Abstract
Bacteria come in a wide variety of shapes and sizes. The true picture of bacterial morphological diversity is likely skewed due to an experimental focus on pathogens and industrially relevant organisms. Indeed, most of the work elucidating the genes and molecular processes involved in maintaining bacterial morphology has been limited to rod- or coccal-shaped model systems. The mechanisms of shape evolution, the molecular processes underlying diverse shapes and growth modes, and how individual cells can dynamically modulate their shape are just beginning to be revealed. Here we discuss recent work aimed at advancing our knowledge of shape diversity and uncovering the molecular basis for shape generation in non-canonical and morphologically complex bacteria.
Keywords: Peptidoglycan, bacterial shape, morphology, pleomorphism, morphological engineering
The Nature of Bacterial Shape
Casting one’s gaze upon the array of observed bacterial shapes and sizes, one wonders about the why and the how of morphogenesis. It is one thing to simply catalog the range of morphologies, from the familiar rods and cocci to the flamboyant star-shaped or appendaged bacteria and including size ranges spanning almost six orders of magnitude, from the nanometer to the millimeter scale [1]. Determining why diverse bacterial shapes exist is far from trivial: a shape might be a compromise between different selective pressures and determining the adaptive value of a particular shape can be difficult [2]. Yet shapes are maintained over evolutionary time scales and, under genetic control, are faithfully reproduced each generation. We can thus infer that at least some shapes are the result of adaptation and selective pressures. For example, pathogenic bacteria with mutations that alter shape often have altered colonization or virulence properties in infection models, suggesting that shape itself is an adaptation for virulence or that the host environment provides a selective pressure driving the adopted morphology [3].
How bacterial shapes are determined is somewhat easier to study than why, and great progress is being made in understanding the molecular mechanisms that can generate different shapes. Understanding how bacteria maintain cell shape through growth and division, how they actively restructure their morphologies in response to environmental conditions, and how morphologies evolve is a worthy endeavor. Such knowledge will benefit, among other things, efforts to impede pathogen persistence, proliferation, and virulence [4], as well as efforts to engineer shape for industrial and agricultural applications.
The major shape determinant of bacteria is the peptidoglycan (PG) cell wall. Some bacteria are able to adopt a spherical or pleomorphic cell wall-deficient state known as the L-form [5]. Much of what we know about the mechanisms of PG synthesis comes from the study of a few model organisms, such as the spherical Staphylococcus aureus, the ovoid Streptococcus pneumoniae, and the rod-shaped Escherichia coli, Bacillus subtilis, Agrobacterium tumefaciens, and Caulobacter crescentus. The PG cell wall is a macromolecular polymer composed of alternating β-1,4-linked N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) sugars, forming strands that are cross-linked by short peptide side chains attached to the MurNAc subunit (Figure 1). While the basic PG precursor subunit (Figure 1, bottom right) is broadly conserved in bacteria, the chemical composition (peptide side chain modification and crosslink type) and structure (degree of crosslinking and chain length) can vary widely [6]. The eventual morphology of a cell arises from a complex interplay between the proteins and regulatory elements that compose the PG biosynthetic machinery (Figure 1). In model rod-shaped bacteria, such as E. coli and B. subtilis, there are at least two PG remodeling complexes that give rise to their shape: the elongasome (Figure 1, bottom left) inserts new PG along the lateral side-wall of the cell as it grows, while the divisome (Figure 1, bottom center) is responsible for the formation of the division septum and cytokinesis [7].
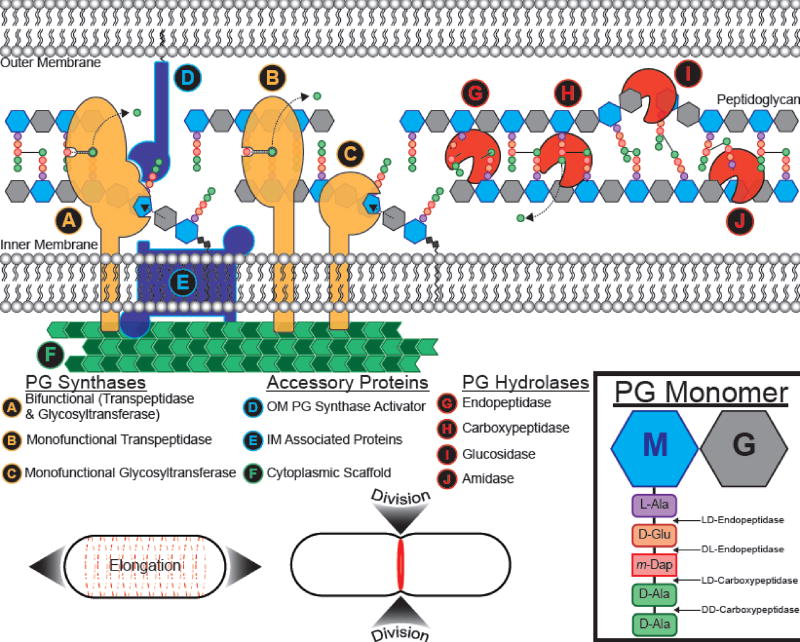
Figure 1. A simplified accounting of peptidoglycan remodeling components in Gram-negative bacteria.
Due to the scope of this review we will only briefly describe common themes in the proteins involved in PG remodeling, using Gram-negatives as an example (for detailed reviews regarding PG remodeling enzymes, start with [7, 11, 148]). Cartoons are not meant to imply an experimentally determined structure for the proteins. Adapted in part from [11]. Inset, right: structure of uncrosslinked PG monomer depicting the disaccharide N-Acetylmuramic acid ("M") and N-Acetylglucosamine ("G") and the pentapeptide stem, from proximal to distal, L-alanine ("L-Ala"), D-glutamic acid ("D-Glu"), meso-diaminopimelic acid ("m-Dap"; a derivative of lysine), and two D-alanines ("D-Ala"). From proximal to distal, the stem peptide isoform pattern is "L-D-L-D-D". Enzymes that break bonds between the stem peptides are prefixed by the isoforms for the peptides they separate. The "makers", or (A–C) PG synthases, assemble the nascent PG meshwork. (A&C) Glycosyltransferases polymerize PG monomers into glycan strands, while (A&B) transpeptidases form crosslinks between the stem peptides to form the sacculus. (D–F) Accessory and SEDS proteins include: (D) Outer membrane anchored PG synthase activators and (E) inner membrane (IM) associated proteins. (E) IM associated proteins include enzymes that synthesize PG monomers in the cytoplasm and flippases that flip the monomers across the IM to the periplasm, SEDS family proteins [13–16], and proteins that help anchor components on the PG synthesis machinery to (F) cytoplasmic scaffolding proteins. (F) Scaffolding proteins recruit various cytoplasmic and IM proteins associated with PG synthesis and localize synthesis activity. The "breakers", or (G–J) PG hydrolases, modify PG after synthesis. (G) Endopeptidases can break crosslinks ("DD-endopeptidases") or peptide linkages ("LD-endopeptidases" or "DL-endopeptidases") of non-terminal amino acids. (H) Carboxypeptidases trim the terminal stem peptide. Shown is removal of the terminal, fifth position D-Ala. (I) Glucosidases, of which there are different classes depending on which bond is broken and the type of catalytic reaction used, cleave the glycan strands. (J) Amidases remove the peptide stem from N-Acetylmuramic acid ("M", inset) in the glycan chain. Bottom, cell growth and division typical of E. coli: Dispersed growth along the long axis elongates the cell (left, dashed red lines), and a division septum (right, solid red ring) is formed at the midcell allowing the daughter cells to recapitulate the initial shape and size of the mother cell.
The elongasome and divisome are large complexes, organized by cytoplasmic, cytoskeleton-like scaffolding proteins (see Glossary). These complexes contain inner membrane (IM) spanning elements and a multitude of periplasmic proteins and enzymes (both membrane bound and soluble), such as PG synthases and PG hydrolases (Figure 1). Due to the scope of this review we will briefly describe common themes involved in cell elongation and division using E. coli as an example (for detailed reviews, start with [7–12]). The actin homolog MreB serves as the scaffolding protein to organize the elongasome. In the model for rod cell elongation, MreB forms filaments that bind to the inner membrane and interact with PG remodeling enzymes, including PG synthases known as penicillin binding proteins (PBPs; see Glossary), PG precursor synthesis enzymes, and PG hydrolases. Cell wall synthesis directionally drives MreB motion, elongating the cell by insertion of new PG in a spiral-like pattern. The tubulin homolog FtsZ is a scaffolding protein that forms a ring-like structure called the "Z-ring", which marks and assembles the site of division or the "divisome". Two negative regulators help position Z-ring assembly at the midcell: First, the Min system inhibits Z-ring formation, and as Min system proteins oscillate from pole to pole, mid-cell Z-ring formation is favored. In the second phenomenon, known as nucleoid occlusion (see Glossary), the E. coli protein SlmA binds to several specific sites on the chromosome and thus blocks cell division over the unreplicated nucleoid by both sequestering free FtsZ and disrupting FtsZ polymers. Once formed at the mid-cell, and throughout the process of septation, the Z-ring recruits and localizes the various divisome proteins. Bifunctional PBPs (Figure 1A) have long been considered the primary enzymes for PG synthesis, yet recent work characterizing SEDS (shape, elongation, division, and sporulation) family proteins (Figure 1E) has challenged this notion. SEDS are a new class of PG glycosyltransferases, distinct from PBPs but functionally, and often genetically, linked to monofunctional PBP transpeptidases, (Figure 1B) [13–16]. The best studied SEDS are RodA and FtsW, which are critical to cell elongation and division, respectively. This exciting discovery of a new class of PG synthases raises question about the primary role of bifunctional PBPs and if SEDS can be targeted for antibiotic development.
While a conserved set of proteins participates in PG synthesis and remodeling (Figure 1) [11, 17], simple shapes such as the rod can arise through a number of distinct mechanisms, including dispersed growth along the length of the cell, elongation from one or both poles, or widening followed by longitudinal division along the long axis of the cell (see Outstanding Questions) [18–20]. How are non-canonical bacterial shapes generated at the molecular level? And how do we identify the proteins and regulatory elements involved in these non-canonical systems? Below we discuss various inroads into the molecular basis of diverse bacterial morphologies. Novel shapes, it is becoming clear, are underpinned by novel strategies for regulating and localizing PG modifying enzymes.
