Abstract
Saccharum spontaneum is a major Saccharum species that contributed to the origin of modern sugarcane cultivars, and due to a high degree of polyploidy is considered to be a plant species with one of the most complex genetics. Fluorescence in situ hybridization (FISH) is a powerful and widely used tool in genome studies. Here, we demonstrated that FISH based on bacterial artificial chromosome (BAC) clones can be used as a specific cytological marker to identify S. spontaneum individual chromosomes and study the relationship between S. spontaneum and other related species. We screened low-copy BACs as probes from the sequences of a high coverage of S. spontaneum BAC library based on BLAST search of the sorghum genome. In total, we isolated 49 positive BAC clones, and identified 27 BAC clones that can give specific signals on the S. spontaneum chromosomes. Of the 27 BAC probes, 18 were confirmed to be able to discriminate the eight basic chromosomes of S. spontaneum. Moreover, BAC-24, BAC-66, BAC-78, BAC-69, BAC-71, BAC-73, and BAC-77 probes were used to construct physical maps of chromosome 1 and chromosome 2 of S. spontaneum, which indicated synteny in Sb01 between S. spontaneum and sorghum. Furthermore, we found that BAC-14 and BAC-19 probes, corresponding to the sorghum chromosomes 2 and 8, respectively, localized to different arms of the same S. spontaneum chromosome, suggesting that there was an inter-chromosomal rearrangement event between S. spontaneum and sorghum. Our study provides the first set of chromosome-specific cytogenetic markers in Saccharum and is critical for future advances in cytogenetics and genome sequencing studies in Saccharum.
Keywords: Saccharum spontaneum, sorghum, polyploidy, bacterial artificial chromosome (BAC), fluorescence in situ hybridization (FISH)
Introduction
Sugarcane (Saccharum spp., Poaceae) is an annual or perennial crop grown in tropical and subtropical regions worldwide. Sugarcane is an important crop for sucrose production, and accounts for 70% of the world sugar production. In addition, as a C4 plant, sugarcane can efficiently convert solar energy into chemical energy, and is therefore an ideal biofuel crop for ethanol production (Lam et al., 2009; Santchurn et al., 2014). The genus Saccharum has six species: S. spontaneum, S. robustum, S. officinarum, S. barberi, S. sinense, and S. edule, among which, S. spontaneum is considered the wild species with a basic chromosome set of x = 8. S. spontaneum is also one of the two main Saccharum species contributing to the modern sugarcane cultivars development (Dhont et al., 1998; Ha et al., 1999). S. spontaneum has a strong environmental adaptability and contains important genetic traits for disease and drought resistance, thus contributing to the stress tolerance of modern cultivar hybrids (Grivet et al., 2004). S. spontaneum has the widest geographic distribution, and its ploidy levels range from 2n = 5x = 40 to 2n = 16x = 128 (Panje and Babu, 1960).
Sugarcane chromosomes have similar morphologies and have a small size of 1–6 μm at the condensed metaphase stage (Ha et al., 1999; Dhont, 2005); hence, it is very challenging to discern between the different chromosomes based on traditional cytogenetic methods. Moreover, as an autopolyploid, its chromosomal structural alterations caused by polyploidization, duplication, deletion, and recombination are very common (Piperidis et al., 2010). In S. spontaneum, genetic linkage maps have been developed (Silva et al., 1995), but development of a cytological genetic map lagged behind other grass species. Reliable cytological tools to identify individual chromosomes can be used for effective genome research and germplasm resource utilization, specifically with the complex genome background of modern sugarcane cultivars. Chromosome-specific bacterial artificial chromosome (BAC) clones are invaluable resource for sugarcane genome researches, and will have many applications in physical mapping, chromosome identification, and marker-assisted breeding of the Saccharum spp., and in assisting sugarcane genome sequencing and assembly projects.
Fluorescence in situ hybridization (FISH) is a powerful tool for molecular cytology (Jiang and Gill, 2006). In Saccharum spp., 45S rDNA and 5S rDNA were used as probes to detect the basic chromosomes of S. spontaneum, S. robustum, and S. officinarum, to identify the different basic chromosomes, x = 8 in S. spontaneum, and x = 10 in S. robustum and S. officinarum (Dhont et al., 1998; Ha et al., 1999). Based on FISH, modern sugarcane cultivars were found to contain 70–80% of the chromosomes derived from S. officinarum, and 10–23% of chromosomes from S. spontaneum, while 5–17% appears to be the product of the recombination between S. spontaneum and S. officinarum (Dhont et al., 1996; Piperidis et al., 2001; Cuadrado et al., 2004). Moreover, chromosome elimination, recombination, and translocation events were detected in some progenitors derived from the hybridization between Saccharum and Erianthus arundinaceus using FISH (Dhont et al., 1995; Huang et al., 2015).
Sorghum has a small diploid genome (730 Mbp) with a low frequency of chromosome recombination events. Sorghum shares high synteny with the sugarcane genome, making it an ideal reference plant for comparative analyses with the sugarcane genome (Ming et al., 1998; Nazeema et al., 2007; Wang et al., 2010; Aitken et al., 2014). In this study, based on the available sequences for high coverage BAC resources, reliable BAC probes were developed for FISH chromosome identification and cytogenetic map construction. The objectives of this study were to: (1) develop a set of chromosome-specific BAC-FISH probes on S. spontaneum chromosome and (2) explore the chromosome rearrangement between S. spontaneum and sorghum.
Materials and Methods
Materials
Saccharum spontaneum SES208 (2n = 8x = 64) was used for cytological analyses in this study. The SES208 plants were grown in the field on the campus of Fujian Agricultural and Forestry University (Fuzhou, China) in February of 2015 and maintained under regular sugarcane growth conditions.
