Abstract
Two-component systems (TCSs) dictate many bacterial responses to environmental change via the activation of a membrane-embedded sensor kinase, which has molecular specificity for a cognate response regulator protein. However, although the major ity of TCSs operate through seemingly strict cognate protein-protein interactions, there have been several reports of TCSs that violate this classical model of signal transduction. Our group has recently demonstrated that some of these cross-interacting TCSs function in a manner that imparts a fitness advantage to bacterial pathogens. In this study, we describe inter-connectivity between the metabolite-sensing TCSs YpdA/YpdB and BtsS/BtsR in uropathogenic Escherichia coli (UPEC). The YpdA/YpdB and BtsS/BtsR TCSs have been previously reported to interact in K12 E. coli, where they alter the expression of putative transporter genes yhjX and yjiY, respectively. These target genes are both up-regulated in UPEC during acute and chronic murine models of urinary tract infection, as well as in response to pyruvate and serine added to growth media in vitro. Here, we show that proper regulation of yhjX in UPEC requires the presence of all components from both of these TCSs. By utilizing plasmid-encoded luciferase reporters tracking the activity of the yhjX and yjiY promoters, we demonstrate that deletions in one TCS substantially alter transcriptional activity of the opposing system's target gene. However, unlike in K12 E. coli, single gene deletions in the YpdA/YpdB system do not alter yjiY gene expression in UPEC, suggesting that niche and lifestyle-specific pressures may be selecting for differential cross-regulation of TCSs in pathogenic and non-pathogenic E. coli.
Keywords: cross-regulation, two-component systems, pyruvate-sensing, Escherichia coli
Introduction
Pathogenic bacteria are becoming increasingly resistant to currently available antibiotics (Andersson & Hughes. 2011; Davies & Davies. 2010; Perry, Westman & Wright. 2014; Gillings, Paulsen & Tetu. 2017), and new strategies to thwart virulence and treat infection are needed. One area of interest for new antimicrobial drugs encompasses the signaling and regulatory networks that are essential for bacterial activity and survival. Signal transduction systems control many bacterial activities such as cell growth and division, the acquisition of vital nutrients, activation of defenses to environmental and host hazards, and the expression of virulence factors needed for pathogenesis. Bacterial two-component systems (TCSs) are an important class of receptor signaling networks that mediate many of the aforementioned processes (Freeman, Dorus & Waterfield. 2013; Hadjifrangiskou et al. 2011; Kostakioti et al. 2009; Skerker et al. 2005; Tipton & Rather. 2016) and have been directly implicated in some forms of antibiotic resistance (Gebhardt & Shuman. 2017; Guckes et al. 2017; Kellogg et al. 2017; Macfarlane, Kwasnicka & Hancock. 2000). Notably, TCSs are absent in mammals, making them an excellent pharmacological target with well-recognized potential (Barrett & Hoch. 1998; Gotoh et al. 2010; Macielag & Goldschmidt. 2000; Okada et al. 2010; Schreiber, Res & Matter. 2009).
In their simplest form, TCSs are composed of a membrane-embedded sensor histidine kinase and a cognate cytosolic response regulator protein. In general, signal detection by the histidine kinase leads to auto-phosphorylation at a conserved histidine residue and subsequent activation of the cognate response regulator via phosphotransfer to and phosphorylation of a conserved aspartate residue on the response regulator protein (Bhate et al. 2015; Casino, Rubio & Marina. 2010; Gao & Stock. 2009; Robinson, Buckler & Stock. 2000). The amino acids at the interaction surface surrounding the conserved histidine and aspartate residues confer a high degree of specificity for cognate partners. System-wide analyses of TCSs have shown that histidine kinases have far greater preference for interaction with their cognate partners than with other possible partners both in vivo and in vitro (Casino et al. 2010; Laub & Goulian. 2007; Podgornaia & Laub. 2013). This cognate partner specificity, TCS stoichiometry, and common spatial localization all work to prevent “signal interference” from aberrant non-partner interactions. Taken together, this led to the general assumption that TCSs function in an insulated fashion: as stimuli-response coupling systems that respond to a particular cue by initiating a particular response, and that during this process, interaction with other TCSs either does not occur or is detrimental to the fidelity and functioning of the system. In recent years, this view of TCSs as linear on/off switches has been challenged, and a clearer image of bacterial signaling network complexity is emerging (Breland et al. 2017; Capra & Laub. 2012; Guckes et al. 2013; Jung et al. 2012; Olivera, Ugalde & Martínez-Antonio. 2010; Salazar & Laub. 2015).
Recent studies by our lab and others have demonstrated that closely related TCSs can cross-interact in a manner that is beneficial for the bacteria, providing benefits such as transient antibiotic resistance or increased flagellar synthesis (Guckes et al. 2017; Guckes et al. 2013; Urano et al. 2017; Wei et al. 2017). Interference with TCS cross-regulation can lead to aberrant “cross-talk” as coined by Mark Goulian and Michael Laub (Laub & Goulian. 2007) and re-wiring of bacterial gene expression. In the case of pathogens such as uropathogenic Escherichia coli (UPEC), this genetic re-wiring can lead to severe attenuation of virulence (Kostakioti et al. 2009). These findings suggest that therapeutics designed to disrupt interacting TCSs have the potential to alter pathogen behavior by shifting bacterial-host interactions to a non-pathogenic state.
In addition to TCSs that contravene traditional fidelity rules, these signaling systems can have additional levels of complexity. For example, many TCSs are encoded by operons that harbor additional genes beyond those coding for the sensor histidine kinase and response regulator. These additional genes may code for “accessory proteins” that can function in several capacities, such as regulating transcription, physically promoting or limiting system protein interactions, or as phosphorelay “connectors”, linking histidine kinase sensors to response regulator proteins (Appleby, Parkinson & Bourret. 1996; Buelow & Raivio. 2010; Jung et al. 2012; Mitrophanov & Groisman. 2008; Tschauner et al. 2014). Although the field is now beginning to appreciate the complexity of TCS physiology, we do not fully understand how bacteria integrate myriad signals and stimuli in a cohesive and orderly fashion.
