Abstract
Mesenchymal stromal cells (MSCs) are a promising cell source to develop cell therapy for many diseases. Human platelet lysate (PLT) is increasingly used as an alternative to foetal calf serum (FCS) for clinical-scale MSC production. To date, the global surface protein expression of PLT-expended MSCs (MSC-PLT) is not known. To investigate this, paired MSC-PLT and MSC-FCS were analysed in parallel using high-throughput flow cytometry for the expression of 356 cell surface proteins. MSC-PLT showed differential surface protein expression compared to their MSC-FCS counterpart. Higher percentage of positive cells was observed in MSC-PLT for 48 surface proteins, of which 13 were significantly enriched on MSC-PLT. This finding was validated using multiparameter flow cytometry and further confirmed by quantitative staining intensity analysis. The enriched surface proteins are relevant to increased proliferation and migration capacity, as well as enhanced chondrogenic and osteogenic differentiation properties. In silico network analysis revealed that these enriched surface proteins are involved in three distinct networks that are associated with inflammatory responses, carbohydrate metabolism and cellular motility. This is the first study reporting differential cell surface protein expression between MSC-PLT and MSC-FSC. Further studies are required to uncover the impact of those enriched proteins on biological functions of MSC-PLT.
Introduction
Mesenchymal stem cells (MSCs) are multipotent progenitor cells bearing a wide variety of biological activities including mesodermal differentiation potential, stromal support and immunomodulation1. MSCs have shown promising therapeutic potential for a wide variety of medical conditions2,3. Due to their low frequency ex vivo expansion is a pre-requisite to achieve clinically relevant cell numbers. Conventional expansion of MSCs relies on the use of foetal calf serum (FCS) as a culture supplement, which provides essential nutrients and growth factors required to support cell growth. Concerns have been raised by regulatory authorities regarding the use of FCS-containing culture media to expand MSCs for clinical use as it is a potential source of unidentified zoonoses and prions that could prompt xenogeneic infections4–7. FCS has also been shown to have high lot-to-lot variability and to elicit immune responses in patients who have received repeated administrations of FCS-expanded MSCs6,7. To eliminate potential risks associated with the use of FCS, intensive research has been carried out to identify a suitable replacement of human origin. Previous findings have demonstrated the superiority of human platelet lysate (PLT) over human AB serum8, human plasma9 and human autologous serum10 for ex vivo expansion of MSCs, owing to its high content of growth factors, low cost and ease in large-scale production11–14. This present study focuses on validating the concept and potential benefit of replacing FCS with PLT in MSC production for clinical use rather than comparing PLT with other human blood products. PLT is a concentrated solution of thrombocyte-derived growth factors produced by mechanical disruption of activated platelets using freeze-thaw procedures15. Currently, industrial scale production of GMP grade PLT utilises the pooling of platelet units derived from a large number of donors, resulting in a product with low lot-to-lot variability, as well as high consistency and purity16. The use of PLT in MSC expansion has been shown to promote proliferation while maintaining multi-lineage differentiation potential and immunomodulatory properties11,12,15,17. In the past decade, MSCs expanded in PLT-containing media have been increasingly used in clinical trials18,19. Despite encouraging clinical outcome, the global surface protein expression of PLT-expanded MSC is yet to be uncovered.
Given the importance of cell surface proteins to a plethora of biological functions this study characterised the phenotypic profile of MSCs expanded in PLT-containing media (MSC-PLT), in comparison to conventional FCS-expanded MSCs (MSC-FCS), using high-throughput flow cytometry. This analysis platform is a reproducible discovery-orientated screening technology that allows for the identification of surface proteins using panels of hundreds of CD markers and enables the use of multiplexed assays