Abstract
The proteinaceous zipper-like structure known as the synaptonemal complex (SC), which forms between pairs of homologous chromosomes during meiosis from yeast to humans, plays important roles in promoting interhomolog crossover formation, regulating cessation of DNA double-strand break formation following crossover designation, and ensuring accurate meiotic chromosome segregation. Recent studies are starting to reveal critical roles for different protein modifications in regulating SC dynamics. Protein SUMOylation, N-terminal acetylation, and phosphorylation have been shown to be essential for the regulated assembly and disassembly of the SC. Moreover, phosphorylation of specific SC components has been found to link changes in SC dynamics with meiotic recombination. This review highlights the latest findings on how protein modifications regulate SC dynamics and functions.
Keywords: Synaptonemal complex, meiosis, SUMOylation, N-terminal acetylation, phosphorylation, meiotic recombination
Meiosis is required for sexual reproduction
Meiosis is a specialized cell division process that generates haploid gametes (i.e. eggs and sperm) from diploid germ cells. It is essential for sexual reproduction and achieves the halving of the genome by following a single round of DNA replication with two consecutive cell divisions referred to as meiosis I (MI) and meiosis II (MII). MI results in the segregation of homologous chromosomes away from each other, whereas during MII, sister chromatids are separated similarly to a mitotic cell division. A remarkable feature of meiosis is a prolonged prophase I, during which a series of events must take place to ensure accurate chromosome segregation at MI. These critical events include pairing between homologous chromosomes, formation of programmed DNA double-strand breaks (DSBs), homologous recombination (HR) resulting in crossover (CO) formation, synaptonemal complex (SC) assembly, and late prophase I chromosome remodeling [1]. Defects in any of these steps can cause errors in meiotic chromosome segregation, which are associated with infertility, miscarriages, birth defects, and tumorigenesis in humans [2].
The SC, a macromolecular structure that assembles between paired homologous chromosomes during early meiotic prophase, is a hallmark feature of meiosis observed in most species from yeast to humans [3, 4]. This scaffold is required to stabilize homologous pairing interactions and promote interhomolog crossover formation. Crossovers not only provide for genetic diversity among offspring, but in conjunction with sister chromatid cohesion, also provide essential physical attachments (chiasmata) between pairs of homologs, allowing for proper metaphase alignment and accurate segregation at MI [5]. Without SC assembly, interhomolog crossovers either fail to occur or are misregulated, suggesting a conserved function for the SC in promoting crossover formation [1, 6]. The importance of functional SC assembly is further supported by the observation that mutations in genes encoding for SC components are linked to miscarriages, infertility and birth defects in humans [7]. Moreover, the SC is not a static structure as it undergoes changes in dynamic state throughout MI (see “SC dynamics are tightly regulated” section). Therefore, understanding the regulation of this complex structure is critical for understanding how changes in SC dynamics are linked to the progression of other meiotic events that together result in accurate chromosome segregation at MI. In this review, we focus on recent studies that have uncovered how different protein modifications regulate the assembly, maintenance and disassembly of this structure and how changes in SC dynamics are linked to the progression of meiotic recombination.
The synaptonemal complex exhibits ultrastructural conservation
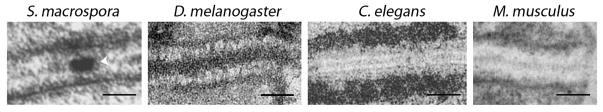
The fully formed SC at pachytene is a tripartite structure, as revealed by electron microscopy, consisting of two lateral elements that run along the electron-dense chromatin and flank the central region, which bridges the paired homologous axes (Figure 1) [3]. In most organisms, the central region consists of two sub-components, the transverse filaments and the central element. The chromatin is organized into a series of loops that are tethered at their bases to the chromosome axes. Sister-chromatids are closely held together by cohesins, thereby forming a single distinguishable unit during early meiotic prophase I. The tripartite configuration of the SC is a conserved feature of this structure throughout most organisms including budding yeast, plants, worms, flies and mammals [4]. Interestingly, the width of the SC is also similar across species (approximately 90–150 nm) (Figure 1), although their genome sizes can span from ~12 Mb in yeast to ~3k Mb in mice and humans.
Figure 1.
EM images of the SC in meiotic nuclei from different organisms. The SCs are visible as zipper-like structures flanked by electron-dense chromatin patches. In the case of the Ascomycete Sordaria macrospora, an electron-dense recombination nodule is observed associated with the central element, as indicate by a white arrowhead. Images were originally published in [4, 87–89]. Scale bars equal 100 nm.
Several components of the SC have been identified in different organisms (Figure 2). Based on their localization, the components can generally be divided into three groups: axial element proteins that assemble onto meiotic chromosome axes (referred to as lateral elements after SC assembly); transverse filament proteins that bridge the parallel homologous axes; and central element proteins that are located along the center of the SC. Transverse filament proteins and central element proteins comprise the central region of the SC.
Figure 2.

(A) Diagram of the SC structure and SC components in different organisms. The C. elegans SYP-2 protein was initially classified as a central region component due to lack of specific information regarding its protein-protein interactions or sublocalization within the SC [87]. Based on recent immuno-EM analysis and protein-protein interaction data [11], we now reclassify SYP-2 as a central element protein. (B) Known modifications for SC components and their functions.
Strikingly, while the SC exhibits a similar ultrastructure across organisms, components of the SC share little, if any, similarity at the amino acid sequence level [8, 9]. However, there are some conserved protein features (protein secondary structure) and similarities in the organization of the SC components that may contribute to the conserved ultrastructure and biological functions observed for the SC. First, central region components in yeast, fly, worms, mouse, and plants contain coiled-coil domains, which are known to promote protein-protein interactions, flanked by globular domains, and can form homodimers [4]. Some lateral element proteins, such as SYCP3 in mice, also contain coiled-coil domains [10, 11]. Moreover, some of these coiled-coil domain-containing proteins, such as SYCP1 and SYCP3 in mammals, SYP-1 in C. elegans, and Zip1 in budding yeast, can self-organize into higher-order structures in the absence of other SC proteins [8, 11, 12], and due to this self-organizing ability they might form the basic structural frame for SC assembly. Finally, transverse filament proteins display conservation on how they are positioned or organized within the SC. For mouse SYCP1, yeast Zip1, fly C(3)G, and worm SYP-1, the N-terminal domain of the transverse filament protein is always located in the middle of the SC, while its C-terminus is located next to the lateral elements [11–14]. Such a conserved organization pattern might be critical for the function of the SC.
Many SC proteins also contain intrinsically disordered or unstructured regions along their sequences (Table 1). Disordered regions, among other functions, have been postulated to promote protein-protein interactions and protein-nucleic acid interactions, as well as regulate protein lifetime [15, 16]. The importance of such regions has gained more attention recently, as shown for example in C. elegans, where the lateral element proteins HTP-1, HTP-2, HTP-3, and HIM-3 all contain such disordered regions in their C-terminal ends, and they mediate the assembly of the meiotic chromosome axes [17]. Most of the known central region proteins also contain disordered regions (Table 1). However, how disordered region-mediated interactions can contribute to the assembly of the highly dynamic SC structure needs further investigation.