I Spy With My Little Eye…Observing Shape Variation
Many of the biological sciences, such as botany or zoology, are rooted in a tradition of natural history, with an emphasis on observation rather than experimentation [21]. Much of the natural history of bacterial diversity, including descriptions of bacteria that are star-shaped, grow prosthecae (see Glossary), or exhibit any number of deviations from the canonical rod or sphere, tends to date from before 1980 [22]. While some current journals do dedicate space to describing new species, the modern approach to non-canonical shapes is often "look, but don’t touch". This is in many ways understandable; model bacteria such as E. coli and B. subtilis are genetically tractable and easy to culture, and these model systems have certainly provided a wealth of knowledge regarding bacterial shape determination at the molecular level.
Stepping outside the realm of model organisms can be challenging, not the least because new genetic systems and culturing methods must often be developed. Take the example of a new Methylococcaceae morphotype hiding in plain sight. Aerobic methanotrophs display a variety of cell shapes, including straight and curved rods, cocci and ovoids, vibrioids and pear-like cells [23]. In an enrichment for methanotrophic cultures sampled from a peat bog, three major cell morphotypes in a mixed population were observed: rods, large cocci, and a shape that had never been observed in methanotrophs: spiral (or helical) shaped cells (Figure 2A) [24]. While the rod and coccal species were readily isolated through standard methods, it took years of continuous purification work to generate a culture enriched for the spirillum, presumably because of its preference for micro-oxic conditions. The discovery of such a morphotype had been elusive in large part due to commonly used culture methods. Widely used culture media, such as LB or NA, are unlikely to be optimal for most bacteria and may mask other morphologies hiding in plain sight, as even among well-studied, "domesticated" bacteria, canonical shapes often vary dramatically depending on the growth conditions. With the advent of high-throughput culturing methods [25] and utilizing high-throughput sequencing data to model metabolic networks for optimal culture conditions [26–29], the future is bright for bypassing this cultivation bottleneck [30]. While developing a true picture of bacterial morphological diversity is important, simply assembling a menagerie at which to marvel should not be the end goal; we should seek to understand how these shapes are generated, both at the molecular level and in the context of microbial diversity and evolution. Let us look at recent advances in our knowledge of shape diversity and the molecular basis for shape generation and modification in non-canonical, morphologically complex bacteria.
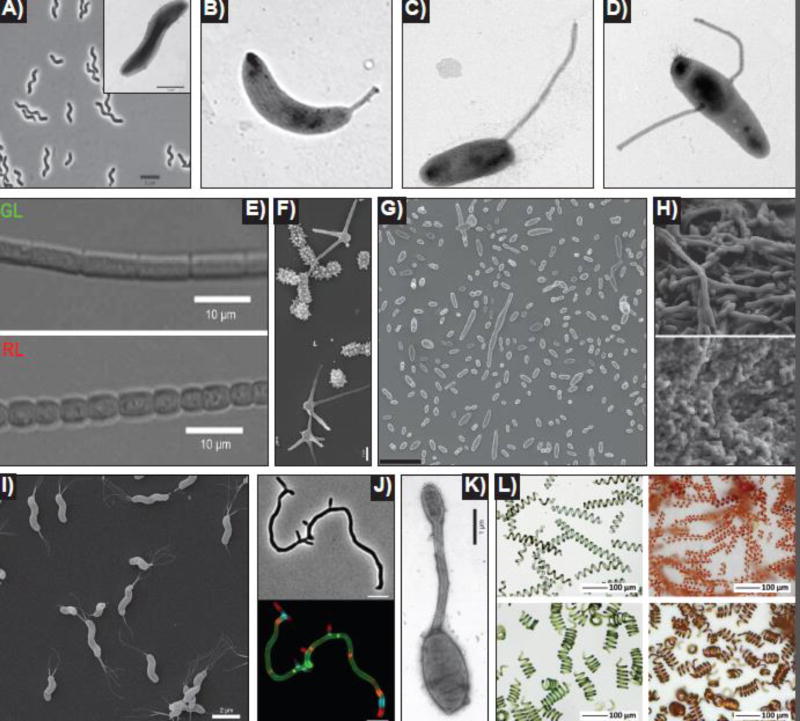
Figure 2. Diverse bacterial morphologies.
A) Uncharacterized spiral-shaped methanotroph. Phase contrast with inset electron micrograph. Adapted from [24]. B)Caulobacter crescentus. Single polar prostheca [32]. C)Asticcacaulis excentricus. Single sub-polar prostheca [32]. D)Asticcacaulis biprosthecum. Two bilateral prosthecae [32]. E)Fremyella diplosiphon. Complimentary chromatic adaptation (CCA) mediated pleomorphism. Cells grown in green light (GL, top) are elongated and rectangular. Cells grown in red light (RL, bottom) are short and rounded. Adapted from [149]. F)Prosthecomicrobium hirschii. Electron micrograph showing short- and long-prosthecate morphotypes. Adapted from [73]. G)Dinoroseobacter shibae. Scanning electron micrograph showing the inherent morphological heterogeneity of wild-type D. shibae. Scale bar = 5 µm. [77]. H)Lactococcus lactis. Scanning electron micrograph showing different regions of the same L. lactis biofilm. The upper region of the biofilm contains elongated rods (top), while the lower region contains ovoid cells (bottom). Adapted from [60]. I)Helicobacter pylori. Scanning electron microscope images of wild-type H. pylori. Adapted from [96]. J)Streptomyces venezuelae. Virtual time-lapse of polarly growing S. venezuelae labeled with a long pulse (cell body) of green fluorescent D-amino acid (FDAA), followed by sequential short pulses of orange, blue, and red FDAAs (apical tips). Top = phase, bottom = fluorescence, scale bar = 5 µm. (Image courtesy of Yen-Pang Hsu, Indiana University). K)Hyphomonas adhaerens (related to H. neptunium). The mother cell (bottom), the prostheca (middle), and the developing daughter bud (top) are visible. Adapted from [150]. L)Spirulina (Arthrospira platensis). Spirulina biotemplated microcoils. The helical pitch of the cyanobacteria can be modified by tuning the culture conditions (left panels). Copper microcoils are produced through a electroless plating technique using Spirulina as the template (right panels). Adapted from [121].
Molecular Basis for Morphological Plasticity and Pleomorphism
Morphological plasticity refers to the ability of a bacterial cell to dynamically change its shape in response to the environment [31]. Pleomorphism, while sometime used interchangeably with morphological plasticity, refers to the population level, where a species can assume multiple forms through processes such as a programmed life cycle or dynamic morphological changes. For the purposes of this review, we will use pleomorphism as a general term for these processes. When describing the shape of a certain bacterial species, we often speak as if shape is immutable (i.e. E. coli are rods, S. aureus are spheres, Helicobacter pylori are helical), yet it is becoming clear that morphological plasticity and pleomorphism are common strategies employed by bacteria. In fact, it may be far more common than we could have imagined (see Outstanding Questions).
One early observation of pleomorphism was bacterial cells during swarming, where several species elongate when transitioning from the motile swimmer state to the swarmer state [3]. Another example is the regulation of prosthecae [32–34]. In the prosthecate Alphaproteobacteria, such as C. crescentus (Figure 2B) or Asticcacaulis spp. (Figure 2C–D), phosphate starvation stimulates prosthecae synthesis, either elongating extant prosthecae or producing prosthecae where previously there were none [32, 35–38]. In addition, the impact of cell morphology on biofilm formation is becoming increasingly clear. Computer modeling and empirical data from E. coli suggests that cell shape affects spatial patterns and composition within microbial communities such as biofilms [39]. Although molecular studies of the shape of bacterial symbionts are rare, in at least two cases host factors controlling symbiont shape have been identified: nodule-specific cysteine rich (NCR) peptides and Coleoptericin A inhibit cell division in symbionts of legumes and weevils, respectively, leading to cell gigantism and polyploidy [40]. Finally, the range of morphologies observed in H. pylori cultures grown from clinical isolates include curved and straight rods of various lengths and helicities [41], suggesting that, despite adopting a predominantly helical form, it may be advantageous for H. pylori to maintain a pleomorphic population and assume alternative morphologies. Below we will discuss several recent observations that not only illustrate the ubiquity of pleomorphism but also identify molecular mechanisms that drive these transitions.
Shedding Light on Cyanobacterial Morphology
Photosynthetic cyanobacteria are found in almost every habitat on earth and display an array of morphologies, including unicellular and filamentous forms [42, 43]. In addition, it has been well documented that cyanobacteria undergo shape changes directly linked to growth and/or development [44]. Vegetative cells change shape in response to light [45] or differentiate into specialized forms: heterocysts capable of fixing nitrogen [46]; sporelike akinetes [47]; or hormogonia, short filaments made of small cells that exhibit gliding motility [48]. Despite an observed range of species-specific morphologies and a propensity for pleomorphism, very little is known about the underlying molecular mechanisms governing shape determination or morphological plasticity in cyanobacteria. The few genes that have been described are referred to as "morphogenes".
The amidase (Figure 1J) AmiC2 directly affects peptidoglycan structure and is pivotal for multicellular development in the cyanobacterium Nostoc punctiforme (Table 1) [49]. N. punctiforme daughter cells do not septate, as in unicellular bacteria such as E. coli, but retain a shared cross wall in the septum. It is thought that the multiprotein complexes which facilitate intercellular communication, and are necessary for cellular differentiation and filamentation, cannot form because the PG of the shared cross wall in amiC2 mutants is so thick [49]. Possibly due to a cell cycle delay, amiC2 mutant cells are also larger than wild-type cells (Table 1) [49].
Table 1.