A BAC library was constructed from the haploid genome of S. spontaneum AP85-441 (4x = 32), which was derived from S. spontaneum SES208 (2n = 8x = 64) via anther in vitro culture. The genome size of AP85-441 is about 3.2 Gbp. The library consisted of 38,400 clones, with an average insert size of 100 kb, covering 6× of the whole genome. This BAC library has been used for identifying the sucrose transporters and fructokinase gene families in S. spontaneum (Zhang et al., 2016; Chen et al., 2017).
Screening the BAC Library
35,156 BAC clones from the AP85-441 libraries were pooled into 701 libraries. Each library contains an average of 50 BAC clones. The DNA libraries were prepared with the PhasePrepTM BAC DNA Kit (Sigma, United States) following the manufacturer’s protocols. BAC DNA libraries were sequenced using Illumina HiSeq 2500 platform with PE250 model. A total of 686 libraries (18 libraries failed for sequencing) were sequenced, and 267.5 Gb of cleaned data were generated after trimming using Trimmomatic version 0.36 (Bankevich et al., 2012). Data from each BAC pool were assembled using SPAdes version 3.09 with default parameter. A total of 2,611,145 contigs were assembled with contig N50 of 7.38 kbp (Zhang and Ming, unpublished data).
The AP85-441 BAC library sequences were BLAST searched against the sorghum genome with an E-value of 1e-4. The sequences that had single BLAST hits in the sorghum genome were selected as candidate FISH probes. To identify low copy number BAC clones from the BAC library, sequence-specific primers were designed using Primer 5.0 software to enable BAC pools to be screened by PCR specifically. Primer lengths of 18–25 bp, Tm values of 55–65°C, and nucleotide compositions of 40–60% cytosine and guanine were selected. A two-step PCR method was used to screen the 3D dimension pools of the BAC library (Asakawa et al., 1997; Crooijmans et al., 2000) with some modifications. Each clone from a 384-well plate was cultured in 80 μl Lysogeny broth + 34 mg/ml chloramphenicol overnight at 37°C in a 384-well culture plate. In total, eight 384-well plates with a total of 3072 BAC clones were cultured. For each 384-well plate, we first constructed 16 row pools (row A to P) and 24 column pools (column 1–24) for each 384-well plate by mixing equal volumes of culture of each individual clone, and then mixed the 16 row pools and 24 column pool together with equal volumes from each row and column pool as one plate pool or superpool. In this study, we focused on screening eight 384-well plates. Thus, eight superpools were prepared for the eight plates separately. The positive 384-well plate from the eight plates was identified and subsequently the positive row among the 16 mixtures was screened. Finally, the PCR positive columns were selected among 24 column mixtures. Twenty-four PCR reactions were performed to identify positive clones from the eight 384-well plates.
PCR amplifications were performed as previously described (Bouzidi et al., 2006). Each reaction included rTaq premix 7.5 μl [10× PCR buffer, MgCl2 (25 mM), dNTPs (2.5 mM), rTaq polymerase (3 U/1.5 μl)], 0.6 μl of each primer (10 μM), 1.0 μl bacteria liquid template, to a final volume of 20 μl. Following initial denaturation at 95°C for 5 min, 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min were performed. PCR products were analyzed on a 1.5% agarose gel.
Metaphase Chromosome Preparation
Chromosome preparation was performed as previously described (Lou et al., 2010) with minor modifications. In brief, S. spontaneum plants were grown in the greenhouse conditions for root tips harvesting. Excised root tips about 1–2 cm were treated with 0.002 mol/l 8-hydroxyquinoline at room temperature for 2–4 h, rinsed in water for 15 min, and fixed in ethanol:acetic acid (3:1) for at least 24 h at room temperature. The root tips were then digested in an enzyme solution (4% cellulose R-10 and 2% pectolyase in 0.1 M citrate buffer) at 37°C for 1 h, washed with ice deionized water for 30 min, and finally incubated in ethanol:acetic acid (3:1) for about 30 min. Slides were prepared according to using the “flame-dried” method (Iovene et al., 2008).
BAC DNA Purification and Probe Labeling
The BAC DNA was extracted with PhasePrepTM BAC DNA Kit according to the manufacturer’s manual. Purified BAC DNA was labeled by standard nick translation reaction, including diluted DNase I, 10× nick translation buffer, DNA Polymerase I, dNTPs, biotin-/digoxigenin-labeled dUTPs, and BAC DNA. The mixture was incubated at 15°C for 1.5 h. The cut products were then examined on a 1.5% agarose gel for the presence of a smear between 300 and 500 bp. The obtained probes were stored at -20°C until used.
In Situ Hybridization and Detection
Probe mix and hybridization: First, the probe mixture (50% deionized formamide, 2× SSC, 80 ng digoxygenin-/biotin-labeled DNA, 10% dextran sulfate, >1 μg C°t-100) was placed in a 90°C hot block for 5 min, then immediately transferred on ice until ready to probe for hybridization. The previously prepared flamed-dried slides were treated with 70% deionized formamide, denatured on a heat block at 80°C for 1.5 min, immediately immersed sequentially in ice cold 70% ethanol, 90% ethanol, and then 100% ethanol each for 5 min, and then air dried on the bench. Finally, denatured probes were added to each slide, and covered with 24 × 32 mm coverslips. Slides were placed in a moist chamber at 37°C overnight.
