Abstract
Plant glutathione peroxidases (GPXs) are non-heme thiol peroxidases that play vital roles in maintaining H2O2 homeostasis and regulating plant response to abiotic stress. Here, we performed a comparative genomic analysis of the GPX gene family in cucumber (Cucumis sativus). As a result, a total of 6 CsGPX genes were identified, which were unevenly located in four out of the seven chromosomes in cucumber genome. Based on the phylogenetic analysis, the GPX genes of cucumber, Arabidopsis and rice could be classified into five groups. Analysis of the distribution of conserved domains of GPX proteins showed that all these proteins contain three highly conserved motifs, as well as other conserved sequences and residues. Gene structure analysis revealed a conserved exon–intron organization pattern of these genes. Through analyzing the promoter regions of CsGPX genes, many hormone-, stress-, and development-responsive cis-elements were identified. Moreover, we also investigated their expression patterns in different tissues and developmental stages as well as in response to abiotic stress and x acid (ABA) treatments. The qRT-PCR results showed that the transcripts of CsGPX genes varied largely under abiotic stress and ABA treatments at different time points. These results demonstrate that cucumber GPX gene family may function in tissue development and plant stress responses.
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1185-3) contains supplementary material, which is available to authorized users.
Keywords: Cucumis sativus, Glutathione peroxidase (GPX), Gene family, Expression analysis, Abiotic stress
Introduction
Plants are often subjected to environmental stresses such as drought, salt, extreme temperature and heavy metals. These stresses can lead to the generation of reactive oxygen species (ROS), which damage cellular components and macromolecules such as DNA, proteins and lipids, and ultimately cause the death of cells (Choudhury et al. 2017; You and Chan 2015; Zhou et al. 2017b). To cope with these stresses, plants have developed enzymatic and non-enzymatic antioxidant defense systems to protect cells from uncontrolled oxidation status. Glutathione peroxidase (GPX, EC 1.11.1.9), an important antioxidant enzyme, can catalyze the conversion of hydrogen peroxide (H2O2) or other organic hydroperoxides into water or the corresponding alcohols using reduced glutathione (GSH), thioredoxin (Trx) or other reducing equivalents as electron donors (Bela et al. 2015; Passaia and Margispinheiro 2015; Ursini et al. 1995).
GPXs were first discovered in mammals and their high antioxidant activity depends on the presence of selenocysteine (SeCys) at the active site. According to the protein structure, substrate specificity and subcellular localization, mammal GPXs can be divided into five distinct groups: cytosolic GPX, gastro-intestinal GPX, plasma GPX, phospholipid hydroperoxide GPX (PHGPX), and seleno-independent epididymis GPX (Herbette et al. 2007; Ursini et al. 1995). However, GPXs from vascular plants identified so far have a preference to Trx than to GSH as a reductant, which enables them to reduce peroxides much more efficiently (Herbette et al. 2005; Iqbal et al. 2006; Navrot et al. 2006).
Numerous GPXs characterized in various plant species have shown their effects in protecting cells from oxidative damage through maintaining the H2O2 homeostasis by acting as efficient ROS scavengers (Miao et al. 2006; Passaia and Margispinheiro 2015). Additionally, a number of previous studies have highlighted that GPXs play critical roles in regulating plant response to abiotic stresses such as metal, cold, drought, salt, and oxidative stress (Islam et al. 2015; Kim et al. 2014; Navrot et al. 2006; Passaia et al. 2013). For example, NnGPX transcript was remarkably increased in response to cold, heat, mechanical damage and salt treatment, and overexpression of NnGPX in rice significantly improved tolerance to salt stress (Diao et al. 2014). Arabidopsis plants expressing two wheat GPX genes showed higher salt and H2O2 tolerance (Zhai et al. 2013). LjGpx1 and LjGpx3 perform major antioxidative functions in nodules and play a protective role against salt stress, oxidative stress, and membrane damage (Matamoros et al. 2015). In rice, five GPX genes were identified, which were upregulated in response to exogenous H2O2 and cold stress, while downregulated in response to drought and UV-B light treatments (Passaia et al. 2013). Among them, OsGPX5 can act as a Trx-dependent glutathione peroxidase and participates in the interaction between ER stress and redox homeostasis, which is required for the normal development of rice under stress conditions (Wang et al. 2017). Knockdown of OsGPX1 or OsGPX3 could severely affect the normal growth and development of rice (Passaia et al. 2013, 2014). A recent study showed that the silencing of mitochondrial GPX1 in rice triggered the impair of photosynthesis in response to salinity (Lima-Melo et al. 2016). These findings suggest that GPXs in different plant species play specific roles in regulating plant growth and development as well as responses to various abiotic stresses.
A previous study examined the GPX homologs from 18 plant species, and identified 6 GPX genes in C. sativus (Ozyigit et al. 2016). To better understand the GPX genes in cucumber, we characterized all the 6 GPX members in cucumber, including their genome distributions, phylogenetic relationships, protein motifs, gene structures and promoter sequences, as well as their expression profiles in different tissues and in response to various abiotic stresses. Our results are expected to establish a solid foundation for further investigating the functional roles of GPX genes in cucumber.
Materials and methods
Identification of CsGPXs and protein properties
To predict the members of the GPX proteins in cucumber, we first collected the full-length GPX protein sequences in different plant species, such as Arabidopsis thaliana (8 members) (Rodriguez Milla et al. 2003), Lotus japonicus (6 members) (Ramos et al. 2009), rice (5 members) (Islam et al. 2015), and Gossypium hirsutum (13 members) (Chen et al. 2017). Subsequently, a BLASTp analysis was performed against the Cucumber Genome Database (http://cucumber.genomics.org.cn/) using these protein sequences as queries. Additionally, the GPX domain (PF00255) downloaded from Pfam (http://pfam.sanger.ac.uk/) was used to search the cucumber database by HMMER software. The candidate GPX protein sequences were aligned to each other to ensure that no member was repeated for multiple times. The remaining GPX proteins were checked using the Pfam (http://www.sanger.ac.uk/software/pfam/) and Simple Modular Architecture Research Tool (SMART) (http://smart.embl-heidelberg.de/), for confirming the presence of the GPX domain (PF00255). The theoretical isoelectric point (pI), molecular weight (MW) and the grand average of hydropathy index (GRAVY) of each deduced CsGPX protein were determined by the ProtParam tool (http://web.expasy.org/protparam/). Prediction of subcellular localization of each deduced CsGPX protein was performed with WoLF PSORT (https://www.genscript.com/wolf-psort.html/) (Horton et al. 2007).