Genes that impact bacterial morphology
| Nostoc punctiforme | amiC2 | Cell wall amidase |
|
| Uropathogenic E. coli (UPEC) | sulA | Binds FtsZ in presence of GFP; prevents Z-ring formation |

|
| damX | SPOR domain containing protein; localizes to the septal ring and binds PG | ||
| Asticcacaulis excentricus and Asticcacaulis biprosthecum | spmX | Phage muramidase domain containing protein; required for stalk synthesis |
|
| Helicobacter pylori | csd1 | LytM (peptidase family M23) DD-endopeptidase |
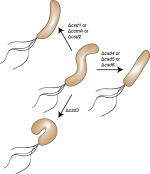
|
| ccmA | Bactofilin; cytoskeletal, polymer-forming, proteins | ||
| csd2 | LytM (peptidase family M23) DD-endopeptidase | ||
| csd3 | DD-endopeptidase and DD-carboxypeptidase activity | ||
| csd4 | DL-carboxypeptidase (note that because Csd4 removes a terminal, uncrosslinked amino acid, it is referred to as a carboxypeptidase and not an endopeptidase) | ||
| csd5 | No known enzymatic domain but may modulate Csd4 activity (directly interacts with Csd4 and the dipeptide product of the Csd4-catalyzed PG reaction) | ||
| csd6 | LD-carboxypeptidase | ||
| Streptomyces | divIVA | Coiled coil-rich protein; assembles polarisome |
|
| scy | Coiled coil-rich protein; component of polarisome | ||
| filP | Coiled coil-rich protein; component of polarisome | ||
| Hyphomonas neptunium | WT | n/a |
|
| dacB | DD-carboxypeptidase |
|
|
| dacL | DD-carboxypeptidase | DacL localizes to the new pole at the start of prostheca synthesis | |
| lmdC | LytM (peptidase family M23) DD-endopeptidase | Could not be deleted. Essential? | |
| lmdE | LytM (peptidase family M23) DD-endopeptidase |
|
|
| amiC | Cell wall amidase |
|
|
| pbp1X | Bifunctional DD-transpeptidase |
|
|
| pbp2 | Monofunctional DD-transpeptidase | Diffuse or patchy localization pattern until the onset of prostheca formation, when it condenses at the prosthecate pole; once a visible bud has formed, the polar complex disappears followed by patchy foci appearing in the mother cell, the prostheca, and the bud. This suggests that PBP2 may contribute to all aspects of H. neptunium growth. Could not be deleted. Essential? | |
| pbp3 | Monofunctional DD-transpeptidase | PBP3 predominantly localizes to the division site of late budding cells where it remains briefly associated with the new pole but then disperses evenly within the cell once a visible prostheca has formed. Consistent with its role in other bacteria, PBP3 thus appears to be an integral part of the divisome involved in septal cell wall remodeling. Could not be deleted. Essential? | |
| mreB | Actin homolog; serves as the scaffolding protein to organize the elongasome |
|
|
| rodZ | Cytoskeletal protein that mediates MreB circumferential movement and couples MreB to cell wall synthesis enzymes | Exhibits similar localization patterns to MreB. |
Many cyanobacteria utilize complementary chromatic acclimation (CCA), a process whereby light-harvesting structures and cellular processes such as morphogenesis are dynamically regulated in response to light conditions such as color and intensity [50]. For example, Arthrospira platensis (Figure 2L) are filamentous, helical cyanobacteria whose cells transition from an elongated, loose helix under low solar ultraviolet (UV) light exposure to a tightly compressed helix when exposed to high solar UV levels [51]. The freshwater, filamentous cyanobacterium Fremyella diplosiphon (Figure 2E), a model organism for CCA and photomorphogenesis, forms short filaments of small, rounded cells under red light and long filaments of rectangular cells under green light (Figure 2E) [45, 52–54]. The F. diplosiphon RcaE photoregulatory CCA sensor kinase increases the levels of a transcriptional regulator, BolA, under red light conditions. BolA binds to the promoter of mreB, repressing transcription of this key element of the elongasome and resulting in small rounded cells (Figure 2E bottom) [53, 54]. Under green light conditions, this repression is released and MreB can accumulate, resulting in cell elongation and rod-shaped cells (Figure 2E top) [53, 54].
Filamentation in Pathogens (Legionella & UPEC)
Pleomorphism is an important strategy employed by pathogens, where morphological transitions are utilized to successfully colonize host tissues or cells, mediate transmission between hosts, or maintain persistence in environmental reservoirs. Several bacterial species transition from rod to filamentous morphologies in response to environmental stress [31]. For example, the filamentous morphology can aid Legionella pneumophila in evading macrophage killing and promoting intracellular replication [55].
During infection, uropathogenic Escherichia coli (UPEC) transitions from nonmotile rods to cocci, then to motile rods, and ultimately to a filamentous form whose size is thought to prevent phagocytosis [56]. While UPEC filamentation had previously been linked to the protein SulA and the SOS response [56–58], a recent study has identified the protein DamX as a key factor by which UPEC mediates the reversible transition between normal rod shape and filamentation through the inhibition of cell division (Table 1) [59]. Both mechanisms are proposed to contribute to UPEC filamentation, but in response to different stimuli: SulA filamentation occurs in response to immune cell attack, while DamX filamentation is triggered by exposure to urine/liquid shear forces upon the shift from intracellular growth to surface growth (Table 1) [56, 59].
Morphological Stratification in Lactococcus Biofilms
Cell morphology is also important to biofilm formation [39]. Lactococcus lactis is a Gram-positive bacterium used extensively in the production of fermented dairy products such as cheese, yogurt, and buttermilk. Until recently, L. lactis have been reported as exclusively ovococcoid shape under common laboratory conditions. During biofilm formation under certain growth conditions, L. lactis produces two stratified and morphologically distinct subpopulations: the base of the biofilm contains ovoid cells, and the upper layers contain rods of various lengths (Figure 3H) [60]. The proposed mechanism for generating elongated cells is that, while FtsZ-rings assemble and recruit components of the divisome, PBP2x-associated septation is arrested. This arrest allows sustained PBP2b-associated peripheral growth to elongate the cell [60]. Cell elongation via inhibition of cell division is an emerging theme in our understanding of morphological transitions.
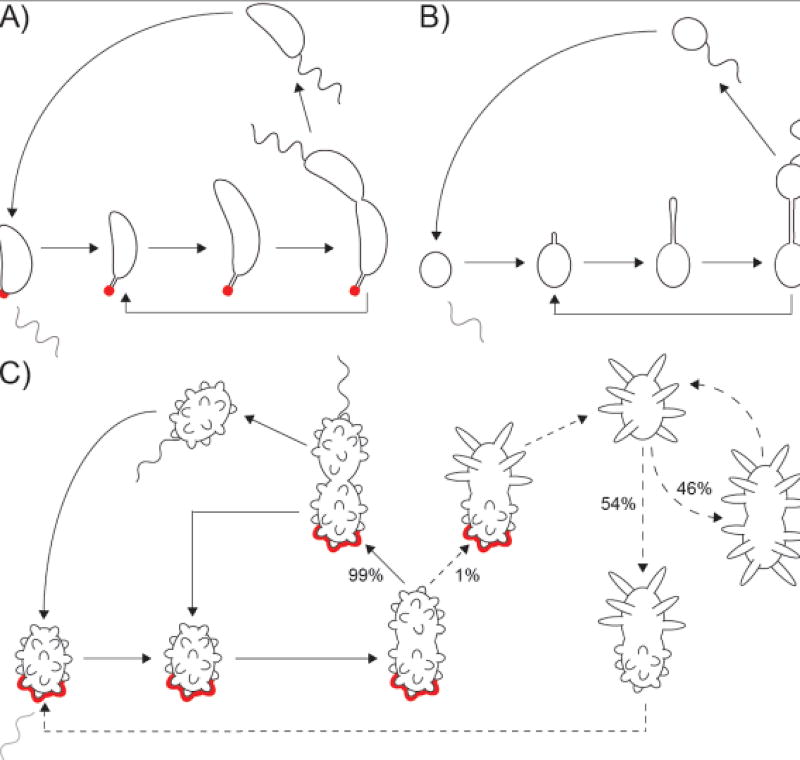
Figure 3. Multimorphic life cycles of prosthecate Alphaproteobacteria.
A) Dimorphic life cycle of Caulobacter crescentus. The prosthecate mother cell produces an adhesive holdfast (shown in red) at the tip of the prostheca. Cell division results in a motile, non-replicating swarmer cell that differentiates into a prosthecate cell. B) Dimorphic life cycle of Hyphomonas neptunium. The prosthecate mother cell produces a bud at the distal end of the prostheca. Upon septation, a motile, non-replicating swarmer cell is released that differentiates into a prosthecate cell. C) Multimorphic life cycle of Prosthecomicrobium hirschii. Most of the time, short-prosthecate P. hirschii cells follow a C. crescentus-like life cycle (solid arrows): a unipolar polysaccharide (UPP, an adhesin similar to C. crescentus holdfast; shown in red) producing mother cell gives rise to a motile, non-replicating swarmer cell that differentiates into a short-prosthecate cell. A rare event, short-prosthecate mother cells can give rise to non-motile, long-prosthecate cells that do not produce UPP (dashed arrow). Long-prosthecate mother cells can produce either long-prosthecate cells or short-prosthecate cells at roughly equal frequency (dashed arrows), though it should be noted that the observed frequency of morphotype conversion was observed on MMB (minimal medium broth) agarose pads and might differ in other conditions.
Prosthecomicrobium hirschii – A Polymorphic Life Cycle
Many Alphaproteobacteria species engage in asymmetric cell division, a characteristic that is shared despite the diverse habitats and lifestyles of these bacteria [61–65]. The dimorphic life cycle, which gives rise to morphologically distinct motile swarmer cells and adherent prosthecate cells (Figure 3A), is the best studied asymmetric cell cycle in Alphaproteobacteria and is exemplified by C. crescentus (Figure 2B) [66]. While the core genes involved in cell cycle regulation are broadly conserved among Alphaproteobacteria [67, 68], their essentiality and how they are regulated varies. Such differences suggest an evolutionary plasticity within this regulatory system which may have aided in adaptation to a species’ respective environment [69–71]. Prosthecomicrobium hirschii (Figure 2F), a member of the Rhizobiales clade, exhibits a complex polymorphic life cycle (Figure 3C) in which two morphologically distinct cell types persist in the same culture: short-prosthecate cells produce numerous short conical prosthecae, whereas long-prosthecate cells typically have fewer than eight long cylindrical prosthecae [72–74]. Short-prosthecate cells undergo Caulobacter-like asymmetric division, almost always producing a motile, short-prosthecate daughter which produces a polar adhesin (Figure 3C) [73]. Long-prosthecate cells, on the other hand, are non-motile, produce no adhesin, and can produce short-prosthecate or long-prosthecate daughter cells (Figure 3C) [73]. In both cases, a mother cell typically produces a daughter of the same morphology for several generations [72, 73]. Does this generational shape patterning represent an "epigenetics of form", whereby the gene expression patterns that regulate the morphological cycle are passed from mother to daughter? Or could it be regulated at the population level via quorum sensing?