Table 1.
Predicted coiled-coil domains and disordered regions among SC components
| Organism | Protein name | Amino acids | Coiled-coil | Disordered |
|---|---|---|---|---|
| S. cerevisiae | Red1 | 827 | 368–388; 767–790 | 363–405; 407–511; 561–700 |
| S. cerevisiae | Zip1 | 857 | 184–326; 403–748 [12] | 1–42; 53–99; 110–179; 310–351; 406–429; 538–597; 615–664; 693–875 |
| S. cerevisiae | Ecm11 | 302 | 250–300 [56] | 1–72; 135–187 |
| S. cerevisiae | Gmc2 | 188 | 100–140; 160–188 [56] | 1–38; |
| C. elegans | HTP-1 | 352 | - | 1–22; 284–352 |
| C. elegans | HTP-2 | 352 | - | 285–352 |
| C. elegans | HTP-3 | 739 | 566–593 | 1–20; 290–319; 327–558; 567–618; 620–642; 691–739 |
| C. elegans | HIM-3 | 291 | - | 259–291 |
| C. elegans | SYP-1 | 489 | 48–402 [11] | 41–81; 108–177; 179–219; 230–254; 286–397; 446–489 |
| C. elegans | SYP-2 | 213 | 98–160 [11] | 1–66; 97–118; |
| C. elegans | SYP-3 | 224 | 61–179 [11] | 43–99; 131–156 |
| C. elegans | SYP-4 | 605 | 115–410 [90] | 231–255; 260–298; 300–981 |
| D. melanogaster | C(3)G | 744 | 158–646 [91] | 1–28; 41–106; 157–196; 231–257; 478–546; 592–633; 660–712 |
| D. melanogaster | corolla | 554 | 20–47; 104–169; 324–351 [92] | 125–179; 244–264; 311–435; 500–554 |
| D. melanogaster | cona | 207 | - | 1–27 |
| M. musculus | SYCP2 | 1500 | 1379–1433 [93] | 446–574; 638–696; 730–833; 911–1015; 1020–1238; 1241–1297 |
| M. musculus | SYCP3 | 254 | 66–223 [94] | 1–78; 219–254 |
| M. musculus | SYCP1 | 993 | 52–752 [95] | 1–20; 119–159; 245–364; 381–434; 436–504; 555–578; 605–650; 660–682; 756–865; 869–890; 893–947; 965–993 |
| M. musculus | SYCE1 | 329 | 55–124; 132–166; 194–214; 242–290 [96] | 1–31; 92–113; 116–162; 244–329 |
| M. musculus | SYCE2 | 171 | 60–87 [97] | 1–49; 58–83 |
| M. musculus | SYCE3 | 88 | 6–39 [98] | 1–28 |
| M. musculus | TEX12 | 123 | - | 1–53 |
| M. musculus | SIX6OS1 | 574 | 199–233 [39] | 218–342; 395–429; 433–461; 470–517; 522–574 |
The coiled-coil domains for S. cerevisiae Red1 and C. elegans HTP-3 were predicted by COILS at http://embnet.vital-it.ch/software/COILS_form.html. This program was run using the MTIDK matrix with a 21-residue window and applying an unweighted scan. Protein regions were predicted to adopt a coiled-coil conformation if the amino acids within those regions had scores of 0.5 or higher. Coiled-coil domains for other proteins were reported in the indicated references. Protein disordered regions were predicted by PONDR-FIT at http://disorder.compbio.iupui.edu/pondr-fit.php. Protein regions (at least 20 amino acids) were predicted as disordered regions if the amino acids within those regions had scores of 0.5 or higher.
SC dynamics are tightly regulated
Three aspects of an SC’s life are dynamic: SC assembly, its highly dynamic steady state, and SC disassembly. Assembly of the SC is mediated through integration of transverse filaments and central elements to connect two axial elements, a process that is poorly understood. The assembly of the SC must be tightly regulated to ensure that it takes place only between pairs of homologous chromosomes and throughout their full lengths. At mid-meiotic prophase, during the sub-stage referred to as pachytene, SC is assembled at the interface of nearly all lengthwise-aligned homologous chromosome pairs. Interestingly, studies from at least two model organisms suggest that the SC structure observed during early prophase is not static. A study in yeast showed that SC components continually come on and off from chromosomes, with higher rates of SC subunit incorporation than dissociation [18]. In C. elegans, after a subset of the programmed meiotic DSBs are designated to be repaired as crossovers, the SC becomes more stable [19, 20]. Finally, after crossover formation, the SC starts to disassemble in an asymmetric manner and SC proteins are retained at specific chromosome subdomains until late prophase I [21, 22]. This sequential disassembly of the SC, which starts upon exit from pachytene and extends through late prophase (diplotene and diakinesis), is accompanied by changes both in chromosome compaction as well as in the composition and localization of additional proteins required for the regulated loss of sister chromatid cohesion at MI [21–23]. This process, termed “chromosome remodeling” occurs during the pachytene to diplotene transition and results in the characteristic cruciform configuration observed for bivalents by late diakinesis. During this process in yeast, flies and mice, SC proteins persist at centromeres where they have been proposed to promote proper centromere bi-orientation leading to proper homolog segregation at MI [24–28]. In C. elegans, which has holocentric chromosomes and therefore lacks a defined single centromere, the central region proteins are retained only on the shorter of the two intersecting axes of different lengths that form the cruciform (the short and long arms of the bivalent), whereas two lateral element HORMA domain-containing proteins (HTP-1/2) are retained only at the long arm of the bivalents [29]. During metaphase I, homologs align at the metaphase plate with the short arms positioned equatorially to the plate and the long arms facing the poles. Short arm-specific loss of sister-chromatid cohesion is required for accurate segregation of homologs at MI [23, 30–33]. Therefore, the coordinated disassembly of the SC plays a critical role in setting the stage for accurate chromosome segregation throughout species.
Both the assembly and disassembly of the SC as well as its changes in dynamic state are likely achieved through multiple layers of regulation so that these processes are precisely coordinated with the HR process. These layers include: 1) transcriptional regulation of the SC genes [34, 35]; 2) translational control of SC mRNAs [36]; 3) association of nonstructural regulators with SC components [37–39]; 4) protein modifications (the main focus of this review); 5) protein stability/degradation [40, 41]; and 6) cytoskeletal-driven chromosome movements [42–45]. While description of the first two layers of regulation has been documented for a relatively long time, recent studies have started to reveal the other layers of regulation including nonstructural regulators and protein modifications. A recent review has also summarized the regulation underlying the assembly and disassembly processes of the SC observed in some model organisms [4], but the regulation by protein modifications was not emphasized due to lack of supporting publications at that time. The rest of this review will focus on the roles of protein modifications in regulating SC dynamics and linking changes in SC dynamics with homologous recombination.