Chromosomal location, sequence alignment and phylogenetic analysis
The MapInspect tool was used to determine the chromosomal locations of the CsGPX genes to identify their detailed chromosomal position provided in the Cucumber Genome Database (Zhou et al. 2017b). Duplication analysis was conducted as previously described (Ozyigit et al. 2016). Multiple sequence alignments of GPX proteins were performed with ClustalW software (Larkin et al. 2007), and the phylogenetic tree was constructed using the neighbor-joining algorithm in MEGA (version 5.0) with 1000 bootstrap replicates (Tamura et al. 2007).
Conserved motif and gene structure predictions
The GPX protein sequences from cucumber, Arabidopsis, and rice were analyzed by the online MEME tool (http://meme-suite.org/tools/meme) with default parameters (Bailey et al. 2009). The open-reading frame (ORF) sequences and corresponding genomic DNA (gDNA) sequences of GPX members in cucumber, Arabidopsis and rice were downloaded from the Cucumber Genome Database, TAIR (http://www.arabidopsis.org/), and TIGR (http://rice.plantbiology.msu.edu/), respectively, and then the gene structures were analyzed by comparing the ORF and gDNA sequences using Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/index.php) (Hu et al. 2015).
Promoter region analysis of CsGPX genes
To analyze the cis-elements in the promoter regions of CsGPX genes, the transcription start site in the 1.0 kb upstream region from translation start code ATG of each gene was obtained from the Cucumber Genome Database. The putative promoter regions of each CsGPX gene were investigated using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al. 2002).
Transcriptome analysis of CsGPX genes in different tissues
For transcriptome analysis, the Illimuna HiSeq readings were retrieved from a public repository database (SRA, Sequence Read Archive) with following accession numbers: SRR351476 (ovary, unexpanded), SRR351489 (expanded ovary, fertilized), SRR351495 (expanded ovary, unfertilized), SRR351499 (root), SRR351905 (stem), SRR351906 (leaf), SRR351908 (male flower), SRR351910 (tendril), SRR351911 (tendril basal tissue), SRR351912 (female flower) (Baloglu et al. 2014). All readings were obtained and analyzed as previously described (Altunoglu et al. 2016), and the heat map and hierarchical clustering were computed with the pheatmap package in R.
Plant materials and treatments
Cucumber (Cucumis sativus L. cv. Chinese long No. 9930) was used as the subject in this research. Two-week-old cucumber seedlings planted in artificial chambers (16 h photoperiod, 400 µmol m−2 s−1 light intensity, and 60–70% relative humidity) were subjected to different abiotic stresses including cold, salt, and PEG, as well as abscisic acid (ABA) treatments following the descriptions in our previous studies (Zhou et al. 2017a, c). The leaf tissues were harvested from seedling samples at 0, 3, 6, and 12 h, frozen immediately in liquid nitrogen, and stored at − 80 °C until RNA extraction.
RNA extraction and quantitative RT-PCR (qRT-PCR)
Total RNA was extracted using Trizol reagent (Tiangen, China) according to the manufacturer’s instruction. Genomic DNA contamination was eliminated with DNase I (Invitrogen, USA), and the first-strand cDNA was synthesized using the Superscript III Reverse Transcriptase kit (Invitrogen, USA) following the manufacturers’ instructions. The qRT-PCR was performed in triplicate with SYBR Green Master Mix (Tiangen, China) as described previously (Zhou et al. 2017a). The reaction conditions were set as follows: 95 °C for 30 s, followed by 40 cycles at 95 °C for 15 s and 60 °C for 60 s. An endogenous actin gene (CsAct3, GenBank accession number: DQ115883) was used as the reference gene for the normalization of gene expression analysis (Wan et al. 2010). The quantitative PCR data were analyzed using the 2−ΔΔCT method (Livak and Schmittgen 2001). The primers used for qRT-PCR are listed in Table S1.
Results
Genome-wide identification and chromosomal distribution of CsGPXs in cucumber
A total of six putative GPX genes were identified in cucumber genome, and were designated as CsGPX1–CsGPX6 according to the order of their location on the chromosomes (Table 1). Table 1 shows the details of gene names, locus names, chromosomal locations, gDNA and open-reading frame (ORF) length, protein length, theoretical molecular weight (MW), isoelectric point (pI), and GRAVY. The predicted gDNA and ORF length varied substantially among members, with that of gDNA ranging from 1721 to 2732 bp, and that of ORF from 504 to 726 bp.
Table 1.