We previously reported in K12 E. coli that YpdA/YpdB and BtsS/BtsR (formerly YehU/YehT) induce the expression of two putative membrane embedded transporters, YhjX and YjiY, respectively. During in vitro growth in lysogeny broth (LB), these targets are naturally induced immediately prior to transition phase, and induction can be altered by the addition of pyruvate or serine to the growth media (Behr, Fried & Jung. 2014; Fried, Behr & Jung. 2013; Kraxenberger et al. 2012). Subsequent ligand-sensor interaction studies have demonstrated that the BtsS sensor histidine kinase binds pyruvate with high affinity (Behr et al. 2017). In this work, we provide evidence that the YpdA/YpdB and BtsS/BtsR TCSs interact in UPEC, providing another example of closely related signaling systems that exhibit affinity for non-partner interactions. Specifically, we demonstrate that induction of the gene yhjX, the known downstream target of the YpdA/YpdB system, in response to a pyruvate signal requires both the sensor BtsS and the response regulator YpdB for proper signaling in UPEC. Similar to previous reports in K12 E. coli, deletion of either downstream target gene alters system signaling, suggesting a contribution of the target genes TCS regulation and function. Additionally, we show that single gene deletions in the YpdA/YpdB TCS do not alter yjiY activity in UPEC, which is contrary to our previous findings in K12 E. coli. In K12 E. coli, deletion of ypdA or ypdB strongly attenuates yjiY expression (Behr et al. 2014), whereas in UPEC deletion of ypdA or ypdB has no effect on yjiY expression. This suggests that there may be niche specific differential evolution of the YpdA/YpdB and BtsS/BtsR TCSs.
Material and Methods
Bacterial Strains, Constructs and Growth Conditions
The parent strain used in all analyses was uropathogenic Escherichia coli cystitis isolate UTI89 (Mulvey, Schilling & Hultgren. 2001). Isogenic deletion strains lacking ypdA (UTI89_C2712), ypdB (UTI89_C2713), btsR (UTI89_C2397), btsS (UTI89_C2398), yhjX (UTI89_C4087), yjiY (UTI89_C5057) and permutations thereof were created using the λ-Red recombinase method of Murphy & Campellone (Murphy & Campellone. 2003). The primer sets used to make the constructs are listed in Table S1. Plasmid-borne promoter-luciferase transcriptional reporters for yhjX and yjiY were previously created (Kraxenberger et al. 2012; Fried et al. 2012) and these constructs were introduced into the different UTI89 strain backgrounds via electroporation. Cells were made “hyper-competent” prior to electroporation, as described in Hadjifrangiskou et al. 2012.
Bacterial cultures were inoculated from freezer stocks (all strains in the Hadjifrangiskou lab are cryopreserved at −80°C in Lysogeny Broth (LB) (Thermo Fisher LB Miller) infused with 25% glycerol). Bacteria were inoculated in LB supplemented with 50 µg/mL gentamycin in order to ensure retention of the transcriptional reporter constructs and incubated at 37°C with shaking (220 rpm). The same growth conditions were used for all the reporter assay experiments. For experiments in which pyruvate was added as the stimulus, a stock concentration of 50 mM sodium-pyruvate was used and added to media as indicated below to a final concentration of 1 mM.
Transcriptional Reporter Assays
Transcriptional reporter assays were conducted in a manner similar to previous in vivo expression studies of the YpdA/YpdB and BtsS/BtsR TCSs (Behr et al. 2014; Fried et al. 2013), with minor modifications. Specifically, overnight cultures were normalized to a starting OD600 of 0.03 and were seeded into sterile polystyrene 96 well black plates with clear flat bottoms. Each well was loaded with 180 µL of sample and a minimum of three technical replicates was conducted per strain per condition. Likewise, three wells were loaded with 180 µL of blank LB media to control for background light readings. Once all strains were seeded into the plate, initial readings of OD600 and luciferase activity were recorded and the plate was subsequently placed into a 37°C incubator shaking at 220 rpm to promote optimal bacterial growth. Luminescence levels are reported as relative light units (RLU). Readings were taken in a Molecular Devices SpectraMax i3 plate reader at 30-minute time intervals, over a period of 5 hours, followed by two additional readings, at 6 h and 7 h respectively. For experiments in which pyruvate was added as a stimulus, pyruvate was introduced directly following the 120-minute time point reading at a final concentration of 1 mM. Luciferase and growth density readings were exported and organized in Microsoft Excel, RLU/OD values were calculated, and then transferred to Graphpad Prism 7 for graphing and analysis. All assays were performed at least three independent times.