Modifications that are required for proper SC formation
In some organisms, the expression of SC components can be observed before meiotic entry, but they do not assemble into a normal SC structure until entry into meiosis [46–48]. In C. elegans, both the SC lateral element components and central region proteins are expressed in proliferating germ cells, but lateral element proteins do not assemble into axial structures and the central region proteins form aggregates [47, 49]. Upon meiotic entry, these proteins assemble into the SC structure. In Drosophila, synapsis can be observed at the centromeres during the premeiotic mitotic divisions [46], However, the SC proteins dissociate from centromeres during mitotic division and SC assembly along chromosomes resumes after meiotic entry. In budding yeast and mammals, SC proteins are abundant in meiotic nuclei but SC assembly occurs only after the initiation of Spo11-mediated meiotic recombination [50, 51]. The regulatory mechanisms keeping these SC proteins in check in yeast, worms and mammals, or prompting the centromere association and loss in flies prior to meiotic entry are not clear, but post-translational modifications might be critical during this process. In fact, recent studies have determined that modifications of SC proteins, including SUMOylation and N-terminal acetylation, are required for SC assembly.
SUMOylation
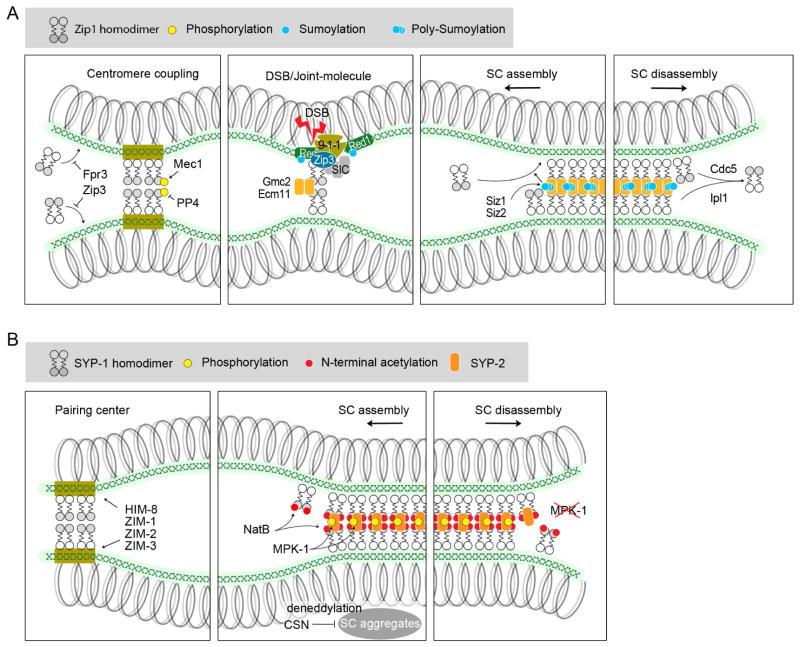
Studies in yeast first revealed SUMOylation as a post-translational modification required for SC assembly [52–56] (Figure 3A, Key Figure). It was shown that the transverse-filament protein Zip1 first loads to coupled centromeres [52], and that this loading is restricted by two proteins: the SUMO E3 ligase Zip3 and the proline isomerase Fpr3. Zip3 regulates centromere-associated Zip1 and Fpr3 regulates unassociated Zip1 in the nucleoplasm [53]. These proteins act in parallel to negatively regulate early SC assembly along chromosome arms. The initiation of synapsis at crossover-designated interhomologous engagements is controlled by a protein structure known as the synapsis initiation complex (SIC), which is composed of at least eight ZMM proteins: Zip1–4, Mer3, Msh4, Msh5 and Spo16. Two of the SIC members, Zip1 and Zip3, are recruited to the DSB sites by the 9-1-1 DNA damage-response complex (Ddc1, Mec3 and Rad17). The interaction between Ddc1 and the lateral-element protein Red1 is essential for SC assembly [55]. Although Red1 can be SUMOylated during this process, the mechanisms underlying Red1 SUMOylation and whether this SUMOylation is required for SC assembly, remain to be determined [54, 55]. A proposed model is that once it is SUMOylated Red1 then interacts with Zip1 and recruits the rest of the SIC proteins [57], followed by recruitment of the central element proteins Gmc2 and Ecm11 [56].
Figure 3, Key Figure.
Summary cartoons for the involvement of protein modifications in SC assembly and disassembly in yeast (A) and worms (B). Note that although Zip1 is depicted as having the same organization at centromeres as it does within the SC, there is some evidence suggesting potentially different configurations at these locations [65]. For simplicity, only SYP-1 and SYP-2 are shown in the central region of the worm SC. CSN, COP9 signalosome.
A role for SUMOylation of Ecm11 in SC assembly has been better defined. SC assembly is abrogated in non-SUMOylatable Ecm11 mutants [56] and the E3 SUMO ligases Siz1 and Siz2 are required for Ecm11 SUMOylation as well as for SC assembly [58]. The transverse filament protein Zip1 can activate SUMOylation of Ecm11 and SUMOylated Ecm11 facilitates the recruitment of more Zip1, forming a positive feedback loop to promote efficient SC assembly [58]. Interestingly, the N-terminal domain of Zip1 is sufficient to activate Ecm11 SUMOylation even in vegetative cells as long as Gmc2 is present. Non-SUMOylated Ecm11 can be incorporated into the SC structure after SC assembly has been initiated, but it cannot support de novo SC assembly, supporting a requirement for SUMOylation in SC assembly [58]. Although the roles of SUMOylation in SC regulation have been well documented in yeast, such regulation in other organisms still needs investigation. In mouse, similar to Red1 in yeast, the lateral element protein SYCP3 can also be SUMOylated [59], but the involvement of this modification in SC assembly remains unclear.
Protein N-terminal acetylation
A recent study in C. elegans revealed a role for protein N-terminal acetylation in SC assembly [60] (Figure 3B, Key Figure). Protein N-terminal acetylation is mediated by N-terminal acetyltransferases (NATs) that catalyze the transfer of an acetyl group from acetyl-CoA to the N-alpha amino group of the proteins. N-terminal acetylation is a prevalent co-translational modification and takes place on the nascent polypeptides on the ribosomes. In C. elegans, the N-terminal acetyltransferase complex B (NatB) is required for the proper assembly of the SC and crossover formation. NatB mutants exhibited delayed and incomplete SC assembly as well as SC assembly on unpaired chromosomes and between non-homologous chromosomes. SYP-1, a central region protein in C. elegans, is a confirmed target of NatB, and interfering with SYP-1 N-terminal acetylation results in delayed and incomplete SC assembly as well as reduced crossovers.
It is interesting that a single small modification can affect the assembly of a large macromolecular structure. This suggests that the N-terminal end of SYP-1 is critical for SC assembly, very likely by mediating protein-protein interactions. Acetylation of the N-terminal amino acid can neutralize the positive charge associated with the unmodified amino group, so that it enhances hydrophobic protein interactions as exemplified by a previous study [61]. In fact, hydrophobic interactions have been proposed as a major requirement for the assembly and disassembly of the SC in multiple organisms including C. elegans [62]. Interestingly, a study of the organization of the SC proteins in C. elegans revealed that the N-terminal domain of SYP-1 is located at the middle of the central region [11]. Moreover, the conservation observed both in the organization pattern and the N-terminal sequences among transverse filament proteins [60] suggests that the requirement of N-terminal acetylation for SC assembly might be a conserved feature of meiosis across organisms.
