Abstract
The cranial neural crest (CNC) explant assay was originally designed to assess the basic requirements for CNC migration in vitro. This protocol describes the key parameters of CNC explants in Xenopus laevis, with a focus on how to extirpate CNC cells and assay their migration in vitro. The protocol can be adapted according to the needs of the experimenter, some examples of which are discussed here.
MATERIALS
It is essential that you consult the appropriate Material Safety Data Sheets and your institution’s Environmental Health and Safety Office for proper handling of equipment and hazardous materials used in this protocol.
RECIPES: Please see the end of this protocol for recipes indicated by <R>. Additional recipes can be found online at http://cshprotocols.cshlp.org/site/recipes.
Reagents
Danilchik medium <R> containing 50 μg/mL of gentamycin
Ethanol (70%)
Fibronectin, bovine (Sigma)
H2O (reverse osmosis [RODI] or distilled [dH2O])
- Modified Barth’s Saline (MBS) (1×) <R>For dissection medium, use 1× MBS containing 50 μg/mL of gentamycin.
- Phosphate-buffered saline (PBS)In addition, prepare PBS containing 1% (w/v) BSA.
Pork gelatin (optional for glass-bottom plates; see Step 2)
Xenopus laevis eggs
Equipment
Dissecting microscope equipped with a gooseneck lighting system
Embryo incubators at the desired temperature (between 15°C and 19°C for X. laevis)
- Eyelash knifeSelect a human eyelash with the desired thickness and curvature and thread it through a 23-gauge needle fitted on a 1-cc syringe. For safety purposes, the tip of the needle can be cut off with scissors before the threading. Fix the eyelash with a drop of nail polish or cyanoacrylate glue.
Forceps (fine, to remove vitelline envelope)
- Glass bead toolThin out the end of a Pasteur pipette under a benzene burner and melt the end into a ball roughly the size of gastrula-stage embryo (~2 mm).
- Hair loopCut a human hair into 3-inch sections. Thread both ends of a section into a 23-gauge needle fitted on a 1-cc syringe. Push the loop into the needle until the desired stiffness is reached. Fix the hair with a drop of nail polish or cyanoacrylate glue.
Petri dishes (60-mm, plastic), coated with a 4-mm layer of 1% agarose
- Petri dishes (60-mm, plastic), coated with plasticineRoll 2 tsp of plasticine (nondrying, toxin free, appropriate for young children) into a ball. Flatten it out into a plastic Petri dish by hand.
- Plates (multiwell, plastic or glass)See Step 2.
Transfer pipette (disposable plastic or glass) with an opening of ≥2 mm, for transferring embryos
METHOD
CNC explantation can be performed as soon as stage 14 and as late as stage 18; see Step 8. Embryos are staged according to Nieuwkoop and Faber (1967).
The following procedure should be conducted between 15°C and 19°C.
- One or two days before explantation, fertilize the eggs in the morning.Embryos kept at 18°C will be ready for explantation in the afternoon the day after fertilization. Alternatively, embryos can be raised at 14°C, in which case they will be ready for explantation two days after fertilization, in the morning.
- On the day before explantation, coat a multiwell plate with fibronectin.
- For plastic plates, dilute fibronectin to 5–10 μg/mL in PBS, distribute the solution into the appropriate wells (e.g., 50 μL per well for a 96-well plate), and let the fibronectin adsorb overnight at 4°C.
- For glass-bottom plates, follow the above procedure but increase the concentration of fibronectin to 50 μg/mL.Alternatively, precoat glass-bottom wells with a solution of 0.1% pork gelatin for 1 h at 37°C, remove the gelatin solution, and then coat with 10 μg/mL of fibronectin overnight at 4°C or 1 h at 37°C. This treatment leads to explants that migrate as well as if plastic plates were used.
On the day of explantation, when the embryos reach stage 14, sterilize a plasticine-coated dish with 70% ethanol for 10 min. Rinse briefly with RODI H2O and then fill with dissection medium.
- Remove the fibronectin from the multiwell plate prepared in Step 2. Block the wells with PBS containing 1% BSA (e.g., 200 μL per well for a 96-well plate) for 1 h at room temperature.This blocking step may be omitted for glass-bottom plates.
Remove the blocking solution from the multiwell plate and replace with Danilchik medium containing 50 μg/mL of gentamicin.
Add Danilchik medium containing 50 μg/mL of gentamicin to an agarose-coated dish.