Breaking the Mold – Pleomorphism in Roseobacter
Polymorphic life cycles in which one morphology utilizes a Caulobacter-like cell cycle have been observed in a number of Alphaproteobacteria [61, 75]. This suggests that variations on Caulobacter-style cell cycle regulation may be widespread and raises questions about the conditions where one form if favored over another. For example, the Roseobacter clade is an abundant, diverse, and ecologically significant group of heterotrophic Alphaproteobacteria commonly found in marine environments [76]. A common morphological trait among many Roseobacters is that cells in a population exhibit pleomorphism [77, 78]. Phaeobacter inhibens, either as single cells or in multicellular rosettes, can range from 1–10 µm long, with cells > 3 µm comprising up to 15% of the population [78]. In Dinoroseobacter shibae, differentiation into a morphologically heterogeneous population (Figure 2G) is regulated via quorum sensing and mediated through the CckA-ChpT-CtrA phosphorelay system [77, 79]. This is the same system that mediates cell cycle control in C. crescentus [80], thus this pathway is utilized to modulate the cell cycle in both species but to antithetical ends; a tightly regulated, dimorphic life cycle in C. crescentus (Figure 3A) becomes chaotic pleomorphism in D. shibae (Figure 2G).
How Do Shapes Evolve?
The earliest attempts to classify bacteria and describe their taxonomic relationships relied on observable characteristics such as morphology, metabolism, locomotion, or mode of cell division [81]. The advent and development of molecular phylogenies invalidated many of these historical relationships [82], but now allows us to map these phenotypes onto robust phylogenies and make inferences regarding the evolution of bacterial shape (see Outstanding Questions) [2, 83]. This approach has been used extensively in eukaryotes for inferring the evolutionary history of traits [84], and it was similarly applied to explore evolutionary transitions from rods to cocci in bacteria [85]. Below we will discuss two recent examples of experimental investigation into morphological evolution.
Evolutionary Transition from Rods to Cocci in Neisseriaceae
Nasopharyngeal (NP) pathogens of the Neisseriaceae family vary in cell shape; some are rods while others are cocci [86]. Recent work suggests that these two shapes are the result of an evolutionary transition [87]. This morphological transition from rod to coccus is correlated with the stepwise loss of first yacF, followed by the elongation machinery (mreBCD, pbpC, rodA, rodZ) [87]. In E. coli, YacF (aka ZapD) promotes FtsZ ring assembly and may help regulate the transition between elongation and division in rod-shaped Neisseria [88–90]. Muropeptide analysis (see Glossary) of PG composition for different species or mutants provides a rough "fingerprint" that can differentiate, for example, Gram positive from Gram negative bacteria or rods from cocci [6]. Concomitant with the evolutionary transition from rods to cocci in Neisseriaceae is an enrichment in the relative abundance of septal and polar PG (pentapeptides) and a decrease in the proportion of lateral PG from the sidewalls (tetrapeptides) in the various lineages [87]. The PG phenotype could be duplicated in vitro in the rod-shaped Neisseria elongata through the stepwise deletion of yacF and the elongation machinery. Moraxella catarrhalis and Neisseria meningitidis are coccal species that occupy the same niche. Analysis of the Moraxellaceae family suggests it has undergone a convergent evolutionary transition, with a common loss of ancestral yacF and elongasome genes leading to a similar transition of coccobacillus-to-coccus shape and accompanying change in PG composition [87]. One can speculate about the forces driving such morphological transitions. For example, rod-shaped NP pathogens are often found in the saliva of the oral cavity, while cocci are usually found attached to the drier NP mucosa. While evidence of the adaptive value of shape requires direct observation of selection acting on phenotype, comparative genomic studies such as this allow for the identification of selective targets associated with specific morphologies [2].
The Evolution of Prosthecate Morphology in Caulobacteraceae
Prosthecae are a common feature in aquatic bacteria living in oligotrophic environments [32–34]. In the Caulobacteraceae, phylogenetic analysis indicates that the single ancestral polar prostheca (exemplified by C. crescentus; Figure 2B) has been repositioned first to a subpolar position (Asticcacaulis excentricus; Figure 2C) and then to a bilateral position (Asticcacaulis biprosthecum; Figure 2D), with two prosthecae located opposite of each other at the midcell [2, 32]. In C. crescentus (polar; Figure 2B), the developmental regulator SpmX is responsible for localizing the histidine kinase DivJ to the prosthecate pole, where it plays a key role in cell cycle regulation [91]. In Asticcacaulis spp., SpmX (Table 1) has been co-opted as a morphogen to position and coordinate the synthesis of prosthecae through zonal PG remodeling [32]. An expanded region within SpmX is responsible for the different localization patterns between A. excentricus (subpolar; Figure 2C) and A. biprosthecum (bilateral; Figure 2D) [32]. The study of these bacteria and how they localize, synthesize, and maintain prosthecae is providing significant understanding about how bacteria can generate novel morphologies through the repositioning of PG synthesis machinery.
Twist and Sprout and Bud: Molecular Factors Underlying Distinct Shapes
Helical Bacteria – Reshaping the Rod
The bacterial helix is a shape that has arisen many times throughout bacterial evolution [2]. Helical bacteria are responsible for many human and animal diseases, although not all helical bacteria cause disease. The best known disease-causing helical bacteria include spirochetes, such as Leptospira spp. (leptospirosis), Treponema spp. (syphilis), and Borrelia spp. (Lyme disease), as well as proteobacteria like Campylobacter jejuni (gastroenteritis) and H. pylori (peptic ulcers and gastric cancer; Figure 2I). Identification of mutations that directly affect peptidoglycan remodeling has proven vital for describing how bacteria establish and maintain their shape. Screens for shape mutants in rod-shaped bacteria such as E. coli and Bacillus spp. have been undertaken since the 1960’s and provided the foundation for our current knowledge of the molecular underpinnings of PG-based shape determination [92]. This approach of screening unbiased transposon libraries or deletion collections (e.g. the E. coli Keio collection) continues to prove useful in revealing the underlying molecular mechanisms involved in complex shape determination [41, 93–95]. Transposon library screens for morphological mutants in H. pylori, paired with fluorescence-activated cell sorting (FACS) to enrich mutant libraries for bacterial cells with altered light scattering profiles that correlate with perturbed cell morphology, have identified several PG modifying enzymes responsible for maintenance of the helical morphology [41, 96, 97]. These screens identified the genes csd1-6 (cell shape determinant) that encode for an array of PG modifying enzymes (Table 1), as well as ccmA (curved cell morphology) (Table 1), a bactofilin (Figure 1F) homolog. Bactofilins are a recently discovered class of cytoskeletal-like scaffolding proteins that are conserved throughout the bacterial kingdom [98] and have been implicated in cell shape determination in a number of species. In the normally rod-shaped P. mirabilis, disruption of the bactofilin homolog results in elongated and curved cells, while overexpression leads to enlarged, rounded cells [99]. In Myxococcus xanthus, deletion of one of its four bactofilins (BacM) leads to a crooked or kinked morphology [100]. In C. crescentus, bactofilins serve as a localization factor for the bifunctional PBP (Figure 1A), PbpC, to the prosthecate pole, and deletion mutants exhibit a reduced rate of prostheca synthesis [98]. Characterization of the H. pylori Csd proteins led to a model in which helical shape is generated through the interaction of two PG modification pathways: 1) DD-endopeptidases generate helical twist by removing tetra–pentapeptide crosslinks, and 2) carboxypeptidases generate curvature through the sequential trimming of monomers from the unlinked peptide side chain. CcmA and Csd5 likely serve localization or regulatory functions in the respective pathways. Csd3, which can perform both crosslink cleavage and trimming of the stem peptide, may act in both pathways. It should be noted that H. pylori Csd4/Csd6 are homologs of C. jejuni Pgp1/Pgp2. C. jejuni Pgp1/Pgp2 have the same catalytic activities as H. pylori Csd4/Csd6, respectively, and are both important for C. jejuni cell shape with similar mutant phenotypes (Table1, H. pylori Csd4/Csd6) [95–97, 101, 102]. These genes and their homologs appear to be exclusive to curved bacteria, thus suggesting a unique and conserved shape generation program for this architecture.
Apical Growth and Hyphal Branching in Streptomyces
Vegetative growth of the filamentous Streptomycetes (Figure 2J) originates from the germination of a single spore and subsequent extension of one or more hyphal branches (see Glossary). Unlike conventional rod-shaped bacteria that elongate via PG synthesis evenly dispersed along the length of the cell, Streptomycetes exhibit apical growth through polar tip extension (Figure 2J). Branches emerge distal to the growing tip, resulting in new hyphae and new branches to form a network of filaments called a mycelium. The morphology of the resulting mycelium is determined in large part by when and where new branches appear [103]. How do Streptomycetes establish cell polarity and the timing and placement of new PG synthesis zones in this striking manifestation of polar growth?
Three scaffolding proteins, DivIVA, Scy, and FilP (Table 1), play different but complementary roles in organizing apical growth. The essential protein DivIVA binds to negative curvature and organizes the PG synthesis machinery and other proteins, such as Scy and FilP, to form the "polarisome" that mediates apical growth [104–109]. It should be noted that the function of DivIVA is different in Firmicutes, where it is required to prevent Z-ring formation at the new cell poles after division in B. subtilis and to coordinate midcell elongation in S. pneumoniae [83]. DivIVA and the cell wall biosynthetic machinery communicate through the serine/threonine kinase AfsK. Phosphorylation of DivIVA by AfsK, at a basal level under normal growth or at higher levels in response to perturbations in cell wall synthesis, causes the disassembly of part of the apical polarisome [110]. This tip-splitting mechanism results in DivIVA foci left behind the growing tip which initiates the formation of new polarisomes to drive growth of new hyphal branches [106, 110, 111]. Scy is thought to regulate the number of DivIVA foci, and therefore the number of polarisomes, by sequestering free DivIVA [108, 112]. As a hypha extends, the tip and newly synthesized cell wall are more flexible and inherently weaker than more highly cross-linked pole distal parts [113, 114]. DivIVA recruits a gradient of FilP as the tip extends to provide a stress-bearing cytoskeletal structure (Table 1).