Results and Discussion
The BtsS/BtsR and YpdA/YpdB Systems Control yjiY and yhjX Expression in UPEC
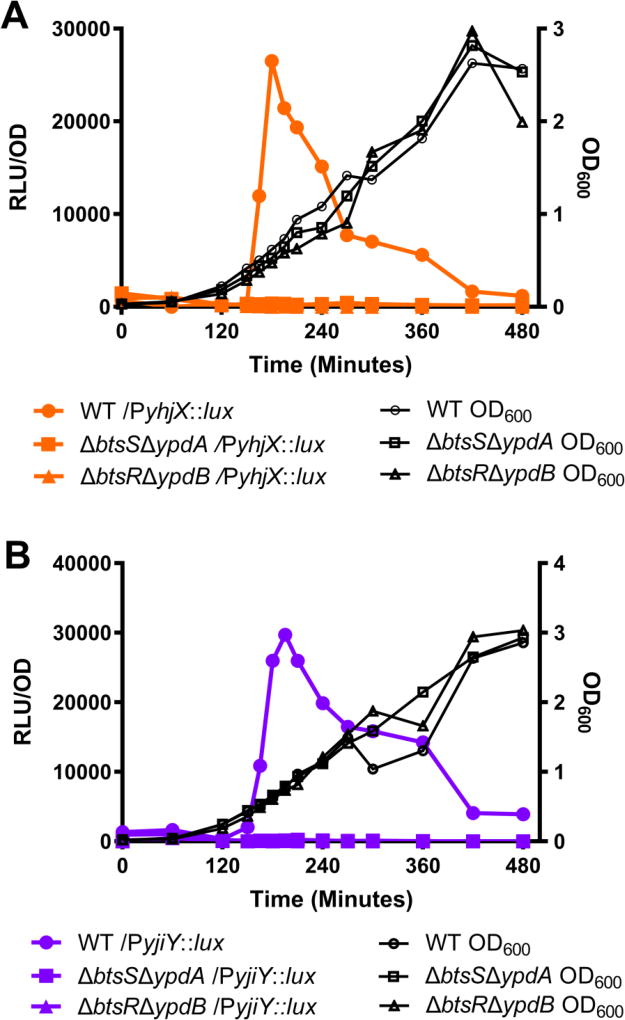
As previously stated, in K12 E. coli the downstream targets of BtsS/BtsR and YpdA/YpdB are naturally induced during in-vitro growth in laboratory media shortly prior to stationary phase (Behr et al. 2014; Fried et al. 2013; Kraxenberger et al. 2012). To determine whether the same was true in UPEC, target gene expression was assayed using previously created transcriptional reporters containing a fusion of either the yjiY or yhjX promoters to the luxCDABE operon (Kraxenberger et al. 2012; Fried et al. 2012). Promoter activity was first monitored during growth in LB under aerobic conditions at 37°C. As with K12 E. coli, UPEC reached maximal yjiY-lux and yhjX-lux reporter activity shortly prior to bacterial post-exponential growth phase, with expression beginning at roughly 120 minutes of growth and peaking at roughly 180 minutes (Fig. 1a–b, filled circles). To determine whether BtsS/BtsR and YpdA/YpdB are the sole regulators of yjiY and yhjX expression under the growth conditions tested, we constructed deletion mutants lacking both sensors (ΔbtsSΔypdA) or lacking both response regulators (ΔbtsRΔypdB). In these deletion mutants, there was no downstream activity of either yhjX or yjiY, indicating that these target genes require signal transduction from one or both two-component systems (Fig. 1a–b, filled squares and filled triangles). This alteration in signaling was not attributed to changes in bacterial growth in either the double sensor kinase or double response regulator deletion mutants (Fig. 1, black lines).
Figure 1. Natural induction of yjiY and yhjX during UPEC growth in laboratory media.
Measurement of optical density and yhjX or yjiY promoter activity in double histidine kinase sensor or double response regulator deletion mutants during in vitro growth in LB. Downstream gene activity is recorded as Relative Light Units (RLUs). RLUs are plotted as a function of OD to normalize for variance in bacterial population. (A) yhjX-lux activity and OD600 of deletion mutants. (B) yjiY-lux activity and OD600 of deletion mutants. WT, wild-type. Experiments were performed at least three independent times and the results of a representative experiment are shown.
Non-Cognate Partner Interactions Regulate the Activity of yhjX
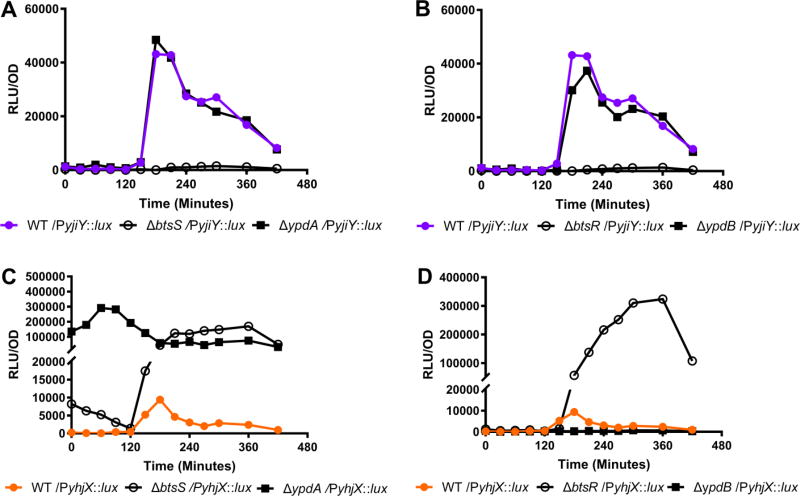
Previous studies suggested that there is interplay between the BtsS/BtsR and YpdA/YpdB TCSs in K12 E. coli (Behr et al. 2014). Given the genetic diversity of the E. coli species, we were interested in exploring the extent to which BtsS/BtsR and YpdA/YpdB interconnectivity might exist or be altered in UPEC. In order to test the interactions between the BtsS/BtsR and YpdA/YpdB systems, we first created single gene deletion mutants lacking only one component from either system. During normal in vitro growth in LB, we found that deletion of either btsS (Fig. 2a) or btsR (Fig. 2b) abolished yjiY target gene expression, while deletion of ypdA or ypdB imparted no effect on yjiY expression (Fig. 2a–b). These results demonstrate that in UPEC, yjiY is under the sole control of the BtsS/BtsR signaling system.
Figure 2. Measurement of yhjX or yjiY promoter luciferase activity in WT UTI89 or isogenic deletion mutants during in vitro growth in LB.
(A) yjiY-lux activity in WT UTI89 or single histidine kinase sensor deletion mutants. (B) yjiY-lux activity in WT UTI89 or single response regulator deletion mutants. (C) yhjX-lux activity in WT UTI89 or single histidine kinase sensor deletion mutants. (D) yhjX-lux activity in WT UTI89 or single response regulator deletions. Experiments were performed at least three times and the results of a representative experiment are shown.
Deletion of either btsS or ypdA sensor genes led to a substantial increase in yhjX promoter activity, but with distinct differences in the transcriptional surge profile (Fig. 2c). Although deletion of btsS led to dramatically increased yhjX driven luciferase activity, the timing of this transcriptional surge during growth was the same as in wild type (WT) UTI89 (Fig. 2c, open circles). Contrarily, deletion of ypdA abolished the transcriptional surge and produced a ubiquitous increase in yhjX expression (Fig. 2c). Testing the individual response regulator deletion mutants revealed that yhjX promoter activity was abolished in ypdB deletion mutants (Fig. 2d, filled squares), while deletion of btsR led to increased levels of yhjX-lux activity at the appropriate surge time (Fig. 2d, open circles). These results suggest that the BtsS/BtsR and YpdA/YpdB TCSs are both needed for proper regulation and activation of yhjX in UPEC.