Protein phosphorylation
It is known that, as in mitosis, the cell cycle regulatory kinases CDK (cyclin-dependent kinase) and DDK (Dbf4-dependent kinase) are also key drivers of meiotic progression [63, 64]. SC proteins could be the direct targets of these kinases, and phosphorylation and dephosphorylation of SC proteins might be involved in SC assembly and disassembly. Indeed, yeast Zip1 interactions at non-homologously coupled centromeres are disrupted by the Mec1 kinase (ATR) and PP4 phosphatase, which regulate the phosphorylation and dephosphorylation of Zip1 on serine 75. Zip1 phosphorylation dynamically destabilizes homology-independent centromere pairing through an unknown mechanism [65]. A proposed model is that phosphorylation of the Zip1 N-terminal domain disrupts the association between Zip1 homodimers. (Figure 3A, Key Figure).
It is known that DSBs trigger activation of multiple kinases including ATM kinase and DNA-protein kinase [66], but it is not clear if DSB-dependent kinase activation is associated with SC protein phosphorylation. In mammals, yeast and plants, assembly of the SC requires DSB formation. However, this requirement is not conserved among organisms that utilize different mechanisms to achieve homologous chromosome pairing such as flies and worms. Interestingly, relatively few DSBs occur in species that can pair chromosomes without recombination (~20–40 per cell on average in D. melanogaster and C. elegans) [67–69]. In contrast, more DSBs are generated in organisms that rely on recombination for efficient pairing, such as S. cerevisiae (~ 50–200 per cell), plants (200–300 in Arabidopsis thaliana and >1500 per cell in lily), or mammals (~200–300 in mouse) [70]. In yeast, SC assembly is initiated at centromeres by unknown mechanisms and at crossover-designated DSB sites by assembly of the SIC complex. However in mice, initiation of synapsis does not specifically depend on crossover-designated DSB sites, and instead requires the overall interhomologous engagements (formation of enough DSBs), which promote homology search and synapsis [71]. Further investigations are still required to identify potential phosphorylation and dephosphorylation of SC proteins that regulate DSB-dependent SC assembly in mammals and other metazoans.
In addition to the modifications mentioned above, there may also be other modifications involved in regulating SC dynamics. The CSN/COP9 signalosome has been shown to regulate SC assembly in C. elegans, and protein neddylation is regulated by the CSN/COP9 signalosome [72]. Moreover, neddylation is required for proper SC assembly and crossover localization in plants [73]. Therefore, other types of modifications, including neddylation, could be involved in SC regulation.
Modifications that link changes in SC dynamics to the HR process
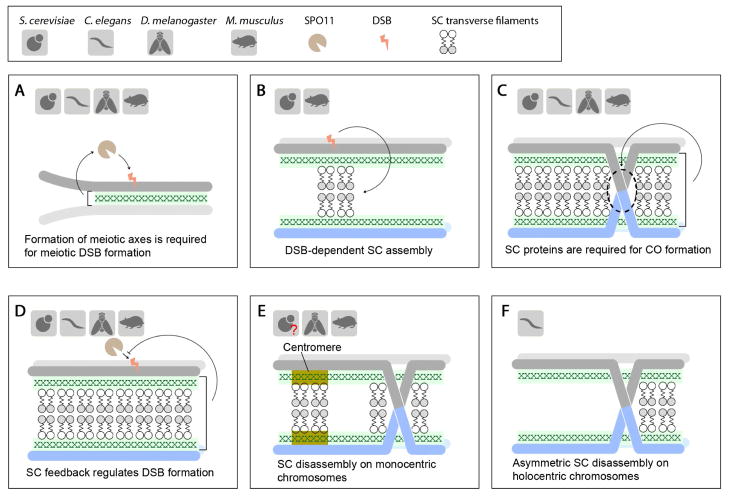
The establishment of meiosis-specific higher-order chromosome structures and the meiotic recombination process are highly integrated events during prophase [6]. Proper formation of axial elements has been found to be essential for the initiation of meiotic recombination across organisms, which involves SPO11-catalyzed formation of DSBs on meiotic chromatin [74] (Figure 4A). In some organisms, initiation of meiotic recombination is in turn required for SC assembly (Figure 4B). The roles of the SC in promoting the completion of recombination and crossover formation, and the reciprocal effects of recombination on the SC, are still poorly understood. Recent findings highlight the roles of post-translational modifications in the communication between SC dynamics and homologous recombination.
Figure 4.
Interactions between the SC and meiotic recombination. (A) Formation of the meiotic axes is required for SPO11-induced DSB formation across organisms. (B) Initiation of SC assembly is DSB-dependent in yeast and mice. (C) The SC is required for crossover (CO) formation among the majority of studied organisms. In budding yeast, some SC proteins, but not necessarily an intact SC structure, are required for CO formation. (D) The SC plays a role in feedback regulation of DSB formation across organisms. (E) SC disassembly in monocentric organisms. In the case of budding yeast, while Zip1 promotes centromere interactions, it is yet unclear if this involves the SC structure. (F) Asymmetric SC disassembly in worms is coupled with CO formation.
A feedback loop regulates DSB formation and SC stability
During pachytene stage, the SC is fully assembled between all pairs of homologous chromosomes and DSBs need to be repaired. After at least one crossover is formed for each pair of homologs, thus fulfilling the requirement for an obligate crossover, further programmed DSB formation must be stopped to reduce potential DNA lesions. Superimposed onto this process is the observation that the SC is not static during early pachytene. First, studies in yeast and worms indicated that the SC is highly dynamic and its components are continuously incorporated and replaced. The addition of postsynaptic SC components occurs at a faster rate than the removal of the same components, resulting in an accumulation of SC components at late pachytene [18, 75]. Second, by using FRAP analysis, two groups revealed that the central region of the SC changes from a more dynamic state in early pachytene to a less dynamic/more stable state in late pachytene in worms [19, 20]. Moreover, the change of SC dynamics is related to the progression of meiotic recombination and is dependent on Polo-like kinases. Specifically, PLK-mediated phosphorylation of the central region protein SYP-4 is critical for the change in SC dynamics in the C. elegans germline, and appearance of this modification on the SC coincides with the timing of CO designation. Furthermore, SYP-4 phosphorylation is also dependent on DSB formation and CO designation. These studies provide novel insights into the regulation of SC dynamics by meiotic recombination.
Studies in C. elegans also revealed a feedback loop linking changes in SC dynamics and meiotic recombination through phosphorylation. Specifically, phosphorylation of SYP-4, which results in a more stable SC state, negatively regulates DSB formation thus providing a mechanism to promote genomic integrity by downregulating DSB formation upon CO designation [20]. The involvement of the SC in feedback regulation of meiotic progression is not limited to worms. In mice, the meiotic chromosome proteins HORMAD1/2 have been shown to be involved in the feedback regulation of DSB formation [76], and the ZMM proteins in yeast (Zip1-4, Msh4-5, Mer3, and others) that promote interhomolog engagement, are also involved in turning off further programmed DSB formation (reviewed in [70]). However, the involvement of specific phosphorylation or other post-translational modification during feedback regulation is not clear in these organisms. Moreover, how meiotic recombination and CO formation can trigger phosphorylation of SC components, are questions that need further investigation.
