abundant bacteria exhibit a pattern of distribution both within (Figs. 3 and 4) and between habitat types (Fig. 1A), with most organisms being found in no more than one assemblage. It is widely recognized that, within individual assemblages, few taxa are abundant and most taxa are rare (Curtis et al. 2002). Here we identify a similar pattern across assemblages: relatively few taxa are cosmopolitan and the vast majority are restricted to individual assemblages. That said, we caution that improved sequencing technology may alter these interpretations in the future. Indeed, Preston’s ‘Veil Line’ concept suggests that many organisms exhibit a normal distribution pattern which can be obscured by undersampling (Preston 1948). He suggests that many organisms with so-called rare distribution patterns will reveal a more intermediate distribution in exhaustively sampled assemblages.
Abundant bacteria are more widely distributed
We observed that, across soil assemblages, abundant organisms were more likely to be widely distributed (Fig. 5). This pattern was observed at two very different spatial scales: within a single hectare of rainforest soil and within a variety of soils sampled across two continents. A positive relationship between abundance and distribution among soil bacteria is also apparent in an examination of the pyrosequencing data from the Fulthorpe et al. (2008) study: 50% of the top 10 most abundant organisms were found in more than one of the four soils that they analyzed. Likewise, Pommier et al. (2007) discovered a positive relationship between OTU abundance and distribution across coastal seawater samples. Spain et al. (2009) reported a similar pattern in the analysis of their large 16S rRNA gene clone library dataset from a grassland soil: they observed that the most abundant orders of Proteobacteria were more highly distributed among other environments. Sloan et al. (2006) also described a positive relationship between abundance in distribution in sewage treatment facilities, estuaries, lakewater and microbiome samples.
As mentioned above, it is difficult to discern a particular organism’s relative abundance from our compiled clone library dataset because of inconsistent reporting and screening methods. However, it is noteworthy that many taxa that are cosmopolitan within habitat types (Fig. 3) have been identified as abundant members of their respective environments in other studies (Janssen 2006; Newton et al. 2007; Rusch et al. 2007). This may be a general feature of all of life, as the positive correlation between the distribution and abundance of macroorganisms has been well documented (Brown 1984). For macroorganisms, distribution varies in geographic scale by more than twelve orders of magnitude, and as techniques to sample microbial communities improve, it will permit us to assess if microbial distribution patterns exhibit a similar level of variation.
What could cause the positive relationship between abundance and distribution? We propose three, non-mutually exclusive possibilities. First, it could simply reflect the fact that these organisms are easier to detect within our current sampling limitations. Another possibility is that higher local population sizes enable wider dispersal potentials. A prevailing hypothesis for microbial biogeography states that population sizes are extremely large and thus dispersal is not limited (Fenchel & Finlay 2004). However, not all organisms are abundant within a community; in fact, most organisms are rare (Sogin et al. 2006; Ashby et al. 2007). Thus, the larger population sizes of the most abundant organisms may facilitate their dispersal and help drive the positive relationship between abundance and distribution. Indeed, some calculations suggest that very rare soil organisms may be present at densities of 1 cell per 27 km2 (Curtis et al. 2002), which would undoubtedly limit their distribution potential. Sloan et al. (2006) described a near-neutral model for microbial community assembly, in which the distribution of taxa is largely determined by immigration and chance. Random assembly processes would lead to a positive relationship between distribution and relative abundance (Sloan et al., 2006) and may partially explain the within-habitat distribution patterns observed here. Finally, the relationship between abundance and distribution may reflect a positive relationship between regional and global distributions, a pattern that has been shown for macroorganisms (reviewed in Prinzing et al. 2004). Because the way that we sample microorganisms (e.g., 1 gram of soil) is far too coarse to permit the examination of single communities, we are actually examining the composition of many communities within a single assemblage (Grundmann 2004). Indeed, high ‘regional’ distribution patterns may cause high abundance values within a sampled assemblage, thus the lognormal-shaped species abundance curves observed within a single community (Curtis et al. 2002) may actually reflect the same phenomena as the distribution patterns that we observed between assemblages (Figs. 1–4).
Conclusion
We emphasize that our results apply to the most abundant organisms that are detected by contemporary sequencing technology. New technological advances are on the horizon, and it is unknown how the patterns that we observed may change when more assemblages can be completely sampled and analyzed. Additionally, methods to assay the entire genomic complement of individual assemblages will become easier in the near future, enabling the analysis of “functional biogeography” (Green et al. 2008) to better understand the process-level implications of the observed patterns of bacterial distribution.