Hyphomonas neptunium – Forever Blowing Bubbles
Prosthecate Alphaproteobacteria of the Hyphomonas spp. (Figure 2K) undergo a dimorphic life cycle but, unlike C. crescentus where division occurs at the non-prosthecate pole (Figure 3A), new offspring of Hyphomonas neptunium bud from the tip of the prostheca (Figure 3B) [115, 116]. Cell division then occurs at the junction between the prostheca tip and the bud neck, resulting in a prosthecate mother cell and a non- prosthecate daughter cell (Figure 3B). A recent comprehensive study of predicted PG biosynthetic and remodeling enzymes, as well as their regulators, showed that H. neptunium cells utilize a complex cell-cycle regulated pattern of PG remodeling to establish shape and reproduce [117]. PG muropeptide analysis suggests an important role for PG hydrolases (Figure 1G-1J) in growth and budding (Table 1). Indeed, the hydrolase LmdC, appears to be essential, and LmdE and AmiC, appear to be necessary for release of the budding daughter from the mother cell (Table 1). Of the biosynthetic PG enzymes, one bifunctional (PBP1X; Figure 1A & Table 1) and two monofunctional DD-transpeptidases (PBP2 and PBP3; Figure 1B & Table 1) may be key factors in H. neptunium growth. Based on their complex spatiotemporal localization patterns, elongasome components MreB, whose inhibition results in morphological defects, and RodZ appear critical for normal development in H. neptunium (Table 1). Collectively, this work provides a tantalizing first look at the molecular underpinnings of a fascinating mode of growth and shape determination that involves the temporal establishment of multiple zones of dispersed and zonal PG modification.
Practical Advantages to Studying Shape and the Molecular Basis for Morphology
Bacteria have long played a critical role in many industrial processes important to humans, and there is a growing appreciation for the impact of cell size and shape during these processes. For example, lactic acid bacteria (LAB) are important industrial microorganisms for the probiotic and dairy industries. Short rod LABs provide higher cell counts and are more stable than elongated rods during processes such as freeze drying or the microencapsulation of bacteria in dough [118]. Another bacterial morphology dependent process is wastewater treatment that uses the activated sludge process, which requires sludge flocs to form and settle. Filamentous bacteria are essential for this by providing a matrix for floc formation, but an excess of filamentous bacteria can result in sludge bulking, where the flocs are so large that they can’t settle. Recent work suggests that high levels of the filamentous Chlorobacterium Kouleothrix aurantiaca is associated with bulking and its presence may provide an indicator for such incidents [119]. This is a systemic problem in wastewater treatment, and an understanding of how bacterial morphology affects the process is allowing the industry to address it.
As we begin to tease out the molecular underpinnings of how bacterial shape is generated, it has become possible to engineer morphologies for practical applications (see Outstanding Questions). For example, microcoils are tiny electrical conductors with a wide variety of applications, including nuclear magnetic resonance (NMR) imaging [120], and are typically produced, one by one, through traditional machining or lithography processes. The cyanobacterium A. platensis, whose helical structure can be experimentally modified, is being developed as a biological template to mass produce microcoils (Figure 2L) [121]. Photosynthetic cyanobacteria also have great potential to produce fuels, chemicals, and biomass, but their successful use depends on developing cost-effective methods to cultivate and harvest cells at large scales. The goal of engineering these cell morphologies is to improve biomass recovery and decrease energetic costs associated with lysing cyanobacterial cells. The obvious approach is to manipulate genes known to be involved in cell shape determination. This is being developed in Synechococcus elongatus [122], where putting MinC (which acts with MinD to inhibit FtsZ assembly) and the DivIVA homolog Cdv3 (which is important for divisome localization and function) under the control of tunable riboswitches causes hyperelongation up to 500% [122]. Hyperelongated cells settle more easily through centrifugation and are more susceptible to lysis, both beneficial traits for reducing harvesting costs. Furthermore, their large size may also prevent predation from grazing protozoan species that might contaminate the culture.
Many bacteria can accumulate inclusion bodies (IB), cytoplasmic granules used to store materials such as sulfur, polyphosphate, carbon sources, or protein aggregates. Many IBs have industrial uses. For example, polyhydroxyalkanoates (PHA), a family of biodegradable plastics, have applications ranging from packaging materials and disposable items (e.g. plastic utensils) [123] to nanoparticles in drug delivery systems [124, 125]. Bacterial production of PHA can be can be costly because "normal" bacterial cells are small, limiting the amount of PHA granules a cell can maintain and making it difficult to efficiently separate the biomass from the growth media at an industrial scale [126]. Manipulation of genes involved in cell morphology can be used to address these production hurdles. Manipulation of SulA and MinCD in Halomonas spp. [127, 128] and EnvC and NlpD, direct regulators of cell septation through the activation of amidases (Figure 1J), and RodZ in E. coli [129] improved cell shape for the production of PHAs. Finally, the CRISPR (clustered regularly interspaced short palindromic repeats) interference system can effectively engineer diverse cell morphologies in E. coli through the tunable repression of ftsZ and/or mreB [130].
Concluding Remarks
Bacteria seem to care very much about cell shape, but why should we? The practical answer is that the ways bacteria most impact our lives, namely though disease and industrial processes, are often impacted by cell morphology. In addition, we probably don’t have a true picture of bacterial morphological diversity because the sampling of phylogenetic diversity historically skews toward these pathogens and industrially relevant organisms [131]. Many bacterial lineages remain undersampled, with few, if any, isolated representatives (see Outstanding Questions) [132]. Probing this "microbial dark matter" [133], may reveal previously undiscovered morphologies.
How do we go about assessing bacterial shape diversity, studying the underlying molecular mechanisms, and understanding morphological evolution? High-content microscopy [134] and image analysis [135–137] of environmental samples can provide a visual accounting of shape diversity. High-throughput culturing methods have been used to isolate previously uncultured bacteria [25] and when that fails, high-throughput sequencing may allow modeling of metabolic networks for culture conditions amenable to these species [26–29, 138]. Once isolated, sequencing the genomes of morphologically interesting species through single-cell genomics [139] allows for comparative genomics approaches to identify genes involved in shape generation and morphological variation. Many genetic tools may not be immediately available to study morphogenesis in newly isolated species, but some do exist. Mutant libraries can be generated via transposon, chemicals, or radiation. Proteins that localize to morphological features can be screened through fluorescent transposon fusions [140]. Combining sequenced genomes and mutant libraries with high-content imaging and analysis allows for efficient forward genetic screens for morphological variants. Fluorescent D-amino acids (FDAAs) allow labeling of PG cell wall biosynthesis in real time (Figure 2J), in live cells, and across a range of bacterial species [141, 142]. Combining advances in microscopy and microfluidics with pulse labeling of wild-type and mutant cells with FDAAs can reveal underlying growth patterns that may not be evident from whole cell imaging.
Because shape mutants often arise through a mutation in PG modifying enzymes or other associated proteins, it stands to reason that the final chemical composition and structure of PG in a mutant may differ from wild type. Another approach to screening for shape mutants is to directly assay PG composition of the mutants through high performance liquid chromatography (HPLC) [143, 144]. Traditional HPLC muropeptide analysis is relatively inefficient, requiring large sample volumes, laborious preparation, and long run times [145]. The recent development of ultra performance liquid chromatography (UPLC) addresses many of these challenges [145, 146] and, along with the advent of robotics/automation, advances in microscope technology, and development of image analysis software [135–137, 147], the potential now exists for quick, high-throughput, and high resolution screens of fully saturated transposon libraries for bacterial shape mutants
Extraordinary and beautiful biological processes underlie even the simplest of shapes. The observable bacterial form is the result of exquisitely controlled expression and repression of genes, metabolic processes that maintain PG precursors and subunits, and the spatio-temporal localization of PG modifying enzymes. The study of shape in model organism has certainly uncovered common themes, such as scaffolding proteins (ex. MreB, FtsZ, DivIVA) that recruit and maintain PG synthesis complexes to specific subcellular locations. What new and wonderful strategies for shape generation will we uncover when we start looking in depth beyond the model organisms currently used? The tools are there to reestablish a natural history tradition in microbiology and apply it to modern molecular biology and genetics. All we need to do is look.
Trends Box.
Bacterial morphology is diverse, yet we are just beginning to understand the molecular basis for shape generation outside of canonical model organisms.
The major determinant of bacterial cell shape is the spatiotemporal regulation of enzymes that modify the peptidoglycan (PG) cell well. Many proteins involved in PG remodeling are conserved, but how these proteins are regulated, modified, and localized can vary, even for shapes that outwardly look the same.
Individual cells can dynamically modulate their shape in response to the environment or through a programmed life cycle.
Combining observable phenotypes with whole genome and single cell sequencing and phylogenomics allow us to make inferences about shape evolution.
Advances in tools such as microscopy and image analysis, PG labeling and chemical analysis, and shape engineering are enhancing the study of bacterial morphology.
Outstanding Questions Box.
What is the true accounting of bacterial morphological diversity? Are rods and cocci the dominant morphology? Or are other morphologies simply undersampled?
-
What are the mechanisms underlying shape evolution?
-
1)
Certain morphologies, such as hyphal branching or prosthecae, often cluster together phylogenetically, suggesting that the morphology (and underlying molecular mechanisms) arose from a common ancestor. Are shapes maintained via selective pressure? Is the ancestral shape iterated upon? If so, how and why does variation among closely related species arise?
-
2)
In other cases, the same morphology, such as curved or helical bacteria, has arisen independently in unrelated species. Do different lineages with similar shapes converge to the same molecular strategies or are unique evolutionary paths forged? Are the selective pressures that result in similar morphologies the same?
-
1)
Work in model organisms has shown that there are many ways to generate even "simple" shapes like the rod, but there seems to be a basic theme of spatiotemporal regulation PG remodeling to generate shape. Is a conserved suite of enzymatic genes utilized, just regulated and deployed in different ways? Are specialized classes of genes associated with specific morphologies? How is the localization of PG remodeling complexes regulated?
How prevalent is pleomorphism? It seems that most bacteria can, at minimum, alter cell shape as part of a stress response. Perhaps the better question is how prevalent is the regulation of pleomorphism? What are the underlying molecular mechanisms of shape change regulation? What are the environmental triggers and how are they sensed? In cases where pleomorphism is precisely timed and/or regulated, how did these systems evolve?
How will new knowledge about bacterial morphologies be harnessed to develop new strategies for shape engineering? How will this affect industry, medicine, and basic research?
Acknowledgments
We wish to thank David Kehoe for discussions about cyanobacterial literature and morphology and Pam Brown for discussions about the P. hirschii life cycle. We are grateful to Jenny Elig and members of the Brun lab: Courtney Ellison, Yen-Pang Hsu Maxime Jacq, Breah LaSarre, and Amelia Randich, for critical reading of and feedback on this manuscript. This work was funded by National Institutes of Health grant R35GM122556 (to YVB).