Non-Cognate Partners BtsS and YpdB Are Sufficient to Induce yhjX
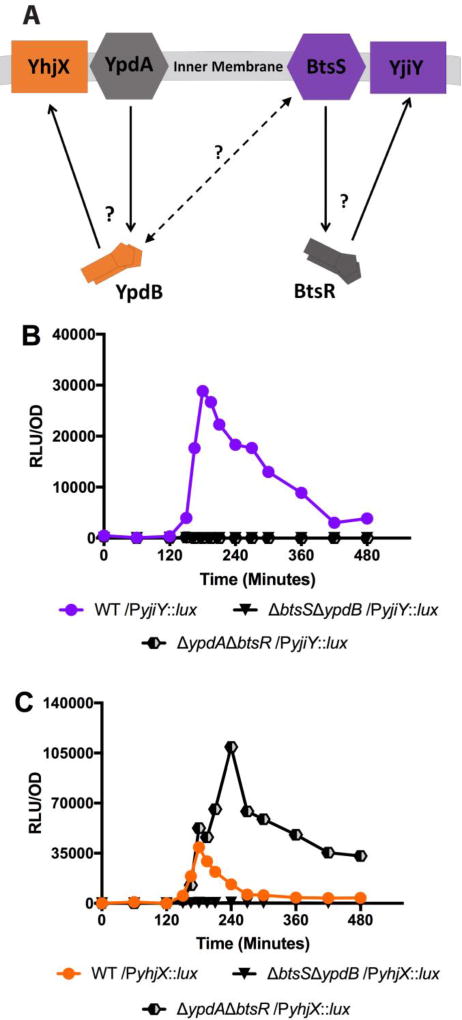
To investigate which components are responsible for the cross-regulation of yhjX expression, we constructed bacterial deletion mutants in which the genes encoding the sensor of one system (ypdA or btsS) and the response regulator of the opposing system (ypdB or btsR) were deleted, leaving only the non-cognate partners (either sensor BtsS with regulator YpdB or sensor YpdA with regulator BtsR). In the case of the BtsS/BtsR system, it appears that both the sensor BtsS and response regulator BtsR are needed for the activation of downstream gene target yjiY, as deletion of either of these genes ablated yjiY promoter activity (Fig. 2a–2b and Fig. 3b). These data demonstrate that BtsS/BtsR mediated regulation of yjiY fits the canonical model of TCS activity under the conditions tested.
Figure 3. Measurement of yhjX or yjiY luciferase activity in non-cognate partner mutants that contain no cognate histidine kinase or response regulator pairs.
(A) Schematic representing a ΔypdAΔbtsR non-cognate partner mutant. Solid lines represent canonical TCS signaling pathways and dashed lines represent potential cross-interactivity. Question marks denote signaling pathway branch points of interest that were monitored for changes in down-stream output between WT and the various mutants. (B) yjiY-lux activity in WT UTI89 and non-cognate partner mutants. (C) yhjX-lux in WT UTI89 and non-cognate partner mutants. Experiments were performed at least three times and the results of a representative experiment are shown.
We next sought to determine how yhjX signaling might be altered in non-cognate partner mutants. We found that the ΔbtsSΔypdB mutant had no yhjX activity (Fig. 3c, black inverted triangles). This was in agreement with the single gene deletion experiments which suggested that ypdB is needed for yhjX activation. Contrarily, the ΔypdAΔbtsR mutant exhibited a noticeable increase in yhjX activity. The ΔypdAΔbtsR mutant fully retains the timing of the WT transcriptional surge but surpasses WT levels of luminescence. Our previous experiments had demonstrated that deletion of both sensors (ΔbtsSΔypdA) (Fig. 1a, filled squares) completely ablated yhjX activity and that deletion of just the YpdA sensor (ΔypdA) led to elevated but unregulated levels of yhjX activity (Fig. 2c, filled squares). Taken together, these data suggest that the sensor BtsS and the response regulator YpdB interact to regulate expression of yhjX. The nature of this coordinated regulation might involve a number of sensor-sensor, sensor-kinase, or even system-target interactions.
Thus far, our data demonstrate an interesting divergence from what has been reported in K12 E. coli: Notably, the single deletion analyses indicate that the expression of yjiY in UPEC is solely controlled by BtsSR. Furthermore, “normal” wild-type like induction and transcription of yhjX in UPEC requires the presence of both the BtsS sensor and the YpdB response regulator suggesting that there may be non-cognate partner interactions between these systems. This deviation between E. coli strains that belong to two different clades and have evolved distinct colonization strategies (Croxen & Finlay. 2010) may reflect different usage of the two TCSs to modulate bacterial homeostasis, as a function of ecological niche. Our previous studies demonstrated that BtsS binds and responds to pyruvate (Behr et al. 2017), a metabolite that is critical for several metabolic processes important for UPEC during urinary tract infections (Shaffer et al. 2017; Floyd et al. 2015; Eberly et al. 2017; Alteri, Smith, & Mobley. 2009; Hadjifrangiskou et al. 2011). It is thus possible that “re-wiring” signal transduction connections in UPEC may be critical for pathogenesis. We therefore next analyzed the transcriptional responses of UPEC yjiY and yhjX in the presence of pyruvate.