A complete list of six CsGPXs identified in this study
| Gene name | Locus name | Chromosomal location | Genomic position (5′–3′) | gDNA (bp) | ORF (bp) | Protein physicochemical characteristics | Subcellular prediction | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Length (aa) | MW (kDa) | pI | GRAVY | |||||||
| CsGPX1 | Csa002969 | 4 | 397887–400248 | 2362 | 510 | 169 | 18.85 | 7.60 | − 0.419 | Chlo/Nucl |
| CsGPX2 | Csa014856 | 4 | 15071913–15074644 | 2732 | 726 | 241 | 26.48 | 9.50 | − 0.126 | Chlo/Mito |
| CsGPX3 | Csa012719 | 5 | 15490251–15492309 | 2059 | 513 | 169 | 19.14 | 5.37 | − 0.327 | Cyto |
| CsGPX4 | Csa012718 | 5 | 15486562–15488282 | 1721 | 504 | 167 | 18.39 | 8.32 | − 0.302 | Chlo |
| CsGPX5 | Csa007936 | 6 | 16648487–16651191 | 2705 | 513 | 170 | 19.02 | 8.66 | − 0.369 | Cyto/Chlo |
| CsGPX6 | Csa001798 | 7 | 13148933–13151151 | 2219 | 531 | 176 | 19.79 | 8.86 | − 0.309 | Chlo/Cyto |
The subcellular localization of CsGPX proteins were predicted using WoLF PSORT (http://www.genscript.com/psort/wolf_psort.html). gDNA genomic DNA, ORF open reading frame; aa, amino acid, MW molecular weight, pI isoelectric point, Chlo chloroplast, Nucl nucleus, Cyto cytosol, Mito mitochondria
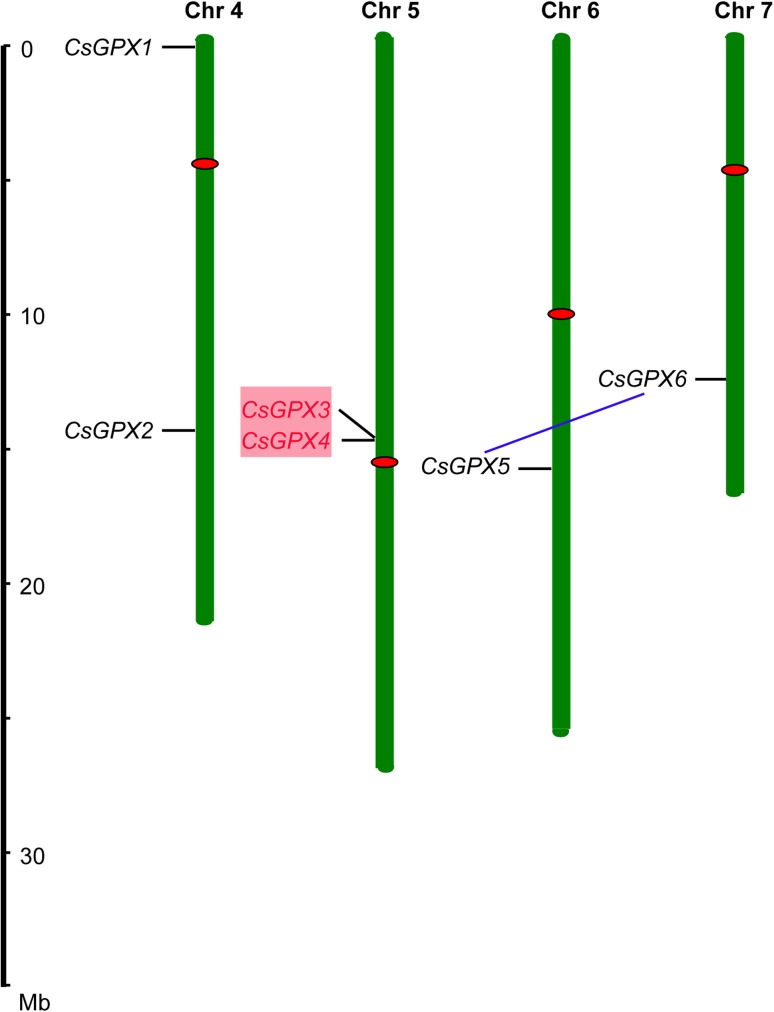
To investigate the chromosomal distribution of CsGPXs, these genes were mapped onto cucumber chromosomes. As shown in Fig. 1, CsGPX1–CsGPX6 were randomly distributed in four out of the total seven chromosomes of cucumber. Each of chromosome 4 and 5 contained two members of CsGPX genes, followed by chromosome 6 and 7 with one member in each. In addition, one segmental duplication event (CsGPX5 and CsGPX6) and one pair of tandem duplicated genes were identified (CsGPX3 and CsGPX4) (Fig. 1).
Fig. 1.
Positions of GPX gene family members on cucumber chromosomes. Gene pairs from tandem duplications highlighted in red. Pairs from segmental duplications are linked with blue lines. Chromosome numbers are indicated at the top of each bar. Scale represents a 10 Mb chromosomal distance
Characterization and comparison of deduced GPX proteins in cucumber
Physicochemical analysis of the predicted CsGPX proteins showed that the lengths, MWs, and pIs of the CsGPX proteins were 167–241 amino acids, 18.39–26.48 kDa, and 5.37–9.50, respectively (Table 1). In addition, the GRAVY of the CsGPX proteins was lower than 0, suggesting that they were hydrophilic. Prediction of protein subcellular localization suggested that the majority of CsGPX proteins were located in chloroplast and cytoplasm, while CsGPX1 and CsGPX2 might be located in other subcellular compartments, such as nucleus and mitochondria.
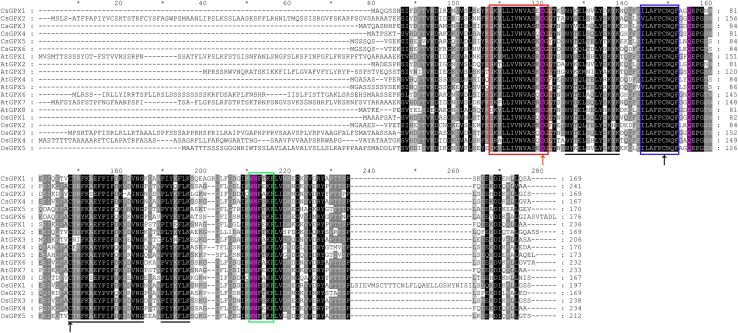
All the putative full-length GPX proteins were aligned using ClustalW to determine the residue conservation patterns within the family from cucumber, Arabidopsis and rice. As shown in Fig. 2, three highly conserved motifs, GKVLLIVNVASXCG (GPX signature 1), ILAFPCNQ (GPX signature 2) and WNFXKF, were present in the members of GPX family, which is consistent with the case of most plant and mammalian GPX sequences (Chen et al. 2017; Gao et al. 2014; Ramos et al. 2009; Zhai et al. 2013). Other evolutionary conserved sequences were also present, such as NYX(E/Q)(L/M)XXLYX(K/R)(Y/H)(K/R), C(T/S/N/E)(R/I)FK(A/S)E(F/Y)P(I/V)F(D/Q)K(V/I)(D/A/R/E)(V/I)NG, and P(I/V/L)Y(K/Q/E/N)FLK. Several highly conserved residues were found in all aligned sequences, including Cys (C), Gln (Q), Trp (W) and Asn (N), suggesting their potential functions in enzyme activity (Fig. 2).
Fig. 2.