The coordination between crossover formation and SC disassembly
Cell-cycle kinases play an important role in regulating SC disassembly. In S. cerevisiae, Cdc5 (Polo-like kinase), Ipl1 (Aurora B), DDK (Dbf4-dependent Cdc7 kinase), and CDK1 are required for SC disassembly [77–79]. However, their targets leading to SC disassembly remain unclear. In mice, PLK1 (Polo-like kinase 1) and Aurora B are also required for SC disassembly. PLK1 is recruited to the SC during meiotic prophase and directly phosphorylates central element proteins SYCP1, TEX12 and SYCE1. Its phosphorylation activity is required for the disassembly of central and lateral elements of the SC and is further required for exit from meiotic prophase [80]. Aurora B and INCENP also localize to meiotic chromosomes during prophase [81]. However, unlike PLK1, Aurora kinase activity seems to be required for disassembly of lateral elements, but not the disassembly of the central elements of the SC [82]. CDK1-Cyclin B is also involved in SC disassembly during late prophase. CDK1 activation during meiotic prophase is controlled by the chaperone protein HSPA2 (formerly HSP70-2), which localizes to the SC during pachytene and might thus recruit CDK1 to the SC [83–85]. Therefore, SC components that contain predicted CDK1-phosphorylation sites, such as SYCP1, might be the targets of CDK1 resulting in mouse SC disassembly [83].
A recent study in C. elegans revealed the involvement of the MAP kinase pathway in regulating SC dynamics during late prophase [86]. Specifically, the mammalian Rho GEF homolog ECT-2 functions through the conserved RAS/ERK MAP kinase signaling pathway in the germline to regulate the disassembly of the SC. The central element protein SYP-2, which was previously classified as a central region component due to lack of specific information regarding its protein-protein interactions or sublocalization within the SC [87], is a target for MPK-1-mediated phosphorylation, and SYP-2 phosphorylation impairs the disassembly of the SC. Inactivation of MAP kinase at late pachytene is critical for timely disassembly of the SC proteins from the long arms of the bivalents, and interestingly, the inactivation of MAP kinase is also dependent on CO promoting factors.
These combined studies suggest the presence of waves of SC phosphorylation and dephosphorylation during meiotic prophase, which can affect SC initiation, dynamics, and late prophase disassembly. However, although the involvement of protein modifications has been found at different steps during prophase in different organisms, further analysis is needed to assess their conservation across organisms.
Concluding Remarks and Future Perspectives
The SC is one of the most complex structures in the cell, whose components undergo various different types of modifications and changes in dynamic state throughout prophase I. Understanding how changes in SC dynamics happen and how these are associated with meiotic recombination is critical for understanding the roles of the SC in promoting accurate chromosome segregation during meiosis. Protein modifications, including co-translational N-terminal acetylation and post-translational modifications such as SUMOylation and phosphorylation, play important roles in regulating SC dynamics during meiosis in different organisms. Interestingly, our current understanding of the roles of protein modifications in regulating SC functions is mainly focused on central region proteins. Further investigation focused on modifications undergone by lateral element proteins might reveal additional levels of regulation. Moreover, phosphorylation of SC components has been found to be a critical link between meiotic recombination and SC dynamics. However, further investigations are needed to completely reveal how regulation of SC dynamics and meiotic recombination interface with each other. Continued studies of the SC in different model organisms will contribute to uncovering both its conserved as well as distinct features and better inform us as to how it impacts human reproductive health (See Outstanding Questions).
Outstanding Questions Box.
What is the complete interaction map of the SC components? For example, the interactions between transverse filament proteins and the lateral element proteins are not clear. Alternatively, do transverse filament proteins interact with chromatin via their C-termini? Technology is now available to probe these and other important questions regarding the organization of this structure.
What are all the modifications undergone by SC proteins in response to meiotic recombination events? How does CO designation trigger the phosphorylation of the SC proteins? Are there any post-translational modifications involved in the asymmetric/uneven disassembly of the SC during late prophase?
Are SC components organized into the same structure/configuration along the entire length of the chromosomes? Studies in yeast and flies suggest that the organization of the SC components may be different at specific subchromosomal regions such as at centromeres and crossover sites. Further investigations will be required to uncover these differences.
Regarding feedback regulation on meiotic recombination exerted by the SC, what is the mechanism by which the SC regulates DBS formation and potentially other meiotic recombination events? Are there any protein modifications that are restricted to specific regions on the SC, which could, for example, be linked to the regulation of CO distribution?
SC components are not highly conserved at the primary sequence level between species, which limits the ability to identify SC components by extrapolating what is found in one organism onto another. What are the common mechanisms that explain the conserved function of the SC across organisms? How does the organization of the SC assist interhomolog recombination and CO formation? Addressing these and other questions pertaining to this structure will provide important insights into the functions and roles of this hallmark of meiosis and how it impacts reproductive health.
Trends Box.
The synaptonemal complex (SC) is a complex protein structure essential for crossover formation and achieving accurate chromosome segregation during meiosis.
Recent studies place multiple protein modifications, including SUMOylation, protein N-terminal acetylation, and phosphorylation, as required for the regulation of SC assembly and disassembly.
Meiotic recombination-dependent phosphorylation of the SC proteins regulates SC dynamics. Phosphorylation and changes in SC dynamics in turn regulate meiotic recombination events, such as DNA double-strand break formation.
A better understanding of the modifications undergone by SC proteins will provide key insights into the regulation of SC dynamics and of the mechanisms linking the SC to meiotic recombination.
Text box. The sub-stages of meiotic prophase I.
Meiosis generates haploid gametes through two consecutive rounds of cell division, meiosis I and II. Prophase I is a prolonged phase of meiosis, and several important events take place during this phase that are critical for accurate segregation of the homologous chromosomes at meiosis I. According to the morphology of the chromosomes, prophase I has historically been divided into five substages: leptotene, zygotene, pachytene, diplotene, and diakinesis. Leptotene is the first stage of prophase I during which duplicated sister chromatids closely associate and meiotic chromosome axes assemble. During the zygotene stage, homologous chromosome pairs align and the central region of the synaptonemal complex (SC) starts to assemble between the paired chromosomes. Chromosomes usually cluster and acquire a characteristic bouquet organization during this stage. At the pachytene stage, the SC is assembled throughout the whole length of the paired homologous chromosomes (except at the non-homologous chromosome arms on the sex chromosomes). Exchange of chromosome segments (crossover formation) occurs through homologous recombination. The SC disassembles upon exit from pachytene and during the diplotene stage, and homologous chromosome arms disassociate. SC components remain at subchromosomal regions. Diakinesis is the last stage of prophase I, during which homologous chromosomes highly condense and remain connected by their paired centromeres and through chiasmata. The nucleolus disappears and the nuclear envelope starts to disassemble.
Acknowledgments
We thank all members of the Colaiacovo lab for helpful discussions and comments. This work was supported by National Natural Science Foundation of China grant 31701176 (to J.G.) and National Institutes of Health grant R01GM072551 (to M.P.C.).
Glossary
- COP9 signalosome (CSN)
a multiprotein complex involved in ubiquitin-proteasome-mediated protein degradation.
- Crossover (CO)
a recombination product that involves the reciprocal exchange of genetic information between homologs.