Probe detection: the coverslips were removed and the slides were washed at room temperature in 2× SSC for 5 min, at 42°C in 2× SSC for 10 min and at room temperature in 1× PBS for 5 min in this sequence. Biotin-labeled probe signals were detected with 2 mg/ml Alexa Fluor 488 streptavidin and digoxygenin-labeled probe signals were detected with 2% anti-digoxigenin-rhodamine from sheep. The antibody cocktail (100 μl TNB buffer, 1 μl Alexa Fluor 488 streptavidin, 1 μl rhodamine anti-dig-sheep) was added to the slides, which were covered with 24 × 32 mm coverslips, incubated for 1 h at 37°C in a moist chamber, and washed at room temperature in 1× PBS three times for 5 min each. Excess liquid was removed and 4′,6-diamidino-2-phenylindole (DAPI) (Sigma, St. Louis, MO, United States) in the antifade solution Vectashield (Vector, Burlingame, CA, United States) was added to counterstain the chromosomes. Images were captured with an Olympus BX63 epifluorescence microscope. FISH signal images were analyzed using the CellSens Dimension software.
Results
Screening Low-Copy BAC Clones
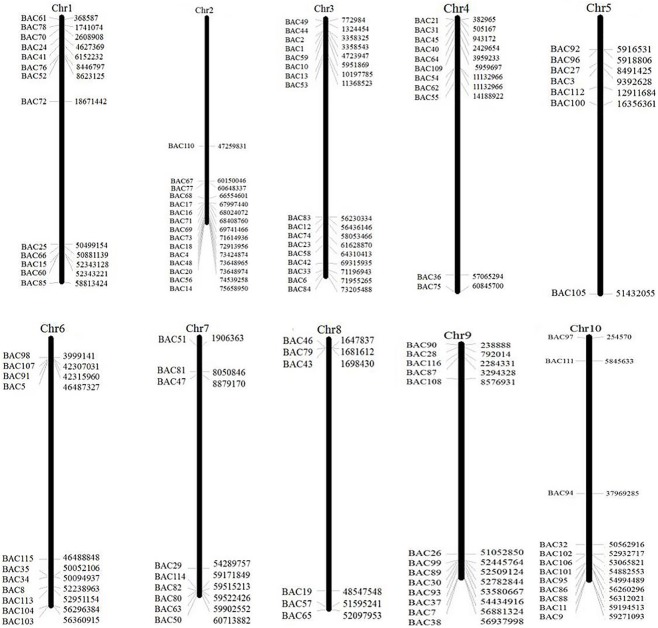
To screen the BAC clones that potentially have low copy number sequences in the S. spontaneum genome, we BLAST searched the sequenced AP85-441 BAC libraries against the sorghum genome. A total of 2000 BAC sequences corresponding to 10 sorghum chromosomes with good collinearity and showing sequence specificity were screened. The 2000 AP85-441 BAC sequences were masked by REPEATMASKER against the high-repeat sorghum DNA database and TIGR gramineae database to filter repeat sequences. Finally, 114 BACs distributed on 10 sorghum chromosomes were selected for FISH analysis (Supplementary Table 1) with 7–16 BACs on each of the 10 sorghum chromosome (Figure 1). To screen positive BAC clones, 114 specific primers were designed based on the BAC clone sequences (Supplementary Table 2) and 49 positive BAC clones were screened from the BAC libraries (Supplementary Table 3).
FIGURE 1.
The distributions of 114 BAC clones from AP85-441 in the chromosomes of sorghum. BAC1-114 represents the BAC number, the data on the right are the physical location of BAC in corresponding, region of the sorghum chromosomes. Unit = bp.
BAC-FISH Signal Strength and Distribution
DNA samples isolated from the 49 positive BAC clones were labeled with biotin or digoxigenin. 5S rDNA and 45S rDNA probes were used as control and were hybridized to S. spontaneum somatic metaphase chromosomes following the FISH procedure. Due to the correlation between signal strength and the variation of repeated sequence content of the BAC sequences, we used C°t-100 as a competitor (Table 1). BAC clones displaying strong and steady hybridization signals were selected for further FISH analysis.
Table 1.
The classification of 27 positive clones with blocking DNA.
| Groups | Blocking DNA | Signal numbers | BAC IDs |
|---|---|---|---|
| I | Not required | 7, 8 | 2, 18, 20, 29, 84 |
| II | 25× | 4, 6,7 | 11, 14, 19, 31, 33, 43, 71, 73,74 |
| III | 75× | 8 | 26, 66, 76, 78 |
| IV | 100× | >8 | 24 |
| V | 150× | 1∼6 | 4, 5, 32, 34, 35, 69, 77, 81 |
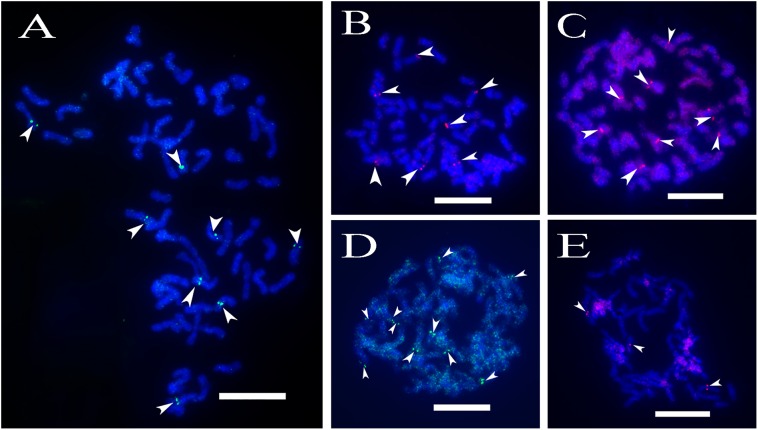
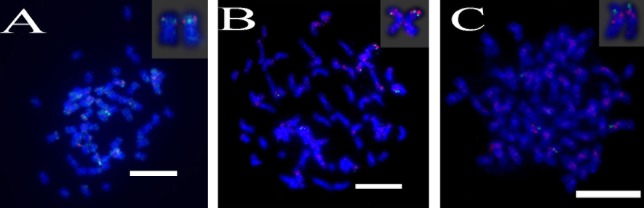