Glossary
- Hypha
A single filament in a bacterial or fungal mycelium. Extends by growth at the hyphal tip also known as apical growth.
- Muropeptide Analysis
Analytic technique used to identify the chemical composition and relative fractions of muropeptides in the peptidoglycan cell wall. The technique typically involves isolation and enzymatic digestion of the peptidoglycan sacculi from a bacterial culture followed by analysis via liquid chromatography and mass spectrometry. Examples of information that can be gleaned from muropeptide analysis include the average length of glycan strands, degree of crosslinking, crosslink types, and stem peptide composition.
- Nucleoid Occlusion
Protective mechanism of the bacterial cell cycle that prevents the chromosome from being severed by the division septum. Negatively regulates Z-ring formation near the nucleoid, which promotes division septum localization to the mid-cell.
- Penicillin Binding Proteins (PBP)
Proteins involved in peptidoglycan synthesis. In general these proteins catalyze the polymerization of (transglycosylation) and the cross-linking between (transpeptidation) glycan chains. The name refers to the fact that PBPs bind to penicillin and most other β-lactam antibiotics.
- Prostheca
A cellular appendage. Prosthecae (also referred to as "stalks") are contiguous with all three layers of the Gram-negative cell envelope (inner membrane peptidoglycan, and outer membrane), contain cytoplasm, and protrude from the cell body.
- Scaffolding Protein
Proteins that physically organize the molecular components of a biological process or pathway.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Young KD. The selective value of bacterial shape. Microbiol Mol Biol Rev. 2006;70:660–703. doi: 10.1128/MMBR.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kysela DT, et al. Diversity Takes Shape: Understanding the Mechanistic and Adaptive Basis of Bacterial Morphology. PLoS Biol. 2016;14:e1002565. doi: 10.1371/journal.pbio.1002565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang DC, et al. Staying in Shape: the Impact of Cell Shape on Bacterial Survival in Diverse Environments. Microbiol Mol Biol Rev. 2016;80:187–203. doi: 10.1128/MMBR.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Teeseling MCF, et al. Determinants of Bacterial Morphology: From Fundamentals to Possibilities for Antimicrobial Targeting. Front Microbiol. 2017;8:1264. doi: 10.3389/fmicb.2017.01264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allan EJ, et al. Bacterial L-forms. Adv Appl Microbiol. 2009;68:1–39. doi: 10.1016/S0065-2164(09)01201-5. [DOI] [PubMed] [Google Scholar]
- 6.Turner RD, et al. Different walls for rods and balls: the diversity of peptidoglycan. Mol Microbiol. 2014;91:862–874. doi: 10.1111/mmi.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Typas A, et al. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2011;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsang MJ, Bernhardt TG. Guiding divisome assembly and controlling its activity. Curr Opin Microbiol. 2015;24:60–65. doi: 10.1016/j.mib.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Errington J. Bacterial morphogenesis and the enigmatic MreB helix. Nat Rev Microbiol. 2015;13:241–248. doi: 10.1038/nrmicro3398. [DOI] [PubMed] [Google Scholar]
- 10.Margolin W. Sculpting the bacterial cell. Curr Biol. 2009;19:R812–822. doi: 10.1016/j.cub.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Randich AM, Brun YV. Molecular mechanisms for the evolution of bacterial morphologies and growth modes. Front Microbiol. 2015;6:580. doi: 10.3389/fmicb.2015.00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams DW, et al. Cell cycle regulation by the bacterial nucleoid. Curr Opin Microbiol. 2014;22:94–101. doi: 10.1016/j.mib.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho H, et al. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat Microbiol. 2016:16172. doi: 10.1038/nmicrobiol.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emami K, et al. RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway. Nat Microbiol. 2017;2:16253. doi: 10.1038/nmicrobiol.2016.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henrichfreise B, et al. Bacterial Surfaces: The Wall that SEDS Built. Curr Biol. 2016;26:R1158–R1160. doi: 10.1016/j.cub.2016.09.028. [DOI] [PubMed] [Google Scholar]
- 16.Leclercq S, et al. Interplay between Penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis. Sci Rep. 2017;7:43306. doi: 10.1038/srep43306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szwedziak P, Lowe J. Do the divisome and elongasome share a common evolutionary past? Curr Opin Microbiol. 2013;16:745–751. doi: 10.1016/j.mib.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 18.Brown PJ, et al. Polarity and the diversity of growth mechanisms in bacteria. Semin Cell Dev Biol. 2011;22:790–798. doi: 10.1016/j.semcdb.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cava F, et al. Modes of cell wall growth differentiation in rod-shaped bacteria. Curr Opin Microbiol. 2013;16:731–737. doi: 10.1016/j.mib.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leisch N, et al. Asynchronous division by non-ring FtsZ in the gammaproteobacterial symbiont of Robbea hypermnestra. Nat Microbiol. 2016;2:16182. doi: 10.1038/nmicrobiol.2016.182. [DOI] [PubMed] [Google Scholar]
- 21.Starr MP, Skerman VB. Bacterial diversity: the natural history of selected morphologically unusual bacteria. Annu Rev Microbiol. 1965;19:407–454. doi: 10.1146/annurev.mi.19.100165.002203. [DOI] [PubMed] [Google Scholar]
- 22.Bergey DH, Holt JG. Bergey’s manual of determinative bacteriology. Williams & Wilkins; 1994. [Google Scholar]
- 23.Bowman J. The Methanotrophs - The Families Methylococcaceae and Methylocystaceae. Prokaryotes: A Handbook on the Biology of Bacteria. (Third) 2006;5:266–289. [Google Scholar]
- 24.Danilova OV, et al. A new cell morphotype among methane oxidizers: a spiral-shaped obligately microaerophilic methanotroph from northern low-oxygen environments. ISME J. 2016;10:2734–2743. doi: 10.1038/ismej.2016.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Connon SA, Giovannoni SJ. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl Environ Microbiol. 2002;68:3878–3885. doi: 10.1128/AEM.68.8.3878-3885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Brien EJ, et al. Using Genome-scale Models to Predict Biological Capabilities. Cell. 2015;161:971–987. doi: 10.1016/j.cell.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oberhardt MA. Harnessing the landscape of microbial culture media to predict new organism-media pairings. Nat Commun. 2015;6:8493. doi: 10.1038/ncomms9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henry CS, et al. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat Biotechnol. 2010;28:977–982. doi: 10.1038/nbt.1672. [DOI] [PubMed] [Google Scholar]
- 29.Hamilton JJ, Reed JL. Software platforms to facilitate reconstructing genome-scale metabolic networks. Environ Microbiol. 2014;16:49–59. doi: 10.1111/1462-2920.12312. [DOI] [PubMed] [Google Scholar]
- 30.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 31.Justice SS, et al. Morphological plasticity as a bacterial survival strategy. Nat Rev Microbiol. 2008;6:162–168. doi: 10.1038/nrmicro1820. [DOI] [PubMed] [Google Scholar]
- 32.Jiang C, et al. Sequential evolution of bacterial morphology by co-option of a developmental regulator. Nature. 2014;506:489–493. doi: 10.1038/nature12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poindexter JS. Biological Properties and Classification of the Caulobacter Group. Bacteriol Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stovepoindexter JL, Cohen-Bazire G. The Fine Structure of Stalked Bacteria Belonging to the Family Caulobacteraceae. J Cell Biol. 1964;23:587–607. doi: 10.1083/jcb.23.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt JM, Stanier RY. The development of cellular stalks in bacteria. J Cell Biol. 1966;28:423–436. doi: 10.1083/jcb.28.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt JM. Stalk Elongation in Mutants of Caulobacter Crescentus. J Gen Microbiol. 1968;53 291-&. [Google Scholar]
- 37.Gonin M, et al. Regulation of stalk elongation by phosphate in Caulobacter crescentus. J Bacteriol. 2000;182:337–347. doi: 10.1128/jb.182.2.337-347.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woldemeskel SA, Goley ED. Shapeshifting to Survive: Shape Determination and Regulation in Caulobacter crescentus. Trends Microbiol. 2017;25:673–687. doi: 10.1016/j.tim.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith WP, et al. Cell morphology drives spatial patterning in microbial communities. Proc Natl Acad Sci U S A. 2017;114:E280–E286. doi: 10.1073/pnas.1613007114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bulgheresi S. Bacterial cell biology outside the streetlight. Environ Microbiol. 2016;18:2305–2318. doi: 10.1111/1462-2920.13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sycuro LK, et al. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori’s helical shape and stomach colonization. Cell. 2010;141:822–833. doi: 10.1016/j.cell.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castenholz RW. Phylum BX. Cyanobacteria. Oxygenic Photosynthetic Bacteria. In: Garrity G, Boone DR, Castenholz RW, editors. Bergey’s Manual of Systematic Bacteriology. Springer-Verlag: 2001. pp. 474–487. [Google Scholar]
- 43.Rajaniemi P, et al. Phylogenetic and morphological evaluation of the genera Anabaena, Aphanizomenon, Trichormus and Nostoc (Nostocales, Cyanobacteria) Int J Syst Evol Microbiol. 2005;55:11–26. doi: 10.1099/ijs.0.63276-0. [DOI] [PubMed] [Google Scholar]
- 44.Singh SP, Montgomery BL. Determining cell shape: adaptive regulation of cyanobacterial cellular differentiation and morphology. Trends Microbiol. 2011;19:278–285. doi: 10.1016/j.tim.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 45.Bennett A, Bogorad L. Complementary chromatic adaptation in a filamentous blue-green alga. J Cell Biol. 1973;58:419–435. doi: 10.1083/jcb.58.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar K, et al. Cyanobacterial heterocysts. Cold Spring Harb Perspect Biol. 2010;2:a000315. doi: 10.1101/cshperspect.a000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaplan-Levy RN, et al. Akinetes: Dormant Cells of Cyanobacteria. Top Curr Genet. 2010;21:5–27. [Google Scholar]
- 48.Rippka R, et al. Generic Assignments, Strain Histories and Properties of Pure Cultures of Cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- 49.Lehner J, et al. The morphogene AmiC2 is pivotal for multicellular development in the cyanobacterium Nostoc punctiforme. Mol Microbiol. 1979;2011;79:1655–1669. doi: 10.1111/j.1365-2958.2011.07554.x. [DOI] [PubMed] [Google Scholar]