Pyruvate Signaling Through Non-Cognate Partners
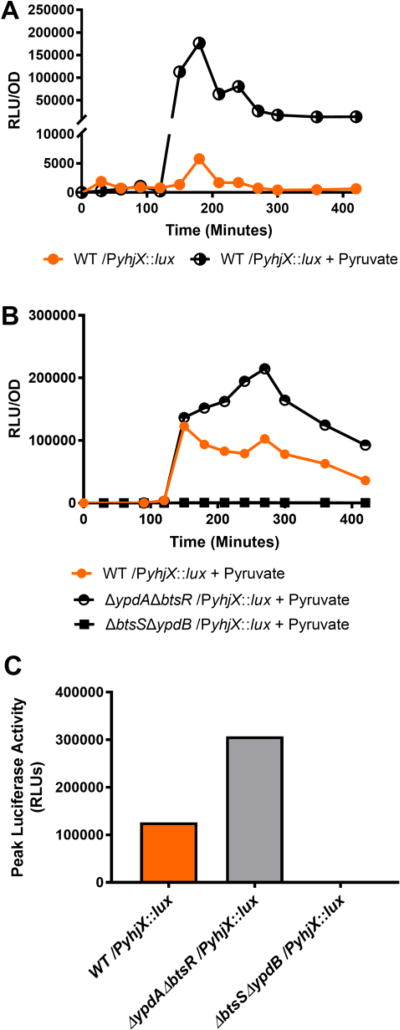
Given the evidence of cross-regulation observed up to this point, we sought to determine how the addition of pyruvate would influence signaling in wild-type UPEC and in isogenic non-cognate partner deletion mutants. In these assays, the growth medium was spiked with sodium pyruvate to a final concentration of 1 mM at 120 minutes of growth, a time that coincides with the natural induction of yhjX and yjiY (Fig. 1). We observed in WT UPEC that pyruvate stimulation led to a considerable increase in yhjX-driven luciferase activity (Fig. 4a), while addition of pyruvate to the ΔbtsSΔypdB mutant elicited no yhjX response (Fig. 4b). Given that BtsS is known to be a high affinity sensor for pyruvate in other E. coli these data suggest that in UPEC BtsS senses pyruvate, becomes activated, and induces the subsequent upregulation of yhjX via the action of YpdB.
Figure 4. Pyruvate induced expression of yhjX-lux activity in WT and in non-cognate partner mutants.
(A) Comparison of yhjX-lux activity in response to 1 mM sodium pyruvate added at 120 minutes during in vitro growth in LB. (B) Comparison of yhjX-lux expression in WT UTI89 or non-cognate partner mutants with 1 mM sodium pyruvate added at 120 minutes. (C) Peak yhjX-lux RLU values observed under the same conditions as (B), data from a separate representative run. All experiments were done in triplicate with representative data sets being shown. (D) Measurement of optical density over time in UTI89 ∆yhjX or ∆yjiY deletion mutants during in vitro growth in LB.
Interestingly, the addition of pyruvate to the ΔypdAΔbtsR mutant led to a sustained increase in yhjX activity beyond that encountered in WT strains (Fig. 4b, half circles, Fig. 4c). This may be explained in part by the fact that removal of YpdA (which is cognate to YpdB) and BtsR (which is cognate to BtsS) removes potential interaction partners, which can potentiate interaction between the remaining non-cognate partners. Similar observations have been made by our group in other TCS interaction studies (Guckes et al. 2013; Guckes & Breland et al. 2017). Together, our findings demonstrate the discovery of physiological interactions across the YpdA/YpdB and BtsS/BtsR two-component systems in UPEC in response to pyruvate, a known BtsS ligand, and demonstrate that all four components of the BtsS/BtsR and YpdA/YpdB systems must be present to properly regulate target gene yhjX.
Deletion of yhjX and yjiY Deregulates Signaling
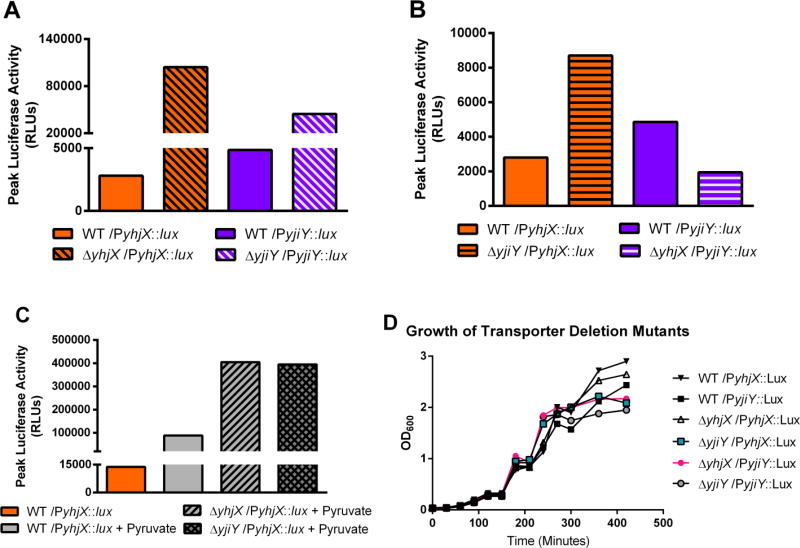
Pyruvate metabolism is central to UPEC pathogenesis; UPEC use pyruvate to fuel the TCA cycle, which is essential during urinary tract infection (Alteri, Smith & Mobley. 2009; Hadjifrangiskou et al. 2011). UPEC also convert pyruvate to serine, which is in turn converted to glycine for use in de-novo purine synthesis, a process essential for UPEC pathogenesis (Shaffer et al. 2017). A signaling network responding to and possibly controlling the acquisition or usage of such metabolites is therefore likely to be important during pathogenesis. In previous studies, we have reported that yhjX and yjiY are significantly upregulated in both acute and long-term murine models of UTI (Behr et al. 2017; Conover et al. 2016), suggesting that these transporters are utilized during infection. Computational analysis of the YhjX and YjiY proteins suggests that they are membrane-embedded transport proteins, the function of which remains uncharacterized. Deletion of either yhjX or yjiY deregulated BtsS/BtsR and YpdA/YpdB mediated induction of yhjX and yjiY (Fig. 5), suggesting the presence of feedback regulation by the target genes to the signaling systems. Although it was not surprising that chromosomal deletion of either downstream gene increased promoter activity of the associated gene in our promoter-luciferase fusions (Fig. 5a), we were excited to see that deletion of yjiY caused an increase in yhjX promoter activity in UPEC (Fig 5b). Indeed, when growth media was spiked with pyruvate as in previous experiments, both the ∆yjiY and ∆yhjX mutants displayed substantially greater yhjX promoter activity than WT under the same conditions (Fig. 5c), suggesting that there may be direct feedback regulation from yjiY to both signaling systems or perhaps that there is some level of functional redundancy between them.