Despite these caveats, our results suggest that while macro-scale environmental factors structure the ecological distribution of bacterial taxa, most bacteria demonstrate a limited distribution within habitat types. We also show a positive relationship between the abundance and distribution of soil bacteria within habitat types. Given the high degree of genetic differentiation between even very closely related lineages of bacteria living in close proximity (Thompson et al. 2005), our results add to the growing body of data that support that the diversity of the overall bacterial metagenome is enormous. The mechanisms governing microbial distribution remain poorly known, but our analyses provide a framework with which to test the importance of macro-ecological environmental gradients, relative abundance, the ecological strategies of individual taxa and neutral processes in structuring microbial communities.
Experimental Procedures
Datasets
To examine the distribution of different bacterial taxa within and between different habitat types, we expanded upon the 16S rRNA gene clone library dataset compiled by Lozupone and Knight (2007) so that it now includes 28,115 16S rRNA sequences, derived from 238 samples taken across 14 different habitat types (Table S1). This dataset was assembled from studies examining the microbial communities of natural environments using 16S rRNA gene cloning and sequencing targeted toward all bacteria. Specifically, sequences were identified from the ENV database of GenBank, reference information was extracted for each record, the sequences that had the same title were grouped, and studies that were associated with the most sequences were selected. Since a single study can report sequences from different assemblages and different habitat types, the sequences were divided into assemblages and habitat types using annotations from the associated publications. Here, habitat types were defined at the macroscale and ranged from soil and seawater to insect and sponge-associated assemblages (Table S1). These clone libraries contained an average of 118 16S rRNA gene sequences per assemblage with a minimum of 20 and a maximum of 836 sequences.
Next, to determine the distribution of bacteria between relatively well-sampled assemblages within soils as well as to examine the relationship between relative abundance and distribution, we examined two pyrosequencing-generated 16S rRNA gene datasets from soils. The first was taken from Lauber et al. (2009) and featured nucleotides 27 to 338 (Escherichia coli numbering) of the 16S rRNA gene (regions V1 and V2). In this study, 88 different soils from across North and South America that were first described by Fierer and Jackson (2006) were analyzed. An average of 1,501 classifiable sequences per soil was obtained with a maximum of 2,167 and a minimum of 1,047 sequences. In addition, we introduce a new pyrosequencing-based dataset of 16S rRNA genes from 27 soil samples obtained from within a single hectare of lowland tropical rainforest soil from the Osa Peninsula in Costa Rica (see Cleveland and Townsend (2006) for site description). Samples were taken from litter removal, litter augmentation, and precipitation exclusion manipulations as well as from control plots (Wieder et al. 2009). Control plots were sampled in April, June and October of 2008, while plots subjected to experimental manipulations were sampled in June and October of 2008. For each treatment at each time point, three replicate plots were obtained for a total of 27 soil samples. For each plot, the top 5 cm of soil were aseptically collected and DNA was extracted using PowerSoil DNA Isolation kits (MO BIO, Carlsbad, CA, USA). Error-corrected bar-coded pyrosequencing was performed as described by Fierer et al. (2008) with the sequencing performed at the Environmental Genomics Core Facility at the University of South Carolina on a Roche FLX 454 pyrosequencing machine. Data were processed as described by Fierer et al. (2008) and Hamady et al. (2008) with an average of 1,384 sequences obtained per soil (range of 1,087 – 2,030).
Data analysis
Taxonomy was assigned to clone library sequences using BLAST with a minimum e-value cutoff of 1e−12, minimum identity of 88%, and a word size of 38 against the Greengenes database and the Hugenholtz taxonomic nomenclature (DeSantis et al. 2006). For the pyrosequencing data, sequences were removed from the analysis if they were <200 or >400 nt, had a quality score <25, contained ambiguous characters, contained an uncorrectable barcode, or did not contain the primer sequence. For the clone library dataset, operational taxonomic units (OTUs) were selected at four different levels of minimum sequence identity: 98%, 95%, 92% and 89% using cd-hit (Li & Godzik 2006). The total number of OTUs at each level of phylogenetic resolution was 14,627 (98%), 9,479 (95%), 6,348 (92%), and 4,383 (89%). For the pyrosequencing datasets we only classified OTUs at the 97% similarity level because of difficulties with consistency when examining shorter pyrosequenced fragments at different levels of phylogenetic resolution as compared to full-length 16S rRNA genes (Elshahed et al. 2008). For the Lauber et al. (2009) dataset, 50,891 OTUs were examined while 10,374 OTUs were obtained from the tropical forest dataset.
For each dataset (clone library data and both of the pyrosequencing datasets) an assemblage by OTU presence-absence matrix was created. For the pyrosequencing datasets, matrices containing the relative sequence abundances of different OTUs in different assemblages were also created. Distribution analysis within and between assemblages and habitats was performed in Microsoft Excel and in MySQL using phpMyAdmin as a graphical user interface. We then tested the significance of macro-scale habitat in structuring the presence/absence of bacteria at all levels of phylogenetic resolution in the clone library data using PERMANOVA analyses in PRIMER v6 (Anderson 2001).