- CO designation
when a DNA double-strand break site is selected/designated to be repaired as a CO and is marked by CO-promoting factors such as COSA-1/CNTD1.
- Diakinesis
the last (5th) stage of meiotic prophase I, where homologous chromosomes are highly condensed and remain associated through centromere pairing and/or chiasmata (physical attachments resulting from earlier crossover events underpinned by flanking sister chromatid cohesion).
- Diplotene
the 4th stage of meiotic prophase I, which occurs between pachytene and diakinesis. The disassembly of the SC initiated in late pachytene continues to progress during diplotene resulting in the separation of some regions between homologous chromosomes.
- Holocentric chromosomes
chromosomes in which the kinetochore protein complex does not assemble at a defined single locus referred to as the centromere, and instead spindle microtubules attach throughout the entire length of chromosomes during mitosis or meiosis.
- Homologous recombination (HR)
a type of genetic recombination that takes place between similar or identical DNA molecules and results in exchange of nucleotide sequences.
- Metaphase I
a phase of of the meiosis I cell division during which pairs of chromosomes (bivalents) are aligned at the equatorial plate of the spindle and bi-oriented by spindle microtubules.
- Neddylation
a post-translational modification process analogous to ubiquitylation in which target proteins are covalently conjugated with the small ubiquitin-like protein NEDD8.
- Pachytene
the 3rd stage of meiotic prophase I when homologous chromosomes are aligned along their whole lengths and held together by the zipper-like structure known as the synaptonemal complex. CO recombination is completed during this stage.
- Polo-like kinases (PLKs)
serine/threonine kinases that control cell cycle progression both in mitosis and in meiosis.
- Premeiotic tip
the distal region of the C. elegans gonad where germ cells undergo mitotic proliferation.
- Prophase I
a phase of meiosis I during which the replicated chromosomes (homologous chromosomes) condense and undergo pairing, synapsis, and recombination. It is comprised of five stages: leptotene, zygotene, pachytene, diplotene and diakinesis.
- SUMOylation
a post-translational modification process in which target proteins are covalently modified with small ubiquitin-like modifier (SUMO) proteins.
- Synapsis
a process during meiotic prophase I by which homologous chromosomes align along their whole lengths and interactions between them are stabilized by assembly of the synaptonemal complex.
- ZMMs
a group of meiotic proteins that promote crossover formation in yeast, including Zip, Msh, and Mer family proteins.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zickler D, Kleckner N. Recombination, Pairing, and Synapsis of Homologs during Meiosis. Cold Spring Harb Perspect Biol. 2015;7(6) doi: 10.1101/cshperspect.a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hassold T, et al. The origin of human aneuploidy: where we have been, where we are going. Hum Mol Genet. 2007;16(2):R203–8. doi: 10.1093/hmg/ddm243. [DOI] [PubMed] [Google Scholar]
- 3.Moses MJ. Structure and function of the synaptonemal complex. Genetics. 1969;61(1) Suppl:41–51. [PubMed] [Google Scholar]
- 4.Cahoon CK, Hawley RS. Regulating the construction and demolition of the synaptonemal complex. Nat Struct Mol Biol. 2016;23(5):369–77. doi: 10.1038/nsmb.3208. [DOI] [PubMed] [Google Scholar]
- 5.Page SL, Hawley RS. Chromosome choreography: the meiotic ballet. Science. 2003;301(5634):785–9. doi: 10.1126/science.1086605. [DOI] [PubMed] [Google Scholar]
- 6.Kleckner N. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma. 2006;115(3):175–94. doi: 10.1007/s00412-006-0055-7. [DOI] [PubMed] [Google Scholar]
- 7.Nagaoka SI, et al. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat Rev Genet. 2012;13(7):493–504. doi: 10.1038/nrg3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraune J, et al. Evolutionary history of the mammalian synaptonemal complex. Chromosoma. 2016;125(3):355–60. doi: 10.1007/s00412-016-0583-8. [DOI] [PubMed] [Google Scholar]
- 9.Grishaeva TM, Bogdanov YF. Conservation and variability of synaptonemal complex proteins in phylogenesis of eukaryotes. Int J Evol Biol. 2014;2014:856230. doi: 10.1155/2014/856230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baier A, et al. Synaptonemal complex protein SYCP3: Conserved polymerization properties among vertebrates. Biochim Biophys Acta. 2007;1774(5):595–602. doi: 10.1016/j.bbapap.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Schild-Prufert K, et al. Organization of the synaptonemal complex during meiosis in Caenorhabditis elegans. Genetics. 2011;189(2):411–21. doi: 10.1534/genetics.111.132431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong H, Roeder GS. Organization of the yeast Zip1 protein within the central region of the synaptonemal complex. J Cell Biol. 2000;148(3):417–26. doi: 10.1083/jcb.148.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson LK, et al. Juxtaposition of C(2)M and the transverse filament protein C(3)G within the central region of Drosophila synaptonemal complex. Proc Natl Acad Sci U S A. 2005;102(12):4482–7. doi: 10.1073/pnas.0500172102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernandez-Hernandez A, et al. The central element of the synaptonemal complex in mice is organized as a bilayered junction structure. J Cell Sci. 2016;129(11):2239–49. doi: 10.1242/jcs.182477. [DOI] [PubMed] [Google Scholar]
- 15.Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem. 2014;83:553–84. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 16.van der Lee R, et al. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;114(13):6589–631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim Y, et al. The chromosome axis controls meiotic events through a hierarchical assembly of HORMA domain proteins. Dev Cell. 2014;31(4):487–502. doi: 10.1016/j.devcel.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voelkel-Meiman K, et al. Full-length synaptonemal complex grows continuously during meiotic prophase in budding yeast. PLoS Genet. 2012;8(10):e1002993. doi: 10.1371/journal.pgen.1002993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pattabiraman D, et al. Meiotic recombination modulates the structure and dynamics of the synaptonemal complex during C. elegans meiosis. PLoS Genet. 2017;13(3):e1006670. doi: 10.1371/journal.pgen.1006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nadarajan S, et al. Polo-like kinase-dependent phosphorylation of the synaptonemal complex protein SYP-4 regulates double-strand break formation through a negative feedback loop. Elife. 2017:6. doi: 10.7554/eLife.23437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Obeso D, et al. Couples, pairs, and clusters: mechanisms and implications of centromere associations in meiosis. Chromosoma. 2014;123(1–2):43–55. doi: 10.1007/s00412-013-0439-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhalla N, et al. ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet. 2008;4(10):e1000235. doi: 10.1371/journal.pgen.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Carvalho CE, et al. LAB-1 antagonizes the Aurora B kinase in C. elegans. Genes Dev. 2008;22(20):2869–85. doi: 10.1101/gad.1691208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nabeshima K, et al. Crossing over is coupled to late meiotic prophase bivalent differentiation through asymmetric disassembly of the SC. J Cell Biol. 2005;168(5):683–9. doi: 10.1083/jcb.200410144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bisig CG, et al. Synaptonemal complex components persist at centromeres and are required for homologous centromere pairing in mouse spermatocytes. PLoS Genet. 