Multiple sequence alignment of GPX protein sequences from cucumber, Arabidopsis and rice. The three conserved domains found in most animal and plant GPXs are boxed by red, blue, and green, respectively. Arrows indicate the conserved three Cys residues of plant GPX proteins, and the red arrow represent the Cys residue replaced by Sec in mammalian phospholipid hydroperoxide GPXs. The amino acid residues (Cys, Gln, Trp and Asn) that form part of the proposed catalytic site of the GPXs are highlighted in purple. Other evolutionary conserved sequences were underlined
Comparison of phylogenesis, conserved motifs and gene structures of GPX gene family between cucumber, Arabidopsis and rice
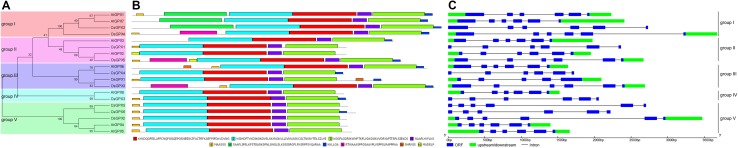
To investigate the evolutionary history GPX gene family, an unrooted neighbor-joining phylogenetic tree was constructed by a multiple sequence alignment of the predicted GPX protein sequences from cucumber, Arabidopsis, and rice (Fig. 3a). These GPX proteins could be divided into five groups: group I, II, III, IV and V. Each of group I–III contained four members, and group IV harbored the least GPXs (2), while group V included the most members (5). AtGPXs and CsGPXs were present in all the five groups, while group IV did not include rice GPX members. According to the phylogenetic tree, several GPX members had paralogs among themselves, such as AtGPX1 and AtGPX7, OsGPX1 and OsGPX3, CsGPX5 and CsGPX6, AtGPX4 and AtGPX5. In addition, there was one pair of closely related orthologous GPXs between cucumber and Arabidopsis (AtGPX1 and CsGPX3), suggesting that they may have similar functions. CsGPXs showed a closer relationship with GPXs in Arabidopsis than with those in rice, implying that the main functions of this family in plants had differentiated before the divergence of dicot and monocot.
Fig. 3.
Phylogenetic relationships, conserved motifs and gene structures of GPX gene family from cucumber, rice and Arabidopsis. a The phylogenetic tree of cucumber, rice and Arabidopsis GPX proteins was constructed from alignment of full-length of amino acid sequences using MEGA 5.0 by the neighbor-joining method with 1000 bootstrap replicates. The groups (I, II, III, IV and V) were marked by colorful backgrounds according to the phylogenetic relationship. b Conserved motifs were identified by MEME with the complete amino acid sequences of GPX proteins from cucumber, rice and Arabidopsis. Lengths of motifs of each GPX protein were displayed proportionally. c Exon–intron structure analysis was carried out by GSDS software. The blue boxes indicate ORFs, the green boxes indicate upstreams or downstreams, and the black lines indicate introns. The information of GPX members were listed in Table S2
To further confirm the evolutionary relationships among the GPXs, MEME server was used to conduct a conserved motif analysis. As shown in Fig. 3b, ten conserved motifs were identified. All of the GPX proteins from cucumber, Arabidopsis, and rice contained motifs 1–4, which include the GPX signatures. In addition, three GPX proteins (AtGPX1, AtGPX7 and CsGPX2) in group I harbored an additional motif 6, which is located in the N-terminal region (Fig. 3b). Motif 5 was found in all the OsGPXs, but not in some GPX members in cucumber (CsGPX2 and CsGPX4) and Arabidopsis (AtGPX2, AtGPX3, AtGPX7 and AtGPX8). Motif 7 in the C-terminal region was present in group I and III members but absent in several members in other groups, such as CsGPX1 and AtGPX2 (group II), AtGPX8 (group IV), CsGPX5, OsGPX2, AtGPX4 and AtGPX5 (group V). It is noteworthy that motif 8 was specific to rice, which was present in OsGPX3, OsGPX4 and OsGPX5. Two members in group III (AtGPX6 and OsGPX1) possessed motif 9. Motif 10 was found only in two group II members (AtGPX2 and OsGPX5). In general, the structure of GPX proteins was conserved throughout the GPX gene family in cucumber, Arabidopsis and rice, and some closely related GPX proteins tended to share similar motif compositions.
To further examine the structural diversity of these GPX genes, a comparison of the genomic sequences with the corresponding ORF sequences was carried out by GSDS software. As shown in Fig. 3c, all of these genes had five introns with the exception of OsGPX1, which harbored four introns. Some GPX genes clustered in the same group had similar gene structures. For example, nearly all GPX genes in group I, II, IV and V had similar lengths of ORFs, although they had variable upstream or downstream lengths. Similar results were also observed in some GPX genes from other plant species, such as Lotus japonicus (Ramos et al. 2009), Triticum aestivum (Zhai et al. 2013), Thellungiella salsuginea (Gao et al. 2014), and Gossypium hirsutum (Chen et al. 2017). These results indicated a high degree of conservation in the exon–intron organization of GPX gene family among plant species.
Bioinformatics analysis of putative CsGPX promoters
To understand the transcriptional regulation of CsGPX genes, 1.0-kb regions upstream of CsGPX genes were extracted and analyzed using the PlantCARE database. A number of cis-acting regulatory elements were found in the promoter regions of CsGPX genes, and these cis-elements could be classified mainly into three groups, including hormone-responsive-, stress-responsive- and development-responsive cis-elements (Table S3; Table 2). Among them, ten types of hormone-responsive cis-elements were identified, and each CsGPX gene harbored at least one type of these cis-elements, implying that CsGPXs are related to the response to hormones (Table 2). In addition, six types of stress-responsive cis-elements, such as heat stress-responsive element (HSE), drought-responsive element (MBS), defense and stress-responsive element (TC-rich repeats), anaerobic induction element (ARE), and elicitor-responsive elements (ELI-box3 and Box-W1), were found to be the major regulatory elements in the promoters of CsGPX genes (Table 2). Moreover, some development-responsive cis-elements, which are involved in circadian control (circadian), endosperm expression (GCN4_motif), zein metabolism regulation (O2-site), endosperm-specific expression (Skn-1 motif), meristem-specific activation (CCGTCC-box), root-specific expression (motif I), and shoot-specific expression (as-2-box), were present in a series of CsGPXs promoters (Table 2). The presence of these cis-elements implied that CsGPX genes might have potential functions in hormone signaling, responses to various stresses, and different developmental processes.
Table 2.