A total of 64 distinct chromosomes can be observed in the metaphase of the S. spontaneum. Each BAC-FISH was performed in four independent experiments (or slides). At least 10 spreads of somatic metaphase chromosomes were analyzed in each slide. The results showed that the 27 specific probes (Supplementary Table 4) can be classified into five groups based on the signal of hybridization (Table 1). In Group I (including BAC-18, BAC-20, BAC-29, and BAC-84), probes displayed eight distinctive sites without competitive in situ suppression (CISS) using C°t-100 (Figure 2A), suggesting few repeated sequences existed in these BAC sequences. In Group II (including BAC-2, BAC-11, BAC-14, BAC-19, BAC-31, BAC-33, BAC-43, BAC-71, BAC-73, and BAC-74), C°t-100 was used as competitor for BAC probe hybridization, and six to seven quite distinct sites were displayed in karyotypes of this group (Figure 2B). The hybridization result was similar to 45S rDNA with seven signal sites, which may be caused by chromosome structure variation in the homologous chromosome. In Group III (including BAC-26, BAC-66, BAC-76, and BAC-78), C°t-100 DNA was used as a competitor and eight distinct sites could be observed in the karyotype, suggesting this group of BAC clones have repetitive DNA sequences (Figure 2C). In Group IV (including BAC-24), more than eight signal sites were observed in the karyotype and appeared in the pericentromeric and telomeric regions (Figure 2D), suggesting these BAC probes had repetitive sequences in S. spontaneum. In group V (including BAC-4, BAC-5, BAC-32, BAC-34, BAC-35, BAC-69, BAC-77, and BAC-81), the probes showed dispersed signals in all chromosomes with limited regions of individual chromosomes displaying specific signals (Figure 2E). The hybridization result indicated that this group of BAC clones contained large amounts of repetitive DNA in S. spontaneum, which may be caused by a rapid evolutionary divergence for the repetitive regions after the split of sorghum and S. spontaneum. Therefore, of the 49 BAC probes, a set of 19 BAC probes (Supplementary Table 3) from Groups I, II, and III showed specific signals on S. spontaneum chromosomes corresponding to nine of the sorghum chromosomes beside Sb05. The signals of the other 22 probes were dispersed in all chromosomes. We selected a set of 19 BAC probes as S. spontaneum chromosome-specific signals (Figure 3). Of the 19 BAC probes, one probe was located on Sb02, Sb04, and Sb09; two probes on Sb07, Sb08, and Sb10; three probes on Sb01 and Sb06; and four probes on Sb03. This probe set provided a useful tool for comparative analysis of S. spontaneum and sorghum (Table 2).
FIGURE 2.
(A–E) The spread of the S. spontaneum chromosome hybridized with chromosome-specific BAC clones using BAC-FISH. (A–E) show hybridization of BAC probes (Table 2) to S. spontaneum chromosomes. (A) BAC-20, (B) BAC-43, (C) BAC-66, (D) BAC-24, and (E) BAC-81 probes labeled with biotin (green) and digoxigenin (red). The arrowheads indicate the signals. Scale bars = 10 μm.
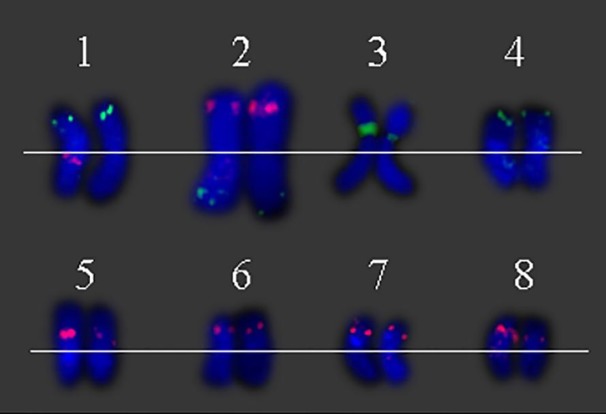
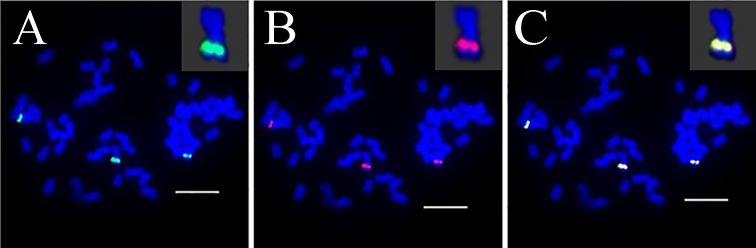
FIGURE 3.
Eight individual S. spontaneum chromosomes with chromosome-specific BAC clones. (1) BAC-24 (green) and BAC-66 (red), (2) BAC-14 (green) and BAC-19 (red), (3) BAC-2 (green), (4) BAC-31 (green), (5) BAC-35 (red), (6) 29 (red), (7) BAC-26 (red), and (8) BAC-32 (red) probes labeled with biotin and digoxigenin.
Table 2.
The distribution of BAC-FISH probes in chromosomes of S. bicolor and S. spontaneum.
|
S. bicolor |
S. spontaneum |
||
|---|---|---|---|
| Chromosome ID | BAC IDs | Chromosome ID | BAC IDs |
| 1 | 24, 66, 78 | 1 | 24∗, 66, 78 |
| 2 | 14 | 2 | 14∗, 19(Sb8) |
| 3 | 2, 33, 74, 84 | 3 | 2∗, 33, 74, 84 |
| 4 | 31 | 4 | 31∗ |
| 5 | N/A | N/A | N/A |
| 6 | 5, 34, 35 | 5 | 35∗, 5, 35 |
| 7 | 29, 81 | 6 | 29∗, 81 |
| 8 | 19, 43 | N/A | N/A |
| 9 | 26 | 7 | 26∗ |
| 10 | 11, 32 | 8 | 32∗, 11 |
∗ The BAC-FISH results were presented in Figure 2.