- 50.Kehoe DM, Gutu A. Responding to color: the regulation of complementary chromatic adaptation. Annu Rev Plant Biol. 2006;57:127–150. doi: 10.1146/annurev.arplant.57.032905.105215. [DOI] [PubMed] [Google Scholar]
- 51.Wu H, et al. Effects of solar UV radiation on morphology and photosynthesis of filamentous cyanobacterium Arthrospira platensis. Appl Environ Microbiol. 2005;71:5004–5013. doi: 10.1128/AEM.71.9.5004-5013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pattanaik B, et al. Light Quantity Affects the Regulation of Cell Shape in Fremyella diplosiphon. Front Microbiol. 2012;3:170. doi: 10.3389/fmicb.2012.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh SP, Montgomery BL. Morphogenes bolA and mreB mediate the photoregulation of cellular morphology during complementary chromatic acclimation in Fremyella diplosiphon. Mol Microbiol. 2014;93:167–182. doi: 10.1111/mmi.12649. [DOI] [PubMed] [Google Scholar]
- 54.Singh SP, Montgomery BL. Regulation of BolA abundance mediates morphogenesis in Fremyella diplosiphon. Front Microbiol. 2015;6:1215. doi: 10.3389/fmicb.2015.01215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prashar A, et al. Filamentous morphology of bacteria delays the timing of phagosome morphogenesis in macrophages. J Cell Biol. 2013;203:1081–1097. doi: 10.1083/jcb.201304095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horvath DJ, Jr, et al. Morphological plasticity promotes resistance to phagocyte killing of uropathogenic Escherichia coli. Microbes Infect. 2011;13:426–437. doi: 10.1016/j.micinf.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Justice SS, et al. Filamentation by Escherichia coli subverts innate defenses during urinary tract infection. Proc Natl Acad Sci U S A. 2006;103:19884–19889. doi: 10.1073/pnas.0606329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li B, et al. SOS regulatory elements are essential for UPEC pathogenesis. Microbes Infect. 2010;12:662–668. doi: 10.1016/j.micinf.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 59.Khandige S, et al. DamX Controls Reversible Cell Morphology Switching in Uropathogenic Escherichia coli. MBio. 2016;7:e00642–00616. doi: 10.1128/mBio.00642-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perez-Nunez D, et al. A new morphogenesis pathway in bacteria: unbalanced activity of cell wall synthesis machineries leads to coccus-to-rod transition and filamentation in ovococci. Mol Microbiol. 2011;79:759–771. doi: 10.1111/j.1365-2958.2010.07483.x. [DOI] [PubMed] [Google Scholar]
- 61.Whittenbury R, Dow CS. Morphogenesis and differentiation in Rhodomicrobium vannielii and other budding and prosthecate bacteria. Bacteriol Rev. 1977;41:754–808. doi: 10.1128/br.41.3.754-808.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lam H, et al. The asymmetric spatial distribution of bacterial signal transduction proteins coordinates cell cycle events. Dev Cell. 2003;5:149–159. doi: 10.1016/s1534-5807(03)00191-6. [DOI] [PubMed] [Google Scholar]
- 63.Hirsch P. Budding bacteria. Annu Rev Microbiol. 1974;28:391–444. doi: 10.1146/annurev.mi.28.100174.002135. [DOI] [PubMed] [Google Scholar]
- 64.Hallez R, et al. Morphological and functional asymmetry in alpha-proteobacteria. Trends Microbiol. 2004;12:361–365. doi: 10.1016/j.tim.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 65.Kysela DT, et al. Biological consequences and advantages of asymmetric bacterial growth. Annu Rev Microbiol. 2013;67:417–435. doi: 10.1146/annurev-micro-092412-155622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Curtis PD, Brun YV. Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol Mol Biol Rev. 2010;74:13–41. doi: 10.1128/MMBR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Panis G, et al. Versatility of global transcriptional regulators in alpha-Proteobacteria: from essential cell cycle control to ancillary functions. FEMS Microbiol Rev. 2015;39:120–133. doi: 10.1093/femsre/fuu002. [DOI] [PubMed] [Google Scholar]
- 68.Brilli M, et al. The diversity and evolution of cell cycle regulation in alpha-proteobacteria: a comparative genomic analysis. BMC Syst Biol. 2010;4:52. doi: 10.1186/1752-0509-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Curtis PD, Brun YV. Identification of essential alphaproteobacterial genes reveals operational variability in conserved developmental and cell cycle systems. Mol Microbiol. 2014;93:713–735. doi: 10.1111/mmi.12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.De Nisco NJ, et al. Global analysis of cell cycle gene expression of the legume symbiont Sinorhizobium meliloti. Proc Natl Acad Sci U S A. 2014;111:3217–3224. doi: 10.1073/pnas.1400421111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Greene SE, et al. Analysis of the CtrA pathway in Magnetospirillum reveals an ancestral role in motility in alphaproteobacteria. J Bacteriol. 2012;194:2973–2986. doi: 10.1128/JB.00170-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Staley JT. Prosthecomicrobium hirschii, a New Species in a Redefined Genus. Int J Syst Bacteriol. 1984;34:304–308. [Google Scholar]
- 73.Williams M, et al. Short-Stalked Prosthecomicrobium hirschii Cells Have a Caulobacter-Like Cell Cycle. J Bacteriol. 2016;198:1149–1159. doi: 10.1128/JB.00896-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Daniel JJ, et al. Draft Genome Sequence of Prosthecomicrobium hirschii ATCC 27832T. Genome Announc. 2015;3:e01355–01315. doi: 10.1128/genomeA.01355-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tyler PA, Marshall KC. Pleomorphy in stalked, budding bacteria. J Bacteriol. 1967;93:1132–1136. doi: 10.1128/jb.93.3.1132-1136.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brinkhoff T, et al. Diversity, ecology, and genomics of the Roseobacter clade: a short overview. Arch Microbiol. 2008;189:531–539. doi: 10.1007/s00203-008-0353-y. [DOI] [PubMed] [Google Scholar]
- 77.Patzelt D, et al. You are what you talk: quorum sensing induces individual morphologies and cell division modes in Dinoroseobacter shibae. ISME J. 2013;7:2274–2286. doi: 10.1038/ismej.2013.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Segev E, et al. Morphological Heterogeneity and Attachment of Phaeobacter inhibens. PLoS One. 2015;10:e0141300. doi: 10.1371/journal.pone.0141300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang H, et al. The CtrA phosphorelay integrates differentiation and communication in the marine alphaproteobacterium Dinoroseobacter shibae. BMC Genomics. 2014;15:130. doi: 10.1186/1471-2164-15-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mann TH, et al. A cell cycle kinase with tandem sensory PAS domains integrates cell fate cues. Nat Commun. 2016;7:11454. doi: 10.1038/ncomms11454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stanier RY, Van Niel CB. The concept of a bacterium. Arch Mikrobiol. 1962;42:17–35. doi: 10.1007/BF00425185. [DOI] [PubMed] [Google Scholar]
- 82.Fox GE, et al. The phylogeny of prokaryotes. Science. 1980;209:457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- 83.Jiang C, et al. Mechanisms of bacterial morphogenesis: evolutionary cell biology approaches provide new insights. Bioessays. 2015;37:413–425. doi: 10.1002/bies.201400098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- 85.Siefert JL, Fox GE. Phylogenetic mapping of bacterial morphology. Microbiology. 1998;144(10):2803–2808. doi: 10.1099/00221287-144-10-2803. [DOI] [PubMed] [Google Scholar]
- 86.Liu G, et al. Non-pathogenic Neisseria: members of an abundant, multi-habitat, diverse genus. Microbiology. 2015;161:1297–1312. doi: 10.1099/mic.0.000086. [DOI] [PubMed] [Google Scholar]
- 87.Veyrier FJ, et al. Common Cell Shape Evolution of Two Nasopharyngeal Pathogens. PLoS Genet. 2015;11:e1005338. doi: 10.1371/journal.pgen.1005338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Durand-Heredia J, et al. Identification of ZapD as a cell division factor that promotes the assembly of FtsZ in Escherichia coli. J Bacteriol. 2012;194:3189–3198. doi: 10.1128/JB.00176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang KH, et al. Characterization of the FtsZ C-Terminal Variable (CTV) Region in Z-Ring Assembly and Interaction with the Z-Ring Stabilizer ZapD in E. coli Cytokinesis. PLoS One. 2016;11:pe0153337. doi: 10.1371/journal.pone.0153337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Roach EJ, et al. Structure and Mutational Analyses of Escherichia coli ZapD Reveal Charged Residues Involved in FtsZ Filament Bundling. J Bacteriol. 2016;198:1683–1693. doi: 10.1128/JB.00969-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Radhakrishnan SK, et al. The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus. Genes Dev. 2008;22:212–225. doi: 10.1101/gad.1601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.de Boer PA. Classic Spotlight: Staying in Shape and Discovery of the mrdAB and mreBCD Operons. J Bacteriol. 2016;198:1479. doi: 10.1128/JB.00180-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.French S, et al. Bacteria Getting into Shape: Genetic Determinants of E. coli Morphology. MBio. 2017;8:e01977–01916. doi: 10.1128/mBio.01977-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fenton AK, et al. CozE is a member of the MreCD complex that directs cell elongation in Streptococcus pneumoniae. Nat Microbiol. 2016;2:16237. doi: 10.1038/nmicrobiol.2016.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Esson D, et al. Genomic variations leading to alterations in cell morphology of Campylobacter spp. Sci Rep. 2016;6:38303. doi: 10.1038/srep38303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sycuro LK, et al. Flow cytometry-based enrichment for cell shape mutants identifies multiple genes that influence Helicobacter pylori morphology. Mol Microbiol. 2013;90:869–883. doi: 10.1111/mmi.12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sycuro LK, et al. Multiple peptidoglycan modification networks modulate Helicobacter pylori’s cell shape, motility, and colonization potential. PLoS Pathog. 2012;8:e1002603. doi: 10.1371/journal.ppat.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kuhn J, et al. Bactofilins, a ubiquitous class of cytoskeletal proteins mediating polar localization of a cell wall synthase in Caulobacter crescentus. EMBO J. 2010;29:327–339. doi: 10.1038/emboj.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hay NA, et al. A novel membrane protein influencing cell shape and multicellular swarming of Proteus mirabilis. J Bacteriol. 1999;181:2008–2016. doi: 10.1128/jb.181.7.2008-2016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Koch MK, et al. BacM, an N-terminally processed bactofilin of Myxococcus xanthus, is crucial for proper cell shape. Mol Microbiol. 2011;80:1031–1051. doi: 10.1111/j.1365-2958.2011.07629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Frirdich E, et al. Peptidoglycan-modifying enzyme Pgp1 is required for helical cell shape and pathogenicity traits in Campylobacter jejuni. PLoS Pathog. 2012;8:e1002602. doi: 10.1371/journal.ppat.1002602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Frirdich E, et al. Peptidoglycan LD-carboxypeptidase Pgp2 influences Campylobacter jejuni helical cell shape and pathogenic properties and provides the substrate for the DL-carboxypeptidase Pgp1. J Biol Chem. 2014;289:8007–8018. doi: 10.1074/jbc.M113.491829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Flardh K, et al. Regulation of apical growth and hyphal branching in Streptomyces. Curr Opin Microbiol. 2012;15:737–743. doi: 10.1016/j.mib.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 104.Flardh K. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2) Mol Microbiol. 2003;49:1523–1536. doi: 10.1046/j.1365-2958.2003.03660.x. [DOI] [PubMed] [Google Scholar]