Kinds and numbers of known cis-elements in the promoter regions of CsGPX genes
| Hormone-responsive cis-elements | Stress-responsive cis-elements | Development-responsive cis-elements | |
|---|---|---|---|
| CsGPX1 | ABRE (1), CGTCA-motif (2), GARE-motif (1), TATC-box(1), TCA-element (4) | MBS (2), TC-rich repeats (1), ARE (2) | circadian (1), GCN4_motif (1), Skn-1_motif (1) |
| CsGPX2 | ABRE (1) | HSE (1), ARE (2), Box-W1 (1) | circadian (1), O2-site (1), Skn-1_motif (4) |
| CsGPX3 | ERE (2), TCA-element (1), TGA-element (1) | HSE (2), ARE (1), Box-W1 (1) | circadian (2), GCN4_motif (1), O2-site (1), Skn-1_motif (6) |
| CsGPX4 | CE3 (1), ABRE (6), P-box (1), TCA-element (1), AuxRR-core (1) | HSE (2), TC-rich repeats (1), ARE (1), ELI-box3 (1) | circadian (3), motif I (1), as-2-box (1) |
| CsGPX5 | TCA-element (2), AuxRR-core (1), TGA-element (1) | HSE (1), TC-rich repeats (2), ARE (1) | circadian (2), Skn-1_motif (1), CCGTCC-box (1) |
| CsGPX6 | ERE (1) | HSE (1), ARE (2) | circadian (1), Skn-1_motif (2) |
The numbers in parentheses indicate the number of the cis-elements in the promoter of CsGPX genes
Tissue-specific expression of CsGPX genes in cucumber
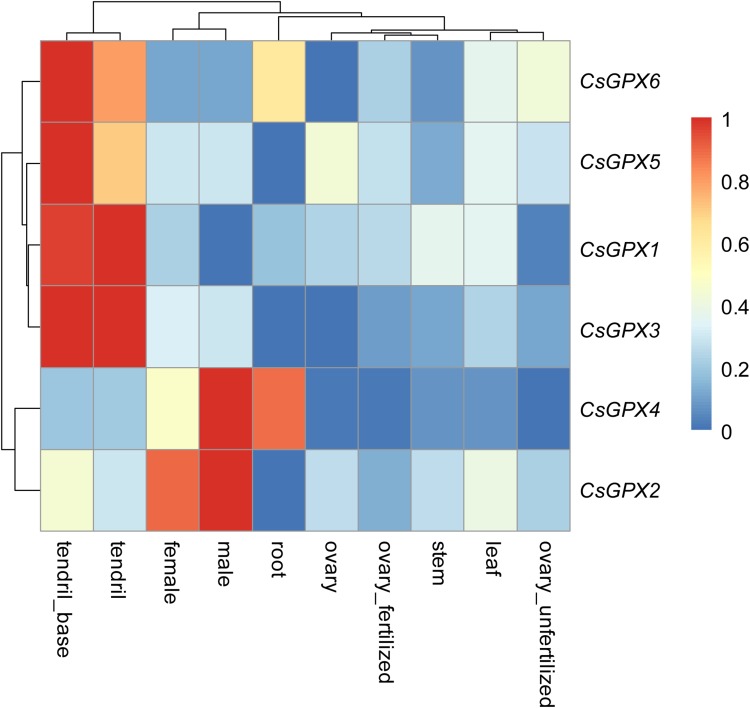
To better understand the functions of CsGPX genes in the growth and development of cucumber, their expression profiles in different tissues were analyzed with publicly available transcriptome data. According to the transcriptomic dataset, all the 6 CsGPX genes were expressed in most tissues at different levels (Fig. 4). CsGPX1, CsGPX3, CsGPX5 and CsGPX6 were highly expressed in the tendril and/or tendril base, suggesting that they may play specific roles in tendril development. On the contrary, the expression of CsGPX1 was not detected in male flowers and unfertilized ovaries, and that of CsGPX3 was not detected in roots and ovaries, while CsGPX6 had no expression in stems and ovaries. CsGPX2 and CsGPX4 showed the highest transcription level in male flowers, and relatively higher expression in female flowers, implying their specific roles in flower development. The expression level of CsGPX4 was high in roots and relatively low in tendrils, and was even undetectable in leaves, stems and ovaries. In roots, both CsGPX2 and CsGPX5 had no expression, while CsGPX6 showed moderate transcription levels. The spatial variations in the expression of CsGPX genes in different tissues revealed that they may participate in different growth and development processes in cucumber.
Fig. 4.
Tissue-specific digital expression profiles of the CsGPX genes in cucumber. The CsGPX genes were listed at the right of the expression array, and the expression values mapped to a color gradient from low (blue) to high expression (red) are shown at the right of the figure
Response of CsGPX gene expression to various abiotic stresses and ABA treatment
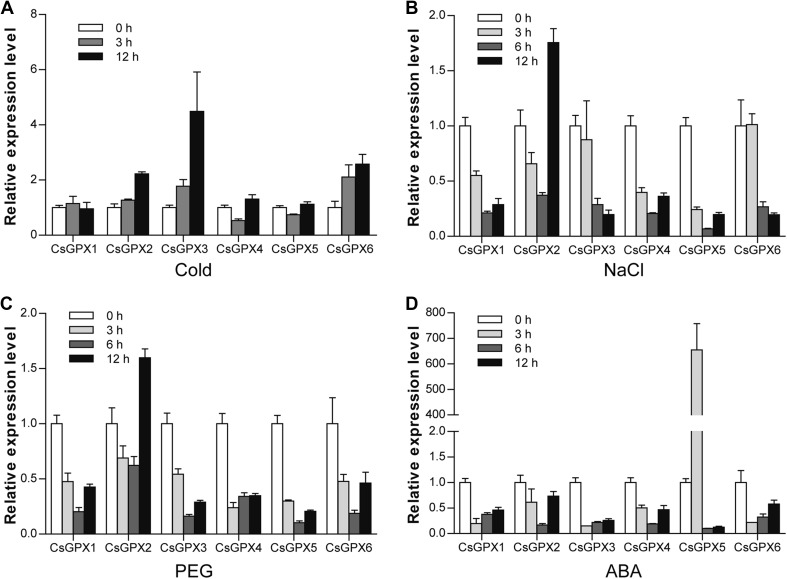
To examine the effects of environmental stresses on CsGPX genes, their expression was analyzed under different conditions (cold, NaCl, PEG, and ABA) by qRT-PCR (Fig. 5). The expression of most CsGPX genes was altered under cold treatment except for CsGPX1. CsGPX2, CsGPX3 and CsGPX6 were significantly induced by cold treatment, while that of CsGPX4 and CsGPX5 was significantly decreased at 3 h after cold treatment, and finally increased at 12 h (Fig. 5a). Under NaCl treatment, nearly all CsGPX genes were downregulated at certain time points except for CsGPX2, whose expression gradually declined at 3 and 6 h, followed by a sharp increase at 12 h (1.76-fold) (Fig. 5b). Similar patterns were also observed under PEG treatment (Fig. 5c). Since ABA plays important roles in the adaptation to environmental stimuli, we checked whether these CsGPX genes are regulated by ABA. As shown in Fig. 5d, most CsGPX genes were downregulated under ABA treatment, except for CsGPX5, which was obviously upregulated at 3 h (654.9-fold), but was downregulated at 6 and 12 h.