Construction of a BAC Library Specific for S. spontaneum Chromosomes
Sb01 and Sb02 are the two largest sorghum chromosomes. To detect the genome synteny between sorghum and S. spontaneum, 10 chromosome-specific BAC clones were selected from Groups I, II, and III. Of these 10 BAC probes, three probes (BAC clone ID: 24, 66, 78) were homologous to Sb01 and seven probes (BAC clone ID: 14, 18, 20, 69, 71, 73, 77) corresponded to Sb02. We utilized dual-color detection of FISH for complementarily labeled BAC pairs and the probes for mapping the chain cytogenetic relationship for the BAC clones. By doing so, we constructed the cytogenetic map of S. spontaneum based on the 10 BAC probes.
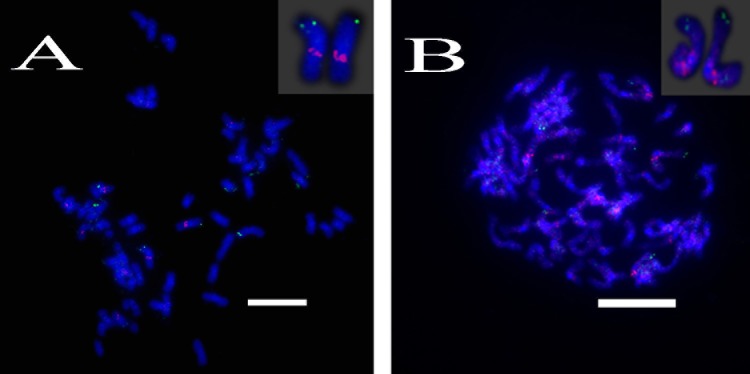
For three probes corresponding to Sb01, FISH analysis of S. spontaneum (SES208) showed that BAC-24 and BAC-78 probes were mainly located on the distal region of S. spontaneum chromosome 1, and BAC-66 was mapped in close proximity to the centromeric region (Figure 4). BAC-24 and BAC-66 located to the same chromosome (S. spontaneum chromosome 1); simultaneously whereas probe 66 and probe 78 also located on the same chromosome, demonstrating that the three BAC probes distributed to the same chromosomes in S. spontaneum thus suggesting synteny between S. spontaneum and sorghum for chromosome 1. FISH results revealed that the seven BAC probes aligned to Sb02; only probes 69, 71, 73, and 77 of the seven BACs generated intense signals on one S. spontaneum chromosome (SsChr2) (Figure 5); while probes 14, 18, and 20 were undetectable on S. spontaneum chromosome 2 but were observed on other S. spontaneum chromosomes. These results demonstrated the chromosome rearrangements occurred in the corresponding Sb02 between S. spontaneum and sorghum.
FIGURE 4.
FISH mapping of BAC-24, BAC-66, BAC-78 on SsChr1 in S. spontaneum. (A) BAC-24 and BAC-66 probes labeled with biotin (green) and digoxigenin (red); (B) BAC-78 and BAC-66 probes labeled with biotin (green) and digoxigenin (red). Scale bars = 10 μm.
FIGURE 5.
FISH mapping of BAC-69, BAC-71, BAC-73, BAC-77 on SsChr2 in S. spontaneum. (A) BAC-71 and BAC-69 probes; (B) BAC-73 and BAC-69 probes; (C) BAC-71 and BAC-77 probes labeled with biotin (green) and digoxigenin (red). Scale bars = 10 μm.
Moreover, the sequence of BAC-2 shared high similarity (97%) to a fragment on chromosome 3 of sorghum. Double color FISH analysis showed that both BAC-2 and 45s rDNA were mapped to the same location of the S. spontaneum chromosome (Figure 6).
FIGURE 6.
FISH mapping of BAC-2, 45srDNA on SsChr3 in S. spontaneum. (A) BAC-2 labeled with biotin (green); (B) 45s rDNA probes labeled with digoxigenin (red). (C) Merged images of (A) and (B). Scale bars = 10 μm.
Chromosome Rearrangement on Sb02 between Sorghum and S. spontaneum
To further investigate the chromosome rearrangement on Sb02 between sorghum and S. spontaneum, chromosome-specific probes corresponding to Sb02 (BAC-14, BAC-18, and BAC-20) and corresponding to Sb08 (BAC-19 and BAC-43) were used for FISH analysis. The results showed that BAC-14 and BAC-19 were mapped to different arms of the same S. spontaneum chromosomes, indicating that chromosome rearrangements took place between chromosomes 2 and 8 in S. spontaneum and sorghum (Figure 7).
FIGURE 7.
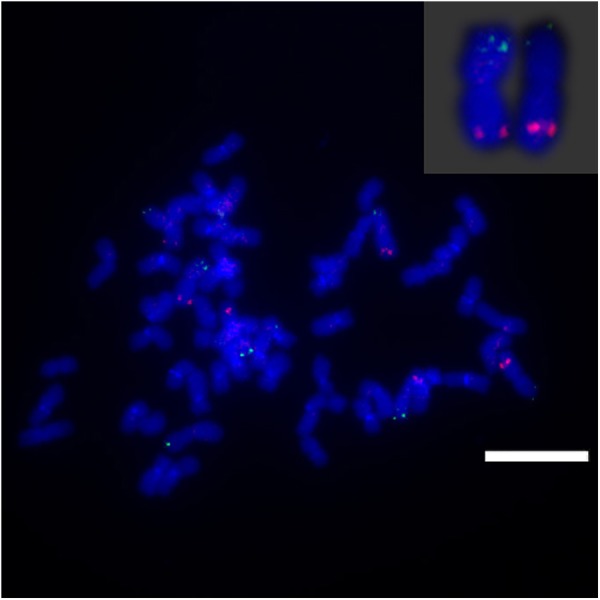
The rearrangement of chromosome in sugarcane and sorghum as revealed by FISH. BAC-14 and BAC-19 probes labeled with biotin (green) and digoxigenin (red). Scale bars = 10 μm.