Figure 5. Analysis of yhjX and yjiY promoter activity in mutants lacking putative transporters yhjX or yjiY.
(A) Comparison of maximum luciferase readings for yhjX and yjiY in WT UPEC vs yhjX activity in ΔyhjX mutants and yjiY activity in ΔyjiY mutants. (B) Comparison of maximum luciferase readings in WT UPEC vs yhjX activity in ΔyjiY mutants and yjiY activity in ΔyhjX mutants; i.e. observing whether deletion of one system’s downstream target will alter promoter activity of the opposing downstream target. (C) Analysis of maximum yhjX-lux activity observed in WT, ΔyhjX, and ΔyjiY strains when exposed to 1 mM sodium pyruvate at 120 minutes during in-vitro growth in LB.
None of the mutants used in this experiment displayed gross growth deficits (Fig. 5d) although the deletion of yjiY and yhjX was correlated with a lower final culture density. It is possible that the loss of these transporters causes UPEC to less efficiently utilize available resources during nutrient limited conditions. In the future, we would like to investigate whether these systems play a role in proper nutrient acquisition or usage in high-density bacterial populations such as the biofilms frequently formed by UPEC during infection. The basis of the observed interconnectivity between these two systems could be due to a number of factors, and in the future we will work to understand whether the results seen here are caused by non-canonical sensor-response regulator interactions, by interactions between the response regulators themselves, or by some other means. Our proposed model of BtsS/BtsR and YpdA/YpdB interconnectivity and functioning in UPEC (Fig. 6) outlines several of these possibilities that we would like to investigate moving forward. Future studies will work to understand the function and importance of YhjX and YjiY and to delineate their role in UPEC physiology and pathogenesis. Moving forward, we will also work to characterize and elucidate the nature of these system cross-interactions and to understand the basis of their differential activity in pathogenic and commensal E. coli strains. It is conceivable that uropathogens, given the limited resources typically available in the bladder environment, have evolved semi-redundant systems that work in concert to allow them to efficiently seek out and capitalize on important resources such as pyruvate. Such systems could provide us with a wealth of knowledge regarding host-pathogen interactions and pathogen competition for resources within the body as well as revealing niche specific resource bottlenecks pertinent to various types of bacteria.
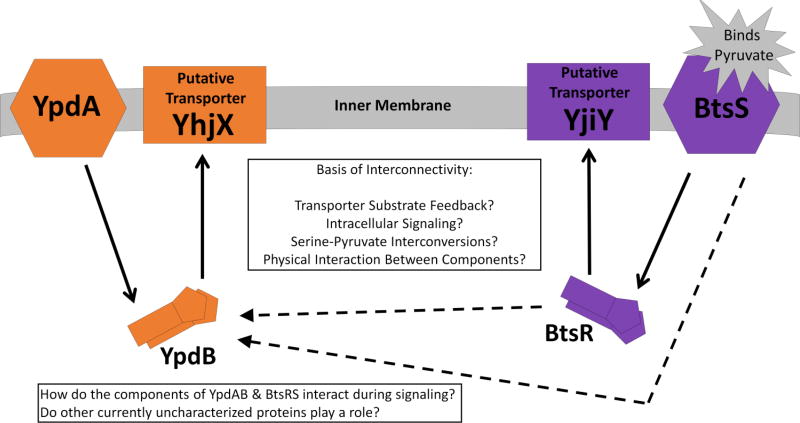
Figure 6. Schematic representing the YpdA/YpdB and BtsS/BtsR TCSs.
Solid lines represent canonical TCS signaling pathways. Dashed lines represent the proposed connectivity between the systems, the basis of which currently remains unknown. The inner textbox proposes several potential mechanisms by which the two systems may influence one another.
Conclusions
Since the description of the first basic TCSs (Ninfa & Magasanik. 1986; Nixon, Ronson & Ausubel. 1986), researchers have found many examples of sophisticated regulation and activity in these bacterial signaling networks. However, despite the field’s growing appreciation of TCS complexity, there are few TCSs for which a true ligand is known and it remains unclear how frequently TCSs interact or how widely distributed these cross-interacting systems are. Additionally, few of these interacting TCSs have had their functionality and biological importance characterized and fewer still have been studied in a pathogen specific context. We propose that the YpdA/YpdB and BtsS/BtsR TCSs represent an interconnected signaling network that functions to regulate uptake and/or usage of pyruvate in UPEC. In this study, we show that UPEC mutants containing deletions of the btsS/btsR genes, coding for the corresponding signaling system components, not only have altered downstream expression of their respective target gene, yjiY, but also have substantial changes in the expression of the YpdA/YpdB system’s target gene, yhjX. These mutations affect yhjX signaling during normal in vitro growth in LB and in response to the addition of pyruvate to growth media. Notably, although deletions in the YpdA/YpdB system have been shown to alter yjiY activity in K12 E. coli, we show here that these mutants do not alter yjiY activity in UPEC. This suggests that interconnected TCSs may evolve differentially in pathogens and non-pathogens. It is therefore possible that interacting TCSs in pathogens can be disrupted to severely attenuate infection and virulence. This could ultimately lead to new therapeutic strategies designed to disrupt vital signaling networks that are only extant in pathogens, thus sparing the natural host microbiota.
Supplementary Material
Acknowledgments
The authors would like to thank the members of the Hadjifrangiskou laboratory for critical review of the manuscript. B.D.S. was supported by the NIH GM07628 T32 training grant. M.N.H. was supported by NIH 5T32GM008554-22 and NIH 2R25GM062459-14 training programs. Financial support for this work was provided by the NIDDK Diabetic Complications Consortium (DiaComp, www.diacomp.org), grant DK076169.
Footnotes
Author Contributions: B.D.S., A.R.E., M.N.H., H.D.G., S.B., K.J., and M.H. conceived and designed experiments. B.D.S., A.R.E., M.N.H., E.Z., H.D.G., and M.H. performed the experiments. K.J. and S.B. provided technical advice and edited the manuscript. B.D.S. and M.H. analyzed the data. B.D.S., A.R.E., M.N.H. and M.H. wrote and edited the manuscript.