Supplementary Material
Acknowledgments
The authors recognize support from the National Science Foundation grants DEB-0136957 and DEB-0852916. Antonio Peña-Gonzales provided support and advice implementing MySQL. The authors also wish to thank an anonymous reviewer for providing a thorough and thoughtful critique of the manuscript.
References
- Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- Ashby MN, Rine J, Mongodin EF, Nelson KE, Dimster-Denk D. Serial analysis of rRNA genes and the unexpected dominance of rare members of microbial communities. Appl Environ Microbiol. 2007;73:4532–4542. doi: 10.1128/AEM.02956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auguet J-C, Barberan A, Casamayor EO. Global ecological patterns in uncultured Archaea. ISME Journal. 2010;4:1–9. doi: 10.1038/ismej.2009.109. [DOI] [PubMed] [Google Scholar]
- Brown JH. On the relationship between abundance and distribution of species. Am Nat. 1984;124:255–279. [Google Scholar]
- Brown JH, Lomolino MV. Biogeography. 2 edn. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- Burtscher MM, Zibuschka F, Mach RL, Lindner G, Farnleitner AH. Heterotrophic plate count vs. in situ bacterial 16S rRNA gene amplicon profiles from drinking water reveal completely different communities with distinct spatial and temporal allocations in a distribution net. Water SA. 2009;35:495–504. [Google Scholar]
- Cho JC, Tiedje JM. Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl Environ Microbiol. 2000;66:5448–5456. doi: 10.1128/aem.66.12.5448-5456.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland CC, Townsend AR. Nutrient additions to a tropical rain forest drive substantial soil carbon dioxide losses to the atmosphere. PNAS. 2006;103:10316–10321. doi: 10.1073/pnas.0600989103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis TP, Sloan WT, Scannell JW. Estimating prokaryotic diversity and its limits. PNAS. 2002;99:10494–10499. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dojka MA, Harris JK, Pace NR. Expanding the known diversity and environmental distribution of an uncultured phylogenetic division of bacteria. Appl Environ Microbiol. 2000;66:1617–1621. doi: 10.1128/aem.66.4.1617-1621.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshahed MS, Youssef NH, Spain AM, Sheik C, Najar FZ, Sukharnikov LO, Roe BA, Davis JP, Schloss PD, Bailey VL, Krumholz LR. Novelty and uniqueness patterns of rare members of the soil biosphere. Appl Environ Microbiol. 2008;74:5422–5428. doi: 10.1128/AEM.00410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenchel T, Finlay BJ. The ubiquity of small species: Patterns of local and global diversity. Bioscience. 2004;54:777–784. [Google Scholar]
- Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. PNAS. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. PNAS. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulthorpe RR, Roesch LFW, Riva A, Triplett EW. Distantly sampled soils carry few species in common. ISME Journal. 2008;2:901–910. doi: 10.1038/ismej.2008.55. [DOI] [PubMed] [Google Scholar]
- Green J, Bohannan BJM. Spatial scaling of microbial biodiversity. Trends Ecol & Evol. 2006;21:501–507. doi: 10.1016/j.tree.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Green JL, Bohannan BJM, Whitaker RJ. Microbial biogeography: From taxonomy to traits. Science. 2008;320:1039–1043. doi: 10.1126/science.1153475. [DOI] [PubMed] [Google Scholar]
- Grundmann GL. Spatial scales of soil bacterial diversity - the size of a clone. FEMS Microbiol Ecol. 2004;48:119–127. doi: 10.1016/j.femsec.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nature Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner-Devine MC, Lage M, Hughes JB, Bohannan BJM. A taxa-area relationship for bacteria. Nature. 2004;432:750–753. doi: 10.1038/nature03073. [DOI] [PubMed] [Google Scholar]
- Horner-Devine MC, Silver JM, Leibold MA, Bohannan BJM, Colwell RK, Fuhrman JA, Green JL, Kuske CR, Martiny JBH, Muyzer G, Ovreas L, Reysenbach AL, Smith VH. A comparison of taxon co-occurrence patterns for macro- and microorganisms. Ecology. 2007;88:1345–1353. doi: 10.1890/06-0286. [DOI] [PubMed] [Google Scholar]
- Hugenholtz P, Goebel BM, Pace NR. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen PH. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol. 2006;72:1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ Microbiol. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WZ, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Knight R. Global patterns in bacterial diversity. PNAS. 2007;104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan M, Martinko J, Parker J. Brock biology of microorganisms. Upper Saddle River, NJ: Prentice-Hall; 2000. [Google Scholar]
- Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, Morin PJ, Naeem S, Ovreas L, Reysenbach AL, Smith VH, Staley JT. Microbial biogeography: putting microorganisms on the map. Nature Rev Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- Newton RJ, Jones SE, Helmus MR, McMahon KD. Phylogenetic ecology of the freshwater Actinobacteria acI lineage. Appl Environ Microbiol. 2007;73:7169–7176. doi: 10.1128/AEM.00794-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguez AM, Arita HT, Escalante AE, Forney LJ, Garcia-Oliva F, Souza V. Microbial macroecology: highly structured prokaryotic soil assemblages in a tropical deciduous forest. Glob Ecol Biogeography. 2005;14:241–248. [Google Scholar]
- Pommier T, Canback B, Riemann L, Bostrom KH, Simu K, Lundberg P, Tunlid A, Hagstrom A. Global patterns of diversity and community structure in marine bacterioplankton. Mol Ecol. 2007;16:867–880. doi: 10.1111/j.1365-294X.2006.03189.x. [DOI] [PubMed] [Google Scholar]
- Preston FW. The commonness, and rarity, of species. Ecology. 1948;29:254–283. [Google Scholar]
- Prinzing A, Ozinga WA, Durka W. The relationship between global and regional distribution diminishes among phylogenetically basal species. Evolution. 2004;58:2622–2633. doi: 10.1111/j.0014-3820.2004.tb01616.x. [DOI] [PubMed] [Google Scholar]
- Prosser JI, Bohannan BJM, Curtis TP, Ellis RJ, Firestone MK, Freckleton RP, Green JL, Green LE, Killham K, Lennon JJ, Osborn AM, Solan M, van der Gast CJ, Young JPW. Essay - The role of ecological theory in microbial ecology. Nature Rev Microbiol. 2007;5:384–392. doi: 10.1038/nrmicro1643. [DOI] [PubMed] [Google Scholar]
- Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD, Daroub SH, Camargo FAO, Farmerie WG, Triplett EW. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME Journal. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu DY, Eisen JA, Hoffman JM, Remington K, Beeson K, Tran B, Smith H, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, Li K, Kravitz S, Heidelberg JF, Utterback T, Rogers YH, Falcon LI, Souza V, Bonilla-Rosso G, Eguiarte LE, Karl DM, Sathyendranath S, Platt T, Bermingham E, Gallardo V, Tamayo-Castillo G, Ferrari MR, Strausberg RL, Nealson K, Friedman R, Frazier M, Venter JC. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through Eastern Tropical Pacific. PLOS Biology. 2007;5:398–431. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Status of the microbial census. Microbiol Mol Biology Rev. 2004;68 doi: 10.1128/MMBR.68.4.686-691.2004. 686-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan WT, Lunn M, Woodcock S, Head IM, Nee S, Curtis TP. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ Microbiol. 2006;8:732–740. doi: 10.1111/j.1462-2920.2005.00956.x. [DOI] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ. Microbial diversity in the deep sea and the underexplored "rare biosphere". PNAS. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain AM, Krumholz LR, Elshahed MS. Abundance, composition, diversity and novelty of soil Proteobacteria. ISME Journal. 2009;3:992–1000. doi: 10.1038/ismej.2009.43. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E, Goebel BM. A place for DNA-DNA Reassociation and 16S ribosomal-RNA sequence analysis in the present species definition in bacteriology. Int J System Bacteriol. 1994;44:846–849. [Google Scholar]
- Tanner MA, Goebel BM, Dojka MA, Pace NR. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl Environ Microbiol. 1998;64:3110–3113. doi: 10.1128/aem.64.8.3110-3113.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JR, Pacocha S, Pharino C, Klepac-Ceraj V, Hunt DE, Benoit J, Sarma-Rupavtarm R, Distel DL, Polz MF. Genotypic diversity within a natural coastal bacterioplankton population. Science. 2005;307:1311–1313. doi: 10.1126/science.1106028. [DOI] [PubMed] [Google Scholar]
- Van der Gucht K, Cottenie K, Muylaert K, Vloemans N, Cousin S, Declerck S, Jeppesen E, Conde-Porcuna JM, Schwenk K, Zwart G, Degans H, Vyverman W, De Meester L. The power of species sorting: Local factors drive bacterial community composition over a wide range of spatial scales. PNAS. 2007;104:20404–20409. doi: 10.1073/pnas.0707200104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrik B, Kudiev D, Abdivasievna UA, Kukor JJ, Zystra GJ, Kerkhof L. Biogeography of actinomycete communities and type II polyketide synthase genes in soils collected in New Jersey and Central Asia. Appl Environ Microbiol. 2007;73:2982–2989. doi: 10.1128/AEM.02611-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker RJ, Grogan DW, Taylor JW. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science. 2003;301:976–978. doi: 10.1126/science.1086909. [DOI] [PubMed] [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: The unseen majority. PNAS. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieder WR, Cleveland CC, Townsend AR. Controls over leaf litter decomposition in wet tropical forests. Ecology. 2009;90:3333–3341. doi: 10.1890/08-2294.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.