Discussion
Saccharum is a complex genus characterized by high polyploidy levels, with small chromosomes (Ha et al., 1999; Dhont, 2005). Distinguishing individual Saccharum chromosomes based on their morphology is very challenging. Taking advantage of sequences that are highly covered by the BAC library of S. spontaneum, we were able to screen for the potential low copy number BAC clones in S. spontaneum using the sorghum genome as a reference. As a high polyploid species, S. spontaneum may contain more genome rearrangements than diploids. The differences of the genomes between S. spontaneum and sorghum may be the cause of inexactitude in the predictions of the low copy number BAC sequences in S. spontaneum genome. In this study, we have initially obtained 114 potential BAC sequences with single-copy based on the sorghum genome. However, only 49 could be identified in the partial BAC libraries, and 27 were found to be chromosome-specific BAC-FISH probes. There are at least two reasons to explain these results. Firstly, the 114 BAC sequences were partial fragments of the BAC insertion due to the limitations of NGS sequence assembly, and may contain repeat sequences in the other regions that have no sequence information. These potentially uncovered repeat sequences could mislead the detection of low-copy sequence through sequence comparison. Secondly, the recent polyploidization in S. spontaneum may cause the variation of repeat sequences between S. spontaneum and sorghum, and thus result in the nonspecific FISH signal in S. spontaneum. In a previous study, sequencing 20 BACs in sugarcane hybrids generated 1.45 Mb contig sequences, the sequences aligning with sorghum genome spanned 0.99 Mb, which accounted for about half of the sugarcane BAC sequences (Wang et al., 2010). Obviously, deletions/insertions existed between sorghum and Saccharum, and the utilization of the sorghum genome as reference cannot replace the unique features of the Saccharum genome, and thus may cause unpredicted FISH results.
Some FISH slides had strong background noise, which may be due to small regions of repeat sequences (such as SSR sequences) in the BAC clone, which consequently produce interference during hybridization. The sorghum genome has a repeat content of approximately 61% (Paterson et al., 2009), whereas about half of the genome is composed of repeat sequences in Saccarrum hybrids (De et al., 2014). In these repeat-rich genomes, it is difficult to develop signal-specific FISH probes, which distinguish chromosomes cytogenetically with similar morphologies. CISS with blocking DNA C°t-100 can efficiently preclude repeat sequences. In this study, blocking DNA C°t-100 was used for FISH analysis of the BAC probes in Groups II and III (Figures 2D–F), which were verified to be the chromosome-specific BAC probes, while the FISH of BAC probes in Groups IV and V produced strong background interference (Figures 2D–F). It is obvious that there are variations of repeat sequences among the examined BAC probes. Therefore, optimization of experiments would be necessary for BAC-FISH analysis in Saccharum. Six to seven sites were displayed in Group II, which could be caused by the homologous chromosome structure variations, such as the fragment deletion in one or two homologous chromosomes. In hexaploid wheat, BAC 676D4 hybridized more strongly to the A-genome chromosomes than to the B- and D-genome chromosomes (Zhang et al., 2004). These undetectable of one to two signals in the homologous chromosomes also could be caused by the technical issues of BAC-FISH for such many chromosomes with small size.
In this study, we identified 27 chromosome-specific BAC-FISH probes that correspond to 9 of the 10 sorghum chromosomes (all except Sb05). In a study of the genetic map derived from a cross between S. officinarum and sugarcane cultivar, Sb05 was merged with Sb06 in sugarcane (Aitken et al., 2014). In our study, a genetic map based on the F1 population of S. spontaneum revealed that Sb05 was divided into two segments, and the two segments were merged with Sb06 and Sb07, respectively (Zhang and Ming, unpublished data). Recently, the genetic map of a member of the Andropogoneae, Miscanthus sinensis, demonstrated that Sb05 has poor collinearity with the corresponding linkage group in M. sinensis (Ma et al., 2012). Similarly, the two ancestral maize chromosomes orthologous to Sb05 retain the smallest number of syntenic orthologs to sorghum genes (Schubert and Lysak, 2011). Therefore, the absence of the BAC-FISH corresponding to Sb05 may indicate chromosome fusion in S. spontaneum.
Sorghum and S. spontaneum diverged 12 million years ago (MYA), the basic chromosome number was reduced from x = 10 to x = 8. In Aitken et al. (2014), the HG2 (homologous group 2) of sugarcane aligned to Sb05 and Sb06, and HG8 (homologous group 8) to Sb02 and Sb08, providing evidence for the basic chromosome reduction event (Aitken et al., 2014). In this study, we observed that the sorghum chromosomes Sb02 and Sb08 had interchromosomal rearrangement in S. spontaneum as demonstrated by the evidence that BAC-14 aligned to Sb02, and BAC-19 aligned to Sb08 (Figure 7). Our study provided the first physical and cytogenetic evidence for the sorghum inter-chromosome rearrangement in S. spontaneum. Unpublished data from our group also revealed that Sb08 is divided into two segments, and were merged with of Sb02 and Sb09 in S. spontaneum, respectively (Zhang and Ming, unpublished data). The probes corresponding to Sb09, Sb08, Sb05, and Sb06 could be further used for investigating the inter-chromosomal events between sorghum and S. spontaneum. S. spontaneum has a wide range of ploidy levels (2n = 40–128) (Irvine, 1999). These BAC probes could be used to confirm the inter-chromosomal rearrangement of S. spontaneum with the different polyploidy levels.