Conflicts of Interest: The authors declare no conflict of interest.
References
- Alteri CJ, Smith SN, Mobley HL. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS Pathog. 2009;5:e1000448. doi: 10.1371/journal.ppat.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson DI, Hughes D. Persistence of antibiotic resistance in bacterial populations. FEMS Microbiol Rev. 2011;35:901–11. doi: 10.1111/j.1574-6976.2011.00289.x. [DOI] [PubMed] [Google Scholar]
- Appleby JL, Parkinson JS, Bourret RB. Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell. 1996;86:845–8. doi: 10.1016/s0092-8674(00)80158-0. [DOI] [PubMed] [Google Scholar]
- Barrett JF, Hoch JA. Two-component signal transduction as a target for microbial anti-infective therapy. Antimicrob Agents Chemother. 1998;42:1529–36. doi: 10.1128/aac.42.7.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behr S, Fried L, Jung K. Identification of a novel nutrient-sensing histidine kinase/response regulator network in Escherichia coli. J Bacteriol. 2014;196:2023–9. doi: 10.1128/JB.01554-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behr S, Kristoficova I, Witting M, Breland EJ, Eberly AR, Sachs C, Schmitt-Kopplin P, Hadjifrangiskou M, Jung K. Identification of a High-Affinity Pyruvate Receptor in Escherichia coli. Sci Rep. 2017;7:1388. doi: 10.1038/s41598-017-01410-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhate MP, Molnar KS, Goulian M, DeGrado WF. Signal transduction in histidine kinases: insights from new structures. Structure. 2015;23:981–94. doi: 10.1016/j.str.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breland EJ, Zhang EW, Bermudez T, Martinez CR, Hadjifrangiskou M. The histidine residue of QseC is required for canonical signaling between QseB and PmrB in uropathogenic Escherichia coli. J Bacteriol. 2017 doi: 10.1128/JB.00060-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buelow DR, Raivio TL. Three (and more) component regulatory systems - auxiliary regulators of bacterial histidine kinases. Mol Microbiol. 2010;75:547–66. doi: 10.1111/j.1365-2958.2009.06982.x. [DOI] [PubMed] [Google Scholar]
- Capra EJ, Laub MT. Evolution of two-component signal transduction systems. Annu Rev Microbiol. 2012;66:325–47. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casino P, Rubio V, Marina A. The mechanism of signal transduction by two-component systems. Curr Opin Struct Biol. 2010;20:763–71. doi: 10.1016/j.sbi.2010.09.010. [DOI] [PubMed] [Google Scholar]
- Conover MS, Hadjifrangiskou M, Palermo JJ, Hibbing ME, Dodson KW, Hultgren SJ. Metabolic Requirements of Escherichia coli in Intracellular Bacterial Communities during Urinary Tract Infection Pathogenesis. MBio. 2016;7:e00104–16. doi: 10.1128/mBio.00104-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxen MA, Finlay BB. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol. 2010;8:26–38. doi: 10.1038/nrmicro2265. [DOI] [PubMed] [Google Scholar]
- Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–33. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberly AR, Floyd KA, Beebout CJ, Colling SJ, Fitzgerald MJ, Stratton CW, Schmitz JE, Hadjifrangiskou M. Biofilm Formation by Uropathogenic Escherichia coli Is Favored under Oxygen Conditions That Mimic the Bladder Environment. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18102077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd KA, Moore JL, Eberly AR, Good JA, Shaffer CL, Zaver H, Almqvist F, Skaar EP, Caprioli RM, Hadjifrangiskou M. Adhesive fiber stratification in uropathogenic Escherichia coli biofilms unveils oxygen-mediated control of type 1 pili. PLoS Pathog. 2015;11:e1004697. doi: 10.1371/journal.ppat.1004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman ZN, Dorus S, Waterfield NR. The KdpD/KdpE two-component system: integrating K+ homeostasis and virulence. PLoS Pathog. 2013;9:e1003201. doi: 10.1371/journal.ppat.1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried L, Behr S, Jung K. Identification of a target gene and activating stimulus for the YpdA/YpdB histidine kinase/response regulator system in Escherichia coli. J Bacteriol. 2013;195:807–15. doi: 10.1128/JB.02051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu Rev Microbiol. 2009;63:133–54. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt MJ, Shuman HA. GigA and GigB are Master Regulators of Antibiotic Resistance, Stress Responses, and Virulence in Acinetobacter baumannii. J Bacteriol. 2017;199 doi: 10.1128/JB.00066-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillings MR, Paulsen IT, Tetu SG. Genomics and the evolution of antibiotic resistance. Ann N Y Acad Sci. 2017;1388:92–107. doi: 10.1111/nyas.13268. [DOI] [PubMed] [Google Scholar]
- Gotoh Y, Eguchi Y, Watanabe T, Okamoto S, Doi A, Utsumi R. Two-component signal transduction as potential drug targets in pathogenic bacteria. Curr Opin Microbiol. 2010;13:232–9. doi: 10.1016/j.mib.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Guckes KR, Breland EJ, Zhang EW, Hanks SC, Gill NK, Algood HM, Schmitz JE, Stratton CW, Hadjifrangiskou M. Signaling by two-component system noncognate partners promotes intrinsic tolerance to polymyxin B in uropathogenic Escherichia coli. Sci Signal. 2017;10 doi: 10.1126/scisignal.aag1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guckes KR, Kostakioti M, Breland EJ, Gu AP, Shaffer CL, Martinez CR, Hultgren SJ, Hadjifrangiskou M. Strong cross-system interactions drive the activation of the QseB response regulator in the absence of its cognate sensor. Proc Natl Acad Sci U S A. 2013;110:16592–7. doi: 10.1073/pnas.1315320110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjifrangiskou M, Gu AP, Pinkner JS, Kostakioti M, Zhang EW, Greene SE, Hultgren SJ. Transposon mutagenesis identifies uropathogenic Escherichia coli biofilm factors. J Bacteriol. 2012;194:6195–205. doi: 10.1128/JB.01012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjifrangiskou M, Kostakioti M, Chen SL, Henderson JP, Greene SE, Hultgren SJ. A central metabolic circuit controlled by QseC in pathogenic Escherichia coli. Mol Microbiol. 2011;80:1516–29. doi: 10.1111/j.1365-2958.2011.07660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K, Fried L, Behr S, Heermann R. Histidine kinases and response regulators in networks. Curr Opin Microbiol. 2012;15:118–24. doi: 10.1016/j.mib.2011.11.009. [DOI] [PubMed] [Google Scholar]