- 105.Flardh K. Cell polarity and the control of apical growth in Streptomyces. Curr Opin Microbiol. 2010;13:758–765. doi: 10.1016/j.mib.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 106.Hempel AM, et al. Assemblies of DivIVA mark sites for hyphal branching and can establish new zones of cell wall growth in Streptomyces coelicolor. J Bacteriol. 2008;190:7579–7583. doi: 10.1128/JB.00839-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bagchi S, et al. Intermediate filament-like proteins in bacteria and a cytoskeletal function in Streptomyces. Mol Microbiol. 2008;70:1037–1050. doi: 10.1111/j.1365-2958.2008.06473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Holmes NA, et al. Coiled-coil protein Scy is a key component of a multiprotein assembly controlling polarized growth in Streptomyces. Proc Natl Acad Sci U S A. 2013;110:E397–406. doi: 10.1073/pnas.1210657110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fuchino K, et al. Dynamic gradients of an intermediate filament-like cytoskeleton are recruited by a polarity landmark during apical growth. Proc Natl Acad Sci U S A. 2013;110:E1889–1897. doi: 10.1073/pnas.1305358110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hempel AM, et al. The Ser/Thr protein kinase AfsK regulates polar growth and hyphal branching in the filamentous bacteria Streptomyces. Proc Natl Acad Sci U S A. 2012;109:E2371–2379. doi: 10.1073/pnas.1207409109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Richards DM, et al. Mechanistic basis of branch-site selection in filamentous bacteria. PLoS Comput Biol. 2012;8:e1002423. doi: 10.1371/journal.pcbi.1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Oliva MA, et al. Features critical for membrane binding revealed by DivIVA crystal structure. EMBO J. 2010;29:1988–2001. doi: 10.1038/emboj.2010.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Goriely A, Tabor M. Biomechanical models of hyphal growth in actinomycetes. J Theor Biol. 2003;222:211–218. doi: 10.1016/s0022-5193(03)00029-8. [DOI] [PubMed] [Google Scholar]
- 114.Koch AL. The Problem of Hyphal Growth in Streptomycetes and Fungi. J Theor Biol. 1994;171:137–150. [Google Scholar]
- 115.Moore RL. The biology of Hyphomicrobium and other prosthecate, budding bacteria. Annu Rev Microbiol. 1981;35:567–594. doi: 10.1146/annurev.mi.35.100181.003031. [DOI] [PubMed] [Google Scholar]
- 116.Wali TM, et al. Timing of swarmer cell cycle morphogenesis and macromolecular synthesis by Hyphomicrobium neptunium in synchronous culture. J Bacteriol. 1980;144:406–412. doi: 10.1128/jb.144.1.406-412.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cserti E, et al. Dynamics of the peptidoglycan biosynthetic machinery in the stalked budding bacterium Hyphomonas neptunium. Mol Microbiol. 2017;103:875–895. doi: 10.1111/mmi.13593. [DOI] [PubMed] [Google Scholar]
- 118.Senz M, et al. Control of cell morphology of probiotic Lactobacillus acidophilus for enhanced cell stability during industrial processing. Int J Food Microbiol. 2015;192:34–42. doi: 10.1016/j.ijfoodmicro.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 119.Nittami T, et al. Quantification of Chloroflexi Eikelboom morphotype 1851 for prediction and control of bulking events in municipal activated sludge plants in Japan. Appl Microbiol Biotechnol. 2017;101:3861–3869. doi: 10.1007/s00253-016-8077-4. [DOI] [PubMed] [Google Scholar]
- 120.Webb AG. Radiofrequency microcoils for magnetic resonance imaging and spectroscopy. J Magn Reson. 2013;229:55–66. doi: 10.1016/j.jmr.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 121.Kamata K, et al. Spirulina-templated metal microcoils with controlled helical structures for THz electromagnetic responses. Sci Rep. 2014;4:4919. doi: 10.1038/srep04919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jordan A, et al. Engineering Cyanobacterial Cell Morphology for Enhanced Recovery and Processing of Biomass. Appl Environ Microbiol. 2017;83:e00053–00017. doi: 10.1128/AEM.00053-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Reddy CS, et al. Polyhydroxyalkanoates: an overview. Bioresour Technol. 2003;87:137–146. doi: 10.1016/s0960-8524(02)00212-2. [DOI] [PubMed] [Google Scholar]
- 124.Rodriguez-Carmona E, Villaverde A. Nanostructured bacterial materials for innovative medicines. Trends Microbiol. 2010;18:423–430. doi: 10.1016/j.tim.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 125.Shrivastav A, et al. Advances in the applications of polyhydroxyalkanoate nanoparticles for novel drug delivery system. Biomed Res Int. 2013;2013:581684. doi: 10.1155/2013/581684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jiang XR, Chen GQ. Morphology engineering of bacteria for bio-production. Biotechnol Adv. 2016;34:435–440. doi: 10.1016/j.biotechadv.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 127.Bi E, Lutkenhaus J. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J Bacteriol. 1993;175:1118–1125. doi: 10.1128/jb.175.4.1118-1125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tan D, et al. Engineering Halomonas TD01 for the low-cost production of polyhydroxyalkanoates. Metab Eng. 2014;26:34–47. doi: 10.1016/j.ymben.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 129.Wu H, et al. Engineering the growth pattern and cell morphology for enhanced PHB production by Escherichia coli. Appl Microbiol Biotechnol. 2016;100:9907–9916. doi: 10.1007/s00253-016-7715-1. [DOI] [PubMed] [Google Scholar]
- 130.Elhadi D, et al. CRISPRi engineering E. coli for morphology diversification. Metab Eng. 2016;38:358–369. doi: 10.1016/j.ymben.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 131.Hugenholtz P. Exploring prokaryotic diversity in the genomic era. Genome Biol. 2002;3:REVIEWS0003. doi: 10.1186/gb-2002-3-2-reviews0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hug LA, et al. A new view of the tree of life. Nat Microbiol. 2016;1:16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- 133.Wu D, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Boutros M, et al. Microscopy-Based High-Content Screening. Cell. 2015;163:1314–1325. doi: 10.1016/j.cell.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 135.Ducret A, et al. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol. 2016;1:16077. doi: 10.1038/nmicrobiol.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Paintdakhi A, et al. Oufti: an integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Mol Microbiol. 2016;99:767–777. doi: 10.1111/mmi.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ursell T, et al. Rapid, precise quantification of bacterial cellular dimensions across a genomic-scale knockout library. BMC Biol. 2017;15:17. doi: 10.1186/s12915-017-0348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.McCloskey D, et al. Basic and applied uses of genome-scale metabolic network reconstructions of Escherichia coli. Mol Syst Biol. 2013;9:661. doi: 10.1038/msb.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gawad C, et al. Single-cell genome sequencing: current state of the science. Nat Rev Genet. 2016;17:175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- 140.Russell JH, Keiler KC. Screen for localized proteins in Caulobacter crescentus. PLoS One. 2008;3:e1756. doi: 10.1371/journal.pone.0001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kuru E, et al. In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed Engl. 2012;51:12519–12523. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kuru E, et al. Synthesis of fluorescent D-amino acids and their use for probing peptidoglycan synthesis and bacterial growth in situ. Nat Protoc. 2015;10:33–52. doi: 10.1038/nprot.2014.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Desmarais SM, et al. Peptidoglycan at its peaks: how chromatographic analyses can reveal bacterial cell wall structure and assembly. Mol Microbiol. 2013;89:1–13. doi: 10.1111/mmi.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Glauner B. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal Biochem. 1988;172:451–464. doi: 10.1016/0003-2697(88)90468-x. [DOI] [PubMed] [Google Scholar]
- 145.Desmarais SM, et al. High-throughput, Highly Sensitive Analyses of Bacterial Morphogenesis Using Ultra Performance Liquid Chromatography. J Biol Chem. 2015;290:31090–31100. doi: 10.1074/jbc.M115.661660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Desmarais SM, et al. Isolation and preparation of bacterial cell walls for compositional analysis by ultra performance liquid chromatography. J Vis Exp. 2014:e51183. doi: 10.3791/51183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shi H, et al. Strain Library Imaging Protocol for high-throughput, automated single-cell microscopy of large bacterial collections arrayed on multiwell plates. Nat Protoc. 2017;12:429–438. doi: 10.1038/nprot.2016.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Egan AJ, et al. Activities and regulation of peptidoglycan synthases. Philos Trans R Soc Lond B Biol Sci. 2015;370 doi: 10.1098/rstb.2015.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bordowitz JR, Montgomery BL. Photoregulation of cellular morphology during complementary chromatic adaptation requires sensor-kinase-class protein RcaE in Fremyella diplosiphon. J Bacteriol. 2008;190:4069–4074. doi: 10.1128/JB.00018-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Weiner RM, et al. Hyphomonas adhaerens sp. nov., Hyphomonas johnsonii sp. nov. and Hyphomonas rosenbergii sp. nov., marine budding and prosthecate bacteria. Int J Syst Evol Microbiol. 2000;50(Pt 2):459–469. doi: 10.1099/00207713-50-2-459. [DOI] [PubMed] [Google Scholar]