Due to different basic chromosome number between sorghum (x = 10) and S. spontaneum (x = 8), the sorghum chromosomes were not a one-to-one correspondence with S. spontaneum. As our genetic mapping study mentioned herein before, Sb08 was divided into two segments which merged with segments of Sb02 and segments of Sb09 in S. spontaneum; Sb05 was divided into two segments which merged with segments of Sb06 and segments of Sb07 S. spontaneum. Therefore, the probes corresponding to Sb08 and Sb05 were not specific to single S. spontaneum chromosomes, whereas the other probes corresponding to the other eight sorghum chromosomes can be used as chromosome-specific cytogenetic BAC-FISH probes for S. spontaneum. Thus, the BAC probes tested were based on the eight sorghum chromosomes (Table 2) were sufficient for chromosome identification using BAC-FISH. A satellite chromosome was found in one S. spontaneum chromosome (chromosome 3) (Ha et al., 1999). Previously, simultaneous FISH revealed that the signals of 45S rDNA were located on the secondary constrictions of the satellite chromosomes within the chromosome 3 from the anther culture-derived S. spontaneum clone (AP85-361) (Ha et al., 1999). In this study, BAC-2 which corresponds to Sb03 and 45s rDNA were mapped to the same location of the S. spontaneum chromosome, thus further supporting previous findings (Ha et al., 1999). Our results also provided direct evidence that chromosome 3 of S. spontaneum named in the previous study is homologous to Sb03.
Conclusion
In this study, we developed chromosome-specific BACs of S. spontaneum as a step toward the development of a simple and reproducible method for chromosome identification using BAC-FISH cytogenetic markers, confirming the feasibility of isolating chromosome-specific BACs based on the sorghum genome to construct a physical map of sugarcane. We also provide the first cytogenetic evidence of inter-chromosomal rearrangement between sorghum and S. spontaneum. The establishment of the sugarcane BAC-FISH technology system offers new opportunities and the means for sugarcane molecular cytogenetics research, including karyotype analysis, gene localization, and physical map construction. These results are essential for assembly of S. spontaneum genome required for whole-genome sequencing.
Author Contributions
GD, JS, and JZ conceived the study and designed the experiments. GD, JS, QZ, JW, QY, RM, KW, and JZ carried out the experiments and analyzed the data. GD and JZ wrote the manuscript. All authors read and approved the final paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Xintan Zhang for assembling the BAC sequences and Dr. Irene Lavagi for editing the English.
Footnotes
Funding. This project was supported by grants from the 863 program (2013AA102604), NSFC (31201260), Program for New Century Excellent Talents in Fujian Province University, and the funding from the Fujian Agriculture and Forestry University.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00218/full#supplementary-material
References
- Aitken K. S., McNeil M. D., Berkman P. J., Hermann S., Kilian A., Bundock P. C., et al. (2014). Comparative mapping in the Poaceae family reveals translocations in the complex polyploid genome of sugarcane. BMC Plant Biol. 14:190. 10.1186/s12870-014-0190-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa S., Abe I., Kudoh Y., Kishi N., Wang Y., Kubota R., et al. (1997). Human BAC library: construction and rapid screening. Gene 191 69–79. 10.1016/S0378-1119(97)00044-9 [DOI] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). Spades: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzidi M. F., Franchel J., Tao Q., Stormo K., Mraz A., Nicolas P., et al. (2006). A sunflower BAC library suitable for PCR screening and physical mapping of targeted genomic regions. Theor. Appl. Genet. 113 81–89. 10.1007/s00122-006-0274-6 [DOI] [PubMed] [Google Scholar]
- Chen Y., Zhang Q., Hu W., Zhang X., Wang L., Hua X., et al. (2017). Evolution and expression of the fructokinase gene family in Saccharum. BMC Genomics 18:197. 10.1186/s12864-017-3535-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooijmans R. P., Vrebalov J., Dijkhof R. J., van der Poel J. J., Groenen M. A. (2000). Two-dimensional screening of the Wageningen chicken BAC library. Mamm. Genome 11 360–363. 10.1007/s003350010068 [DOI] [PubMed] [Google Scholar]
- Cuadrado A., Acevedo R., de la Espina M. D. S., Jouve N., Torre C. D. L. (2004). Genome remodelling in three modern S. officinarum × S. spontaneum sugarcane cultivars. J. Exp. Bot. 55 847–854. 10.1093/jxb/erh093 [DOI] [PubMed] [Google Scholar]
- De S. N., Monteiro-Vitorello C. B., Metcalfe C. J., Cruz G. M., Del Bem L. E., Vicentini R., et al. (2014). Building the sugarcane genome for biotechnology and identifying evolutionary trends. BMC Genomics 15:540. 10.1186/1471-2164-15-540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhont A. (2005). Unraveling the genome structure of polyploids using FISH and GISH; examples of sugarcane and banana. Cytogenet. Genome Res. 109 27–33. 10.1159/000082378 [DOI] [PubMed] [Google Scholar]
- Dhont A., Grivet L., Feldmann P., Rao S., Berding N., Glaszmann J. (1996). Characterisation of the double genome structure of modern sugarcane cultivars (Saccharum spp.) by molecular cytogenetics. Mol. Genet. Genomics 250 405–413. 10.1007/BF02174028 [DOI] [PubMed] [Google Scholar]
- Dhont A., Ison D., Alix K., Roux C., Glaszmann J. (1998). Determination of basic chromosome numbers in the genus Saccharum by physical mapping of ribosomal RNA genes. Genome 41 221–225. 10.1139/g98-023 [DOI] [Google Scholar]