- Kellogg SL, Little JL, Hoff JS, Kristich CJ. Requirement of the CroRS Two-Component System for Resistance to Cell Wall-Targeting Antimicrobials in Enterococcus faecium. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.02461-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostakioti M, Hadjifrangiskou M, Pinkner JS, Hultgren SJ. QseC-mediated dephosphorylation of QseB is required for expression of genes associated with virulence in uropathogenic Escherichia coli. Mol Microbiol. 2009;73:1020–31. doi: 10.1111/j.1365-2958.2009.06826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraxenberger T, Fried L, Behr S, Jung K. First insights into the unexplored two-component system YehU/YehT in Escherichia coli. J Bacteriol. 2012;194:4272–84. doi: 10.1128/JB.00409-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genet. 2007;41:121–45. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- Macfarlane EL, Kwasnicka A, Hancock RE. Role of Pseudomonas aeruginosa PhoP-phoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology. 2000;146(Pt 10):2543–54. doi: 10.1099/00221287-146-10-2543. [DOI] [PubMed] [Google Scholar]
- Macielag MJ, Goldschmidt R. Inhibitors of bacterial two-component signalling systems. Expert Opin Investig Drugs. 2000;9:2351–69. doi: 10.1517/13543784.9.10.2351. [DOI] [PubMed] [Google Scholar]
- Mitrophanov AY, Groisman EA. Signal integration in bacterial two-component regulatory systems. Genes Dev. 2008;22:2601–11. doi: 10.1101/gad.1700308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey MA, Schilling JD, Hultgren SJ. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect Immun. 2001;69:4572–9. doi: 10.1128/IAI.69.7.4572-4579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KC, Campellone KG. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol Biol. 2003;4:11. doi: 10.1186/1471-2199-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninfa AJ, Magasanik B. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc Natl Acad Sci U S A. 1986;83:5909–13. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon BT, Ronson CW, Ausubel FM. Two-component regulatory systems responsive to environmental stimuli share strongly conserved domains with the nitrogen assimilation regulatory genes ntrB and ntrC. Proc Natl Acad Sci U S A. 1986;83:7850–4. doi: 10.1073/pnas.83.20.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada A, Igarashi M, Okajima T, Kinoshita N, Umekita M, Sawa R, Inoue K, Watanabe T, Doi A, Martin A, Quinn J, Nishimura Y, Utsumi R. Walkmycin B targets WalK (YycG), a histidine kinase essential for bacterial cell growth. J Antibiot (Tokyo) 2010;63:89–94. doi: 10.1038/ja.2009.128. [DOI] [PubMed] [Google Scholar]
- Olivera BC, Ugalde E, Martínez-Antonio A. Regulatory dynamics of standard two-component systems in bacteria. J Theor Biol. 2010;264:560–9. doi: 10.1016/j.jtbi.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Perry JA, Westman EL, Wright GD. The antibiotic resistome: what's new? Curr Opin Microbiol. 2014;21:45–50. doi: 10.1016/j.mib.2014.09.002. [DOI] [PubMed] [Google Scholar]
- Podgornaia AI, Laub MT. Determinants of specificity in two-component signal transduction. Curr Opin Microbiol. 2013;16:156–62. doi: 10.1016/j.mib.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Robinson VL, Buckler DR, Stock AM. A tale of two components: a novel kinase and a regulatory switch. Nat Struct Biol. 2000;7:626–33. doi: 10.1038/77915. [DOI] [PubMed] [Google Scholar]
- Salazar ME, Laub MT. Temporal and evolutionary dynamics of two-component signaling pathways. Curr Opin Microbiol. 2015;24:7–14. doi: 10.1016/j.mib.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber M, Res I, Matter A. Protein kinases as antibacterial targets. Curr Opin Cell Biol. 2009;21:325–30. doi: 10.1016/j.ceb.2009.01.026. [DOI] [PubMed] [Google Scholar]
- Shaffer CL, Zhang EW, Dudley AG, Dixon BR, Guckes KR, Breland EJ, Floyd KA, Casella DP, Algood HM, Clayton DB, Hadjifrangiskou M. Purine Biosynthesis Metabolically Constrains Intracellular Survival of Uropathogenic Escherichia coli. Infect Immun. 2017;85 doi: 10.1128/IAI.00471-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005;3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton KA, Rather PN. An ompR/envZ Two-Component System Ortholog Regulates Phase Variation, Osmotic Tolerance, Motility, and Virulence in Acinetobacter baumannii strain AB5075. J Bacteriol. 2016 doi: 10.1128/JB.00705-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschauner K, Hörnschemeyer P, Müller VS, Hunke S. Dynamic interaction between the CpxA sensor kinase and the periplasmic accessory protein CpxP mediates signal recognition in E. coli. PLoS One. 2014;9:e107383. doi: 10.1371/journal.pone.0107383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano H, Yoshida M, Ogawa A, Yamamoto K, Ishihama A, Ogasawara H. Cross-regulation between two common ancestral response regulators, HprR and CusR, in Escherichia coli. Microbiology. 2017;163:243–252. doi: 10.1099/mic.0.000410. [DOI] [PubMed] [Google Scholar]
- Wei CF, Tsai YH, Tsai SH, Lin CS, Chang CJ, Lu CC, Huang HC, Lai HC. Cross-talk between bacterial two-component systems drives stepwise regulation of flagellar biosynthesis in swarming development. Biochem Biophys Res Commun. 2017;489:70–75. doi: 10.1016/j.bbrc.2017.05.077. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.