Using a glass bead tool, dig trenches in the plasticine-coated dish from Step 3 corresponding to the length of the embryos: The depth of each trench should correspond to the two-thirds of the embryos’ width. The depression should be perpendicular relative to the experimenter or slightly angled toward the bottom left (3° to 5°).
- Select an embryo between stages 14 and 15 and transfer the embryo to the plasticine-coated dish. Remove the vitelline envelope using fine forceps. Ensure that the embryo does not breach the liquid surface once the vitelline envelope is removed.If one wishes to observe the migration of all the CNC, stage 14 or 15 embryos are ideal as the segments (mandibular hyoid and branchial) have not yet formed and the entirety of the tissue can be explanted without losing any of the segments in the process. However, stage 14 CNC may be difficult to distinguish from the surrounding neuroepithelium and placodal ectoderm. In that case, CNC explants can be performed at a later stage. However, by stage 17, the segments are already formed and the loss of one of the segments will likely occur during the dissection.See Troubleshooting.
- Move the embryo into a trench and orient it according to the experimenter’s preference.The goal is to orient the embryo so that the CNC directly faces the experimenter. In Figure 1A, the embryo is positioned in a ¾ anterior view with the anterior neural plate slanted slightly to the left.Being able to recognize the location of the CNC before immobilizing the embryos is critical but sometimes difficult for the novice. The experimenter should be familiar with the Xenopus developmental table. The CNC makes up the bulk of the anterior neural plate borders. They appear as two bulges on each side of the anterior neural plate and can therefore be easily identified if the gooseneck guide lights are oriented in a manner that highlights the three-dimensional conformation of the embryo (Fig. 1A, anterior neural fold bulge).
- To prevent the embryo from moving during the surgery, tighten the plasticine around it by pushing gently on the plasticine with the glass bead.This step is optional for experimenters that prefer performing microdissection on free-moving embryos.
- Insert the tip of the eyelash knife under the ectoderm at the ventral and posterior end of the anterior neural fold (Movie 1). Thread the knife under the ectoderm, move it slightly anteriorly, and cut the ectoderm by lifting the knife swiftly. Repeat the motion anteriorly along the ventral edge of the anterior neural fold until the ventral edge of the CNC is completely uncovered (Movie 1).The goal of this step is to cut the ectoderm ventrally to the anterior neural fold to peel it dorsally (i.e., toward the neural plate) in Step 12.
Peel off the ectoderm dorsally, to expose the underlying neural fold, and drape it over the other side of the embryo using the hair loop (Fig. 1B, Movie 1).
- Identify the location of the CNC (Sadaghiani and Thiebaud 1987). Using the knife tip, cut the CNC outline. Cut superficially to avoid damaging or contaminating your explant with the underlying mesoderm and endoderm (Movie 1).In some cases, the CNC inherits some of the pigment deposited in the egg during oogenesis, which makes it easy to distinguish from the neural plate and mesoderm. However, CNC are unpigmented the majority of the time. The experimenter should be familiar with landscape of the embryos and learn to recognize the bulge formed by the neural folds. Adequate lighting (described in Step 9) is critical.
- Lift the explant off the embryo with the knife and hair loop and inspect it for any contaminating mesoderm and endoderm (Fig. 1C). Scrape off contaminating cells with the hair loop and knife.CNC explants are made of small cells which typically give the tissue a blue hue. The large size and high yolk content of mesoderm and endoderm cells give them a white to yellow coloration.
- Prepare a P20 micropipette fitted with a yellow tip for explant transfer. To prevent explants from sticking to the walls of the tip, aspirate 10 μL of Danilchik medium or PBS containing 1% BSA through the tip a few times to coat the plastic.This step should be performed only once.
Use the pipette to transfer the explant to the agarose-coated dish from Step 6, taking care that the explant remains submerged in the medium and does not breach the liquid surface at any time. Leave the explant in the dish until all the explants needed for the experiment have been dissected and transferred (20 min).
- Remove any debris from the explants by moving them around the dish.Gentle flushing of medium out of the P20 pipette is recommended for moving the explants.
Transfer each explant, one at a time, into a fibronectin-coated well from Step 5. To avoid diluting the medium in the wells (which may contain a drug or other compound) and for better control over where the explant will land, do not pipette the explant into the well. Instead, insert the pipette containing the explant into the medium and let the explant fall out of the pipette tip.