Fig. 5.
qRT-PCR analysis of CsGPX genes under different conditions including cold (a), NaCl (b), PEG (c), and ABA (d). Data were normalized to level of CsAct3 expression and expressed relative to the values at 0 h. Data represent the mean ± SD (standard deviation) of three independent biological replicates
Discussion
In previous studies, genome-wide identification and characterization of GPX gene family have been performed in various plant species, such as Arabidopsis thaliana (8 members) (Rodriguez Milla et al. 2003), Lotus japonicus (6 members) (Ramos et al. 2009), Thellungiella salsuginea (8 members) (Gao et al. 2014), Oryza sativa (5 members) (Islam et al. 2015; Passaia et al. 2013), and Gossypium hirsutum (13 members) (Chen et al. 2017). In this study, a total of 6 GPX family members were identified in cucumber (Table 1), and the number of family members is not correlated with the increase in genome size, which may be due to gene- and whole-genome duplications (WGD) including tandem and segmental duplications at different points of evolution (Cao et al. 2016; Flagel and Wendel 2009; Zhou et al. 2017b). For example, one and two segmental duplication events were observed in rice (OsGPX1 and OsGPX3) and Arabidopsis (AtGPX1 and AtGPX7, AtGPX4 and AtGPX5), respectively (Ozyigit et al. 2016). In the present study, one segmental duplication event (CsGPX5 and CsGPX6) was also identified. In addition, chromosome 5 possesses one tandem duplication event (CsGPX3 and CsGPX4), which has not been observed in other plant species so far. Therefore, these tandem and segmental duplications may play an important role in the expansion of CsGPX gene family.
The six CsGPX proteins possess three highly conserved motifs including two GPX signatures (Fig. 2), which are also present in other plant species (Chen et al. 2017; Gao et al. 2014; Ramos et al. 2009; Zhai et al. 2013) as well as in mammalian GPX members (Toppo et al. 2008). Furthermore, other evolutionarily conserved sequences and residues were also identified in GPX proteins (Fig. 2). To further understand the conserved protein motifs in individual GPX proteins, the GPX proteins were analyzed using MEME server. As a result, the evolutionarily conserved motifs were also identified in all the GPX proteins. Notably, some conserved motifs were restricted to specific groups of GPX proteins, or particular species (Fig. 3b), implying that these motifs may contribute to particular functions of GPX proteins.
The phylogenetic analysis showed that GPX proteins can be divided into five major groups, and CsGPXs and AtGPXs were present in every group, while group IV contained no OsGPXs (Fig. 3a). Each CsGPX protein has a homologue in Arabidopsis, suggesting that the GPX genes in cucumber and Arabidopsis are more closely related to each other than to those in rice (Fig. 3a). The exon–intron organization of genes plays an important role in the evolution of multi-gene families (Cao et al. 2016). Gene structure analysis revealed that almost all GPX genes in cucumber, Arabidopsis and rice have five introns, and some closely related GPX genes have similar lengths of ORFs (Fig. 3c), suggesting that these genes are highly conserved during evolution and may have similar functions.
As an efficient ROS scavenger, GPX is a type of stress-responsive protein in response to various abiotic stresses. Previous studies have shown that plant GPXs play important regulatory roles in the responses to various abiotic stresses, including salinity, drought, and cold (Fu 2014; Islam et al. 2015; Passaia et al. 2013; Rodriguez Milla et al. 2003; Zhai et al. 2013). However, the members of GPX gene family may display differential expression patterns in response to various abiotic stresses. For example, in Thellungiella salsuginea, different GPX members were coordinately regulated under various abiotic stress conditions (Gao et al. 2014). In rice, both OsGPX2 and OsGPX4 were upregulated in response to drought and oxidative stress, while were downregulated in response to salinity, heat and cold (Islam et al. 2015). In the present study, each of the six CsGPX genes possesses at least two types of stress-responsive cis-elements (HSE, MBS, TC-rich repeats, ARE, ELI-box3, and Box-W1) in their promoter regions, implying their potential roles in stress response. We then investigated the responses of these GPX genes to different stresses including cold, salt, and drought. Three genes were upregulated by cold treatment, including CsGPX2, CsGPX3, and CsGPX6 (Fig. 5a), suggesting their possible roles in the response to cold stress. For other two treatments (NaCl and PEG), most CsGPX genes showed a decrease in expression levels, indicating their possibly negative regulatory roles. Similarly, the transcript levels of GhGPX1, − 3, − 5, − 8, and − 13 decreased in leaves after treatment with 400 mM of NaCl for 3 h (Chen et al. 2017). Some members of TsGPX family genes were also repressed by salt stress at least at one time point during salt treatment (Gao et al. 2014).
ABA is a key phytohormone that plays important roles in plant growth and development as well as in response to various stresses (Kayum et al. 2017; Li et al. 2017; Vishwakarma et al. 2017). Previous studies have shown that some members of GPX gene family can be regulated by an ABA-dependent signaling pathway (Miao et al. 2006; Rodriguez Milla et al. 2003; Zhai et al. 2013). In this study, almost all the six CsGPX genes were differentially downregulated by ABA except for CsGPX5, which was markedly upregulated at 3 h after ABA treatment (Fig. 5d). Besides, almost all CsGPX genes were responsive to one or more abiotic stresses including cold, salinity and drought (Fig. 5a–c), suggesting that these genes may play regulatory roles in stress responses by modulating ABA signaling. In fact, AtGPX3 acts as a scavenger of H2O2 and specifically relays the H2O2 signaling as an oxidative signal transducer in ABA and drought stress signaling (Miao et al. 2006). Two GPX genes in wheat (W69 and W106) also play important roles in salt and ABA signaling to regulate abiotic stress responses (Zhai et al. 2013).