- Dhont A., Rao P. S., Feldmann P., Grivet L., Islamfaridi N., Taylor P. W. J., et al. (1995). Identification and characterisation of sugarcane intergeneric hybrids, Saccharum officinarum x Erianthus arundinaceus, with molecular markers and DNA in situ hybridisation. Theor. Appl. Genet. 91 320–326. 10.1007/BF00220894 [DOI] [PubMed] [Google Scholar]
- Grivet L., Daniels C., Glaszmann J. C., D’Hont A. (2004). A review of recent molecular genetics evidence for sugarcane evolution and domestication. Ethnobot. Res. Appl. 2 9–17. 10.17348/era.2.0.9-17 [DOI] [Google Scholar]
- Ha S., Moore P. H., Heinz D. J., Kato S., Ohmido N., Fukui K. (1999). Quantitative chromosome map of the polyploid Saccharum spontaneum by multicolor fluorescence in situ hybridization and imaging methods. Plant Mol. Biol. 39 1165–1173. 10.1023/A:1006133804170 [DOI] [PubMed] [Google Scholar]
- Huang Y., Wu J., Wang P., Lin Y., Fu C., Deng Z., et al. (2015). Characterization of chromosome inheritance of the intergeneric BC2 and BC3 progeny between Saccharum spp. and Erianthus arundinaceus. PLoS One 10:e0133722. 10.1371/journal.pone.0133722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovene M., Wielgus S. M., Simon P. W., Buell C. R., Jiang J. (2008). Chromatin structure and physical mapping of chromosome 6 of potato and comparative analyses with tomato. Genetics 180 1307–1317. 10.1534/genetics.108.093179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine J. E. (1999). Saccharum species as horticultural classes. Theor. Appl. Genet. 98 186–194. 10.1007/s001220051057 [DOI] [Google Scholar]
- Jiang J., Gill B. S. (2006). Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 49 1057–1068. 10.1139/g06-076 [DOI] [PubMed] [Google Scholar]
- Lam E., Shine J., Silva J. D., Lawton M., Bonos S. A., Calvino M., et al. (2009). Improving sugarcane for biofuel: engineering for an even better feedstock. Glob. Change Biol. Bioenergy 1 251–255. 10.1111/j.1757-1707.2009.01016.x [DOI] [Google Scholar]
- Lou Q., Iovene M., Spooner D. M., Buell C. R., Jiang J. (2010). Evolution of chromosome 6 of Solanum species revealed by comparative fluorescence in situ hybridization mapping. Chromosoma 119 435–442. 10.1007/s00412-010-0269-6 [DOI] [PubMed] [Google Scholar]
- Ma X. F., Jensen E., Alexandrov N., Troukhan M., Zhang L., Thomas-Jones S., et al. (2012). High resolution genetic mapping by genome sequencing reveals genome duplication and tetraploid genetic structure of the diploid Miscanthus sinensis. PLoS One 7:e33821. 10.1371/journal.pone.0033821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R., Liu S., Lin Y., Silva J. D., Wilson W. A., Braga D. G., et al. (1998). Detailed alignment of Saccharum and sorghum chromosomes: comparative organization of closely related diploid and polyploid genomes. Genetics 150 1663–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazeema J., Laurent G., Nathalie C., Olivier G., Christophe G. J., Paulo A., et al. (2007). Orthologous comparison in a gene-rich region among grasses reveals stability in the sugarcane polyploid genome. Plant J. 50 574–585. 10.1111/j.1365-313X.2007.03082.x [DOI] [PubMed] [Google Scholar]
- Panje R. R., Babu C. N. (1960). Studies in Saccharum spontaneum distribution and geographical association of chromosome numbers. Cytologia 25 152–172. 10.1508/cytologia.25.152 [DOI] [Google Scholar]
- Paterson A. H., Bowers J. E., Bruggmann R., Dubchak I., Grimwood J., Gundlach H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457 551–556. 10.1038/nature07723 [DOI] [PubMed] [Google Scholar]
- Piperidis G., D’Hont A., Hogarth D. M. (2001). “Chromosome composition analysis of various Saccharum interspecific hybrids by genomic in situ (hybridisation) (GISH),” in Proceedings of the International Society of Sugar Cane Technologists, XXIV Congress (Brisbane, QLD: ASSCT; ), 565–566. [Google Scholar]
- Piperidis G., Piperidis N., Dhont A. (2010). Molecular cytogenetic investigation of chromosome composition and transmission in sugarcane. Mol. Genet. Genomics 284 65–73. 10.1007/s00438-010-0546-3 [DOI] [PubMed] [Google Scholar]
- Santchurn D., Ramdoyal K., Badaloo M. G. H., Labuschagne M. T. (2014). From sugar industry to cane industry: evaluation and simultaneous selection of different types of high biomass canes. Biomass and Bioenergy 61 82–92. 10.1016/j.biombioe.2013.11.023 [DOI] [Google Scholar]
- Schubert I., Lysak M. A. (2011). Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet. 27 207–216. 10.1016/j.tig.2011.03.004 [DOI] [PubMed] [Google Scholar]
- Silva J. D., Honeycutt R. J., Burnquist W. L., Aljanabi S. M., Sorrells M. E., Tanksley S. D., et al. (1995). Saccharum spontaneum L. ‘SES 208’ genetic linkage map combining RFLP- and PCR-based markers. Mol. Breed. 1 165–179. 10.1007/BF01249701 [DOI] [Google Scholar]
- Wang J., Roe B. A., Macmil S. L., Yu Q., Murray J. E., Tang H., et al. (2010). Microcollinearity between autopolyploid sugarcane and diploid sorghum genomes. BMC Genomics 11:261. 10.1186/1471-2164-11-261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Li W., Friebe B., Gill B. S. (2004). Simultaneous painting of three genomes in hexaploid wheat by BAC-FISH. Genome 47 979–987. 10.1139/G04-042 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Hu W., Zhu F., Wang L., Yu Q., Ming R., et al. (2016). Structure, phylogeny, allelic haplotypes and expression of sucrose transporter gene families in Saccharum. BMC Genomics 17:88. 10.1186/s12864-016-2419-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.