- Allow the CNC cells to adhere to the substrate for 15–30 min at 18°C.See Troubleshooting.
- Record a time-lapse movie between 18°C and 20°C.Typically, one frame every 3 min for 8 to 10 h provides a good compromise between file size and temporal resolution (Movie 2). Magnification can vary between 50× and 250×, depending on the cellular resolution required and on whether or not one wishes to observe all the individual cells once they migrate out of the explants.See Troubleshooting.
FIGURE 1.
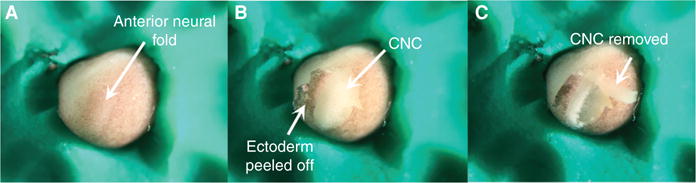
Key steps of the CNC explantation procedure.
TROUBLESHOOTING
Problem (Step 8)
The plasticine is sticky and wounds the embryo once the vitelline envelope is removed.
Solution
This problem will occur if the plasticine is new, but will disappear after a few uses. Until then, 1% BSA (w/v) can be added to the dissection medium to coat the surface of the plasticine. Alternatively, dishes coated with agarose can be used.
Problem (Step 19)
The CNC does not attach to the fibronectin-coated wells.
Solution
It is possible that the explanted tissue is not the CNC. Ensure that the correct territory is dissected (Sadaghiani and Thiebaud 1987; Alfandari et al. 2003). Another possibility is that the well was not coated properly with fibronectin (often a problem on glass-bottom dishes). Try coating with fibronectin at a higher concentration, or coat the wells with 0.1% pork gelatin before coating with fibronectin.
Problem (Step 20)
The CNC attaches to the fibronectin-coated wells, but cells do not emigrate or emigrate very poorly.
Solution
Ensure that the Danilchik medium has the correct pH and has not been adjusted with HCl. This medium contains low chlorine ions and was specifically developed to match the physiology of Xenopus nonepithelial tissue. Alternatively, try another type of Danilchik solution, in particular DFA (Danilchik for Amy), which was developed for culturing other type of explants (Sater et al. 1993).
DISCUSSION
The Use of the CNC explant assay (Alfandari et al. 2003) has led to many discoveries, including the findings that CNC cells use fibronectin and integrin α5β1 to migrate, and that they perform the majority of their migration by keeping cell–cell contact, a phenomenon called collective cell migration (Carmona-Fontaine et al. 2008; Friedl and Mayor 2017). Today, this assay is used to investigate the basic cellular and molecular mechanisms of CNC migration and of collective migration in general (Friedl and Mayor 2017). While an incredibly powerful system, experimenters should be aware that it may not always be useful for studying the functions of certain genes during CNC migration. For example, the metalloprotease ADAM13 is required for CNC migration in the context of the embryo but is dispensable in vitro (Cousin et al. 2012).
This protocol can be used with Xenopus tropicalis as well as X. laevis, provided the experimenter adapts the timing to that of X. tropicalis development. X. tropicalis tissues are slightly stiffer than those of X. laevis, and while our eyelash knife still works fine, the thickness and tapered end of human eyelashes vary widely. If cutting through the tissues proves troublesome, we suggest experimenting with various hair as the knife. For example, eyebrow hairs tend to be stiffer but have blunter ends, which may not be ideal for cutting X. tropicalis embryonic tissues.