2012;8(6):e1002701. doi: 10.1371/journal.pgen.1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiao H, et al. Interplay between synaptonemal complex, homologous recombination, and centromeres during mammalian meiosis. PLoS Genet. 2012;8(6):e1002790. doi: 10.1371/journal.pgen.1002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newnham L, et al. The synaptonemal complex protein, Zip1, promotes the segregation of nonexchange chromosomes at meiosis I. Proc Natl Acad Sci U S A. 2010;107(2):781–5. doi: 10.1073/pnas.0913435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gladstone MN, et al. The synaptonemal complex protein Zip1 promotes bi-orientation of centromeres at meiosis I. PLoS Genet. 2009;5(12):e1000771. doi: 10.1371/journal.pgen.1000771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Perez E, et al. Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes Dev. 2008;22(20):2886–901. doi: 10.1101/gad.1694108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tzur YB, et al. LAB-1 targets PP1 and restricts Aurora B kinase upon entrance into meiosis to promote sister chromatid cohesion. PLoS Biol. 2012;10(8):e1001378. doi: 10.1371/journal.pbio.1001378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe Y. Shugoshin: guardian spirit at the centromere. Curr Opin Cell Biol. 2005;17(6):590–5. doi: 10.1016/j.ceb.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Rabitsch KP, et al. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr Biol. 2004;14(4):287–301. doi: 10.1016/j.cub.2004.01.051. [DOI] [PubMed] [Google Scholar]
- 33.Rogers E, et al. The aurora kinase AIR-2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J Cell Biol. 2002;157(2):219–29. doi: 10.1083/jcb.200110045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kershner A, et al. Germline stem cells and their regulation in the nematode Caenorhabditis elegans. Adv Exp Med Biol. 2013;786:29–46. doi: 10.1007/978-94-007-6621-1_3. [DOI] [PubMed] [Google Scholar]
- 35.Honigberg SM, Purnapatre K. Signal pathway integration in the switch from the mitotic cell cycle to meiosis in yeast. J Cell Sci. 2003;116(Pt 11):2137–47. doi: 10.1242/jcs.00460. [DOI] [PubMed] [Google Scholar]
- 36.Merritt C, Seydoux G. The Puf RNA-binding proteins FBF-1 and FBF-2 inhibit the expression of synaptonemal complex proteins in germline stem cells. Development. 2010;137(11):1787–98. doi: 10.1242/dev.050799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang W, et al. HAL-2 promotes homologous pairing during Caenorhabditis elegans meiosis by antagonizing inhibitory effects of synaptonemal complex precursors. PLoS Genet. 2012;8(8):e1002880. doi: 10.1371/journal.pgen.1002880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gomez R, et al. Sororin loads to the synaptonemal complex central region independently of meiotic cohesin complexes. EMBO Rep. 2016;17(5):695–707. doi: 10.15252/embr.201541060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gomez HL, et al. C14ORF39/SIX6OS1 is a constituent of the synaptonemal complex and is essential for mouse fertility. Nat Commun. 2016;7:13298. doi: 10.1038/ncomms13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahuja JS, et al. Control of meiotic pairing and recombination by chromosomally tethered 26S proteasome. Science. 2017;355(6323):408–411. doi: 10.1126/science.aaf4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rao HB, et al. A SUMO-ubiquitin relay recruits proteasomes to chromosome axes to regulate meiotic recombination. Science. 2017;355(6323):403–407. doi: 10.1126/science.aaf6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato A, et al. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 2009;139(5):907–19. doi: 10.1016/j.cell.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horn HF, et al. A mammalian KASH domain protein coupling meiotic chromosomes to the cytoskeleton. J Cell Biol. 2013;202(7):1023–39. doi: 10.1083/jcb.201304004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christophorou N, et al. Microtubule-driven nuclear rotations promote meiotic chromosome dynamics. Nat Cell Biol. 2015;17(11):1388–400. doi: 10.1038/ncb3249. [DOI] [PubMed] [Google Scholar]
- 45.Scherthan H, et al. Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2007;104(43):16934–9. doi: 10.1073/pnas.0704860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christophorou N, et al. Synaptonemal complex components promote centromere pairing in pre-meiotic germ cells. PLoS Genet. 2013;9(12):e1004012. doi: 10.1371/journal.pgen.1004012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacQueen AJ, et al. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 2002;16(18):2428–42. doi: 10.1101/gad.1011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanchez-Moran E, et al. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 2007;21(17):2220–33. doi: 10.1101/gad.439007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodyer W, et al. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev Cell. 2008;14(2):263–74. doi: 10.1016/j.devcel.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 50.Padmore R, et al. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell. 1991;66(6):1239–56. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- 51.Dietrich AJ, et al. The sequential appearance of components of the synaptonemal complex during meiosis of the female rat. Genome. 1992;35(3):492–7. doi: 10.1139/g92-072. [DOI] [PubMed] [Google Scholar]
- 52.Tsubouchi T, et al. Initiation of meiotic chromosome synapsis at centromeres in budding yeast. Genes Dev. 2008;22(22):3217–26. doi: 10.1101/gad.1709408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Macqueen AJ, Roeder GS. Fpr3 and Zip3 ensure that initiation of meiotic recombination precedes chromosome synapsis in budding yeast. Curr Biol. 2009;19(18):1519–26. doi: 10.1016/j.cub.2009.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng CH, et al. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 2006;20(15):2067–81. doi: 10.1101/gad.1430406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eichinger CS, Jentsch S. Synaptonemal complex formation and meiotic checkpoint signaling are linked to the lateral element protein Red1. Proc Natl Acad Sci U S A. 2010;107(25):11370–5. doi: 10.1073/pnas.1004248107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Humphryes N, et al. The Ecm11-Gmc2 complex promotes synaptonemal complex formation through assembly of transverse filaments in budding yeast. PLoS Genet. 2013;9(1):e1003194. doi: 10.1371/journal.pgen.1003194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shinohara M, et al. Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat Genet. 2008;40(3):299–309. doi: 10.1038/ng.83. [DOI] [PubMed] [Google Scholar]
- 58.Leung WK, et al. The synaptonemal complex is assembled by a polySUMOylation-driven feedback mechanism in yeast. J Cell Biol. 2015;211(4):785–93. doi: 10.1083/jcb.201506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiao Y, et al. Identification of cell-specific targets of sumoylation during mouse spermatogenesis. Reproduction. 2016;151(2):149–66. doi: 10.1530/REP-15-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao J, et al. N-terminal acetylation promotes synaptonemal complex assembly in C. elegans. Genes Dev. 2016;30(21):2404–2416. doi: 10.1101/gad.277350.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scott DC, et al. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science. 2011;334(6056):674–8. doi: 10.1126/science.1209307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rog O, et al. The synaptonemal complex has liquid crystalline properties and spatially regulates meiotic recombination factors. Elife. 2017:6. doi: 10.7554/eLife.21455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marston AL, Amon A. Meiosis: cell-cycle controls shuffle and deal. Nat Rev Mol Cell Biol. 2004;5(12):983–97. doi: 10.1038/nrm1526. [DOI] [PubMed] [Google Scholar]