Conclusions
In this study, a total of six CsGPX genes were identified in C. sativus through genome-wide analysis. Their genome distributions, promoter sequences, and expression profiles in different tissues and in response to various abiotic stresses were analyzed. A comparison of phylogenetic relationships, protein motifs, and gene structures between cucumber, Arabidopsis and rice GPX genes was also carried out. The expression profiling of CsGPX genes under different abiotic stresses and ABA treatments indicated that one or more CsGPX genes might be involved in ABA signaling pathway and stress responses. Our results provide a preliminary exploration of CsGPX genes, and facilitate future investigation of the biological functions of GPX proteins in cucumber.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was funded by the Key Project of Youth Science Foundation of Jiangxi Province (20171ACB21025), and the National Natural Science Foundation of China (31460522 and 31660578).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Yong Zhou and Lifang Hu contributed equally to this work.
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1185-3) contains supplementary material, which is available to authorized users.
References
- Altunoglu YC, Baloglu P, Yer EN, Pekol S, Baloglu MC. Identification and expression analysis of LEA gene family members in cucumber genome. Plant Growth Regul. 2016;80:225–241. doi: 10.1007/s10725-016-0160-4. [DOI] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloglu MC, Eldem V, Hajyzadeh M, Unver T. Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS ONE. 2014;9:e96014. doi: 10.1371/journal.pone.0096014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bela K, Horvath E, Galle A, Szabados L, Tari I, Csiszar J. Plant glutathione peroxidases: emerging role of the antioxidant enzymes in plant development and stress responses. J Plant Physiol. 2015;176:192–201. doi: 10.1016/j.jplph.2014.12.014. [DOI] [PubMed] [Google Scholar]
- Cao Y, Han Y, Jin Q, Lin Y, Cai Y. Comparative genomic analysis of the GRF genes in Chinese pear (Pyrus bretschneideri Rehd), poplar (Populous), grape (Vitis vinifera), Arabidopsis and rice (Oryza sativa) Front Plant Sci. 2016;7:1750. doi: 10.3389/fpls.2016.01750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Li K, Li H, Song CP, Miao Y. The glutathione peroxidase gene family in Gossypium hirsutum: genome-wide identification, classification, gene expression and functional analysis. Sci Rep. 2017;7:44743. doi: 10.1038/srep44743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury FK, Rivero RM, Blumwald E, Mittler R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017;90:856–867. doi: 10.1111/tpj.13299. [DOI] [PubMed] [Google Scholar]
- Diao Y, Xu H, Li G, Yu A, Yu X, Hu W, Zheng X, Li S, Wang Y, Hu Z. Cloning a glutathione peroxidase gene from Nelumbo nucifera and enhanced salt tolerance by overexpressing in rice. Mol Biol Rep. 2014;41:4919–4927. doi: 10.1007/s11033-014-3358-4. [DOI] [PubMed] [Google Scholar]
- Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants. New Phytol. 2009;183:557–564. doi: 10.1111/j.1469-8137.2009.02923.x. [DOI] [PubMed] [Google Scholar]
- Fu JY. Cloning of a new glutathione peroxidase gene from tea plant (Camellia sinensis) and expression analysis under biotic and abiotic stresses. Bot Stud. 2014;55:7. doi: 10.1186/1999-3110-55-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Chen J, Ma T, Li H, Wang N, Li Z, Zhang Z, Zhou Y. The glutathione peroxidase gene family in Thellungiella salsuginea: genome-wide identification, classification, and gene and protein expression analysis under stress conditions. Int J Mol Sci. 2014;15:3319–3335. doi: 10.3390/ijms15023319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbette S, Menn AL, Rousselle P, Ameglio T, Faltin Z, Branlard G, Eshdat Y, Julien JL, Drevet JR, Roeckel-Drevet P. Modification of photosynthetic regulation in tomato overexpressing glutathione peroxidase. Biochim Biophys Acta. 2005;1724:108–118. doi: 10.1016/j.bbagen.2005.04.018. [DOI] [PubMed] [Google Scholar]
- Herbette S, Roeckel-Drevet P, Drevet JR. Seleno-independent glutathione peroxidases. More than simple antioxidant scavengers. FEBS J. 2007;274:2163–2180. doi: 10.1111/j.1742-4658.2007.05774.x. [DOI] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal A, Yabuta Y, Takeda T, Nakano Y, Shigeoka S. Hydroperoxide reduction by thioredoxin-specific glutathione peroxidase isoenzymes of Arabidopsis thaliana. FEBS J. 2006;273:5589–5597. doi: 10.1111/j.1742-4658.2006.05548.x. [DOI] [PubMed] [Google Scholar]
- Islam T, Manna M, Kaul T, Pandey S, Reddy CS, Reddy MK. Genome-wide dissection of Arabidopsis and rice for the identification and expression analysis of glutathione peroxidases reveals their stress-specific and overlapping response patterns. Plant Mol Biol Rep. 2015;33:1413–1427. doi: 10.1007/s11105-014-0846-6. [DOI] [Google Scholar]
- Kayum MA, Park JI, Nath UK, Biswas MK, Kim HT, Nou IS. Genome-wide expression profiling of aquaporin genes confer responses to abiotic and biotic stresses in Brassica rapa. BMC Plant Biol. 2017;17:23. doi: 10.1186/s12870-017-0979-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Jang MG, Noh HY, Lee HJ, Sukweenadhi J, Kim JH, Kim SY, Kwon WS, Yang DC. Molecular characterization of two glutathione peroxidase genes of Panax ginseng and their expression analysis against environmental stresses. Gene. 2014;535:33–41. doi: 10.1016/j.gene.2013.10.071. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Han H, Chen M, Yang W, Liu L, Li N, Ding X, Chu Z. Overexpression of OsDT11, which encodes a novel cysteine-rich peptide, enhances drought tolerance and increases ABA concentration in rice. Plant Mol Biol. 2017;93:21–34. doi: 10.1007/s11103-016-0544-x. [DOI] [PubMed] [Google Scholar]