Time–lapse movies of CNC explants can yield a large amount of quantifiable information on the effects of the loss or gain of function of a particular gene on CNC migration. Loss- or gain-of-function experiments can be performed by injecting embryos with antisense morpholino, or with mRNA encoding either a constitutively active or dominant negative version of the gene of interest. In addition, experimenters can treat the explants with inhibitors using concentrations similar to what has been described in tissue culture experiments. Various assays can be done to test CNC fitness to migrate. For example, experimenters can assess the trajectory, persistence and speed of the migrating cells using software such as FIJI (Carmona-Fontaine et al. 2008). CNC are also known to perform Contact Inhibition of Locomotion (CIL), a phenomenon whereby two CNC cells colliding with one another will reverse the direction of their migration. CIL is essential for CNC migration both in vivo and in vitro. To assess CIL, one can perform a collision assay, in which two explants are placed in close proximity to one another (Becker et al. 2013). CNC have been shown to be attracted by the growth factor Sdf-1, which is secreted by placodal ectoderm and is essential for their migration in vivo (Theveneau et al. 2013). To assay the ability of the CNC to respond to this chemoattractant, the experimenter can add a source of Sdf-1, such as a bead coated with Sdf-1 or an explant of placodal ectoderm (Theveneau and Mayor 2011; Cousin et al. 2012; Theveneau et al. 2013), in proximity to the explant. Attraction to Sdf-1 can be used to test the fluidity of the CNC explants. For example, explant and Sdf-1 can be deposited in two separate microchambers connected only through openings of defined sizes. By varying the size of these openings, one can test the ability of the CNC to squeeze through openings and therefore challenge the fluidity of the explant (Cousin et al. 2012). The concept of fluidity of tissues undergoing collective cell migration is important, and CNC cells are one of the few models that allow it to be tested and quantified (Friedl and Mayor 2017).
RECIPES
Danilchik Medium
| Bicine | 17.5 mM |
| Bovine serum albumin | 1 mg/mL |
| CaCl2 | 1 mM |
| MgSO4 | 2 mM |
| Na2CO3 | 11.7 mM |
| NaCl | 53 mM |
| Potassium gluconate | 4.25 mM |
| Adjust pH to 8.3. Store at Room temperature for up to 1 month. |
Modified Barth’s Saline (MBS) (1×)
| CaCl2 | 0.41 mM |
| CaNO3 | 0.3 mM |
| HEPES-NaOH | 15 mM |
| KCl | 1 mM |
| MgSO4 | 0.82 mM |
| NaCl | 88 mM |
| NaHCO3 | 2.4 mM |
| Adjust pH to 7.6. Store at XX for up to XXX. |
Supplementary Material
Acknowledgments
H.C. and D.A. are supported by NIH/NIDCR DE025691 and DE016289, respectively.
Footnotes
From the Xenopus collection, edited by Hazel L. Sive.
References
- Alfandari D, Cousin H, Gaultier A, Hoffstrom BG, DeSimone DW. Integrin alpha5beta1 supports the migration of Xenopus cranial neural crest on fibronectin. Dev Biol. 2003;260:449–64. doi: 10.1016/s0012-1606(03)00277-x. [DOI] [PubMed] [Google Scholar]
- Becker SF, Mayor R, Kashef J. Cadherin-11 mediates contact inhibition of locomotion during Xenopus neural crest cell migration. PLoS One. 2013;8:e85717. doi: 10.1371/journal.pone.0085717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Fontaine C, Matthews HK, Kuriyama S, Moreno M, Dunn GA, Parsons M, Stern CD, Mayor R. Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature. 2008;456:957–61. doi: 10.1038/nature07441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousin H, Abbruzzese G, McCusker C, Alfandari D. ADAM13 function is required in the 3 dimensional context of the embryo during cranial neural crest cell migration in Xenopus laevis. Dev Biol. 2012;368:335–44. doi: 10.1016/j.ydbio.2012.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Mayor R. Tuning collective cell migration by cell-cell junction regulation. Cold Spring Harb Perspect Biol. 2017;9:a029199. doi: 10.1101/cshperspect.a029199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus Laevis (Daudin) 2nd. North-Holland, Amsterdam: 1967. [Google Scholar]
- Sadaghiani B, Thiebaud CH. Neural crest development in the Xenopus laevis embryo, studied by interspecific transplantation and scanning electron microscopy. Dev Biol. 1987;124:91–110. doi: 10.1016/0012-1606(87)90463-5. [DOI] [PubMed] [Google Scholar]
- Sater AK, Steinhardt RA, Keller R. Induction of neuronal differentiation by planar signals in Xenopus embryos. Dev Dyn. 1993;197:268–80. doi: 10.1002/aja.1001970405. [DOI] [PubMed] [Google Scholar]
- Theveneau E, Mayor R. Beads on the run: Beads as alternative tools for chemotaxis assays. Methods Mol Biol. 2011;769:449–60. doi: 10.1007/978-1-61779-207-6_30. [DOI] [PubMed] [Google Scholar]
- Theveneau E, Steventon B, Scarpa E, Garcia S, Trepat X, Streit A, Mayor R. Chase-and-run between adjacent cell populations promotes directional collective migration. Nat Cell Biol. 2013;15:763–72. doi: 10.1038/ncb2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