- 64.Matos J, et al. Dbf4-dependent CDC7 kinase links DNA replication to the segregation of homologous chromosomes in meiosis I. Cell. 2008;135(4):662–78. doi: 10.1016/j.cell.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 65.Falk JE, et al. A Mec1- and PP4-dependent checkpoint couples centromere pairing to meiotic recombination. Dev Cell. 2010;19(4):599–611. doi: 10.1016/j.devcel.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 66.Hustedt N, Durocher D. The control of DNA repair by the cell cycle. Nat Cell Biol. 2016;19(1):1–9. doi: 10.1038/ncb3452. [DOI] [PubMed] [Google Scholar]
- 67.Mehrotra S, McKim KS. Temporal analysis of meiotic DNA double10 strand break formation and repair in Drosophila females. PLoS Genet. 2006;2(11):e200. doi: 10.1371/journal.pgen.0020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gao J, et al. NatB domain-containing CRA-1 antagonizes hydrolase ACER-1 linking acetyl-CoA metabolism to the initiation of recombination during C. elegans meiosis. PLoS Genet. 2015;11(3):e1005029. doi: 10.1371/journal.pgen.1005029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saito TT, et al. SLX-1 is required for maintaining genomic integrity and promoting meiotic noncrossovers in the Caenorhabditis elegans germline. PLoS Genet. 2012;8(8):e1002888. doi: 10.1371/journal.pgen.1002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Keeney S, et al. Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annu Rev Genet. 2014;48:187–214. doi: 10.1146/annurev-genet-120213-092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kauppi L, et al. Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes Dev. 2013;27(8):873–86. doi: 10.1101/gad.213652.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brockway H, et al. The CSN/COP9 signalosome regulates synaptonemal complex assembly during meiotic prophase I of Caenorhabditis elegans. PLoS Genet. 2014;10(11):e1004757. doi: 10.1371/journal.pgen.1004757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jahns MT, et al. Crossover localisation is regulated by the neddylation posttranslational regulatory pathway. PLoS Biol. 2014;12(8):e1001930. doi: 10.1371/journal.pbio.1001930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Keeney S, et al. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88(3):375–84. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 75.Voelkel-Meiman K, et al. SUMO localizes to the central element of synaptonemal complex and is required for the full synapsis of meiotic chromosomes in budding yeast. PLoS Genet. 2013;9(10):e1003837. doi: 10.1371/journal.pgen.1003837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wojtasz L, et al. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 2009;5(10):e1000702. doi: 10.1371/journal.pgen.1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sourirajan A, Lichten M. Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes Dev. 2008;22(19):2627–32. doi: 10.1101/gad.1711408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jordan P, et al. Ipl1/Aurora B kinase coordinates synaptonemal complex disassembly with cell cycle progression and crossover formation in budding yeast meiosis. Genes Dev. 2009;23(18):2237–51. doi: 10.1101/gad.536109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Argunhan B, et al. Fundamental cell cycle kinases collaborate to ensure timely destruction of the synaptonemal complex during meiosis. EMBO J. 2017;36(17):2488–2509. doi: 10.15252/embj.201695895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jordan PW, et al. Polo-like kinase is required for synaptonemal complex disassembly and phosphorylation in mouse spermatocytes. J Cell Sci. 2012;125(Pt 21):5061–72. doi: 10.1242/jcs.105015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parra MT, et al. Dynamic relocalization of the chromosomal passenger complex proteins inner centromere protein (INCENP) and aurora-B kinase during male mouse meiosis. J Cell Sci. 2003;116(Pt 6):961–74. doi: 10.1242/jcs.00330. [DOI] [PubMed] [Google Scholar]
- 82.Sun F, Handel MA. Regulation of the meiotic prophase I to metaphase I transition in mouse spermatocytes. Chromosoma. 2008;117(5):471–85. doi: 10.1007/s00412-008-0167-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dix DJ, et al. HSP70–2 is required for desynapsis of synaptonemal complexes during meiotic prophase in juvenile and adult mouse spermatocytes. Development. 1997;124(22):4595–603. doi: 10.1242/dev.124.22.4595. [DOI] [PubMed] [Google Scholar]
- 84.Zhu D, et al. HSP70–2 is required for CDC2 kinase activity in meiosis I of mouse spermatocytes. Development. 1997;124(15):3007–14. doi: 10.1242/dev.124.15.3007. [DOI] [PubMed] [Google Scholar]
- 85.Allen JW, et al. HSP70–2 is part of the synaptonemal complex in mouse and hamster spermatocytes. Chromosoma. 1996;104(6):414–21. doi: 10.1007/BF00352265. [DOI] [PubMed] [Google Scholar]
- 86.Nadarajan S, et al. The MAP kinase pathway coordinates crossover designation with disassembly of synaptonemal complex proteins during meiosis. Elife. 2016;5:e12039. doi: 10.7554/eLife.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Colaiacovo MP, et al. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev Cell. 2003;5(3):463–74. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- 88.Ortiz R, et al. The width of the lateral element of the synaptonemal complex is determined by a multilayered organization of its components. Exp Cell Res. 2016;344(1):22–9. doi: 10.1016/j.yexcr.2016.03.025. [DOI] [PubMed] [Google Scholar]
- 89.De Muyt A, et al. E3 ligase Hei10: a multifaceted structure-based signaling molecule with roles within and beyond meiosis. Genes Dev. 2014;28(10):1111–23. doi: 10.1101/gad.240408.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Smolikov S, et al. A yeast two-hybrid screen for SYP-3 interactors identifies SYP-4, a component required for synaptonemal complex assembly and chiasma formation in Caenorhabditis elegans meiosis. PLoS Genet. 2009;5(10):e1000669. doi: 10.1371/journal.pgen.1000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Page SL, Hawley RS. c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev. 2001;15(23):3130–43. doi: 10.1101/gad.935001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Collins KA, et al. Corolla is a novel protein that contributes to the architecture of the synaptonemal complex of Drosophila. Genetics. 2014;198(1):219–28. doi: 10.1534/genetics.114.165290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang F, et al. Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J Cell Biol. 2006;173(4):497–507. doi: 10.1083/jcb.200603063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Syrjanen JL, et al. A molecular model for the role of SYCP3 in meiotic chromosome organisation. Elife. 2014:3. doi: 10.7554/eLife.02963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Meuwissen RL, et al. A coiled-coil related protein specific for synapsed regions of meiotic prophase chromosomes. EMBO J. 1992;11(13):5091–100. doi: 10.1002/j.1460-2075.1992.tb05616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Costa Y, et al. Two novel proteins recruited by synaptonemal complex protein 1 (SYCP1) are at the centre of meiosis. J Cell Sci. 2005;118(Pt 12):2755–62. doi: 10.1242/jcs.02402. [DOI] [PubMed] [Google Scholar]
- 97.Davies OR, et al. Structural analysis of the human SYCE2-TEX12 complex provides molecular insights into synaptonemal complex assembly. Open Biol. 2012;2(7):120099. doi: 10.1098/rsob.120099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schramm S, et al. A novel mouse synaptonemal complex protein is essential for loading of central element proteins, recombination, and fertility. PLoS Genet. 2011;7(5):e1002088. doi: 10.1371/journal.pgen.1002088. [DOI] [PMC free article] [PubMed] [Google Scholar]