- Lima-Melo Y, Carvalho FE, Martins MO, Passaia G, Sousa RH, Neto MC, Margis-Pinheiro M, Silveira JA. Mitochondrial GPX1 silencing triggers differential photosynthesis impairment in response to salinity in rice plants. J Integr Plant Biol. 2016;58:737–748. doi: 10.1111/jipb.12464. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Matamoros MA, Saiz A, Peñuelas M, Bustos-Sanmamed P, Mulet JM, Barja MV, Rouhier N, Moore M, James EK, Dietz K-J, Becana M. Function of glutathione peroxidases in legume root nodules. J Exp Bot. 2015;66:2979–2990. doi: 10.1093/jxb/erv066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Lv D, Wang P, Wang XC, Chen J, Miao C, Song CP. An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell. 2006;18:2749–2766. doi: 10.1105/tpc.106.044230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navrot N, Collin V, Gualberto J, Gelhaye E, Hirasawa M, Rey P, Knaff DB, Issakidis E, Jacquot JP, Rouhier N. Plant glutathione peroxidases are functional peroxiredoxins distributed in several subcellular compartments and regulated during biotic and abiotic stresses. Plant Physiol. 2006;142:1364–1379. doi: 10.1104/pp.106.089458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozyigit II, Filiz E, Vatansever R, Kurtoglu KY, Koc I, Öztürk MX, Anjum NA. Identification and comparative analysis of H2O2-scavenging enzymes (ascorbate peroxidase and glutathione peroxidase) in selected plants employing bioinformatics approaches. Front Plant Sci. 2016;7:301. doi: 10.3389/fpls.2016.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passaia G, Margispinheiro M. Glutathione peroxidases as redox sensor proteins in plant cells. Plant Sci. 2015;234:22–26. doi: 10.1016/j.plantsci.2015.01.017. [DOI] [PubMed] [Google Scholar]
- Passaia G, Fonini LS, Caverzan A, Jardim-Messeder D, Christoff AP, Gaeta ML, Je DAM, Margis R, Margis-Pinheiro M. The mitochondrial glutathione peroxidase GPX3 is essential for H2O2 homeostasis and root and shoot development in rice. Plant Sci. 2013;208:93–101. doi: 10.1016/j.plantsci.2013.03.017. [DOI] [PubMed] [Google Scholar]
- Passaia G, Caverzan A, Fonini LS, Carvalho FEL, Silveira JAG, Margis-Pinheiro M. Chloroplastic and mitochondrial GPX genes play a critical role in rice development. Biol Plant. 2014;58:375–378. doi: 10.1007/s10535-014-0394-9. [DOI] [Google Scholar]
- Ramos J, Matamoros MA, Naya L, James EK, Rouhier N, Sato S, Tabata S, Becana M. The glutathione peroxidase gene family of Lotus japonicus: characterization of genomic clones, expression analyses and immunolocalization in legumes. New Phytol. 2009;181:103–114. doi: 10.1111/j.1469-8137.2008.02629.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez Milla MA, Maurer A, Rodriguez Huete A, Gustafson JP. Glutathione peroxidase genes in Arabidopsis are ubiquitous and regulated by abiotic stresses through diverse signaling pathways. Plant J. 2003;36:602–615. doi: 10.1046/j.1365-313X.2003.01901.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Toppo S, Vanin S, Bosello V, Tosatto SC. Evolutionary and structural insights into the multifaceted glutathione peroxidase (Gpx) superfamily. Antioxid Redox Signal. 2008;10:1501–1514. doi: 10.1089/ars.2008.2057. [DOI] [PubMed] [Google Scholar]
- Ursini F, Maiorino M, Brigelius-Flohe R, Aumann KD, Roveri A, Schomburg D, Flohe L. Diversity of glutathione peroxidases. Methods Enzymol. 1995;252:38–53. doi: 10.1016/0076-6879(95)52007-4. [DOI] [PubMed] [Google Scholar]
- Vishwakarma K, Upadhyay N, Kumar N, Yadav G, Singh J, Mishra RK, Kumar V, Verma R, Upadhyay RG, Pandey M, Sharma S. Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Front Plant Sci. 2017;8:161. doi: 10.3389/fpls.2017.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Zhao Z, Qian C, Sui Y, Malik AA, Chen J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem. 2010;399:257–261. doi: 10.1016/j.ab.2009.12.008. [DOI] [PubMed] [Google Scholar]
- Wang X, Fang G, Yang J, Li Y. A thioredoxin-dependent glutathione peroxidase (OsGPX5) is required for rice normal development and salt stress tolerance. Plant Mol Biol Rep. 2017;35:333–342. doi: 10.1007/s11105-017-1026-2. [DOI] [Google Scholar]
- You J, Chan Z. ROS regulation during abiotic stress responses in crop plants. Front Plant Sci. 2015;6:1092. doi: 10.3389/fpls.2015.01092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai CZ, Zhao L, Yin LJ, Chen M, Wang QY, Li LC, Xu ZS, Ma YZ. Two wheat glutathione peroxidase genes whose products are located in chloroplasts improve salt and H2O2 tolerances in Arabidopsis. PLoS ONE. 2013;8:e73989. doi: 10.1371/journal.pone.0073989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Hu L, Jiang L, Liu H, Liu S. Molecular cloning and characterization of an ASR gene from Cucumis sativus. Plant Cell Tissue Organ Cult. 2017;130:553–565. doi: 10.1007/s11240-017-1246-z. [DOI] [Google Scholar]
- Zhou Y, Hu L, Wu H, Jiang L, Liu S. Genome-wide identification and transcriptional expression analysis of cucumber superoxide dismutase (SOD) family in response to various abiotic stresses. Int J Genom. 2017;2017:7243973. doi: 10.1155/2017/7243973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liu S, Yang Z, Yang Y, Jiang L, Hu L. CsCAT3, a catalase gene from Cucumis sativus, confers resistance to a variety of stresses to Escherichia coli. Biotechnol Biotechnol Equip. 2017;31:886–896. doi: 10.1080/13102818.2017.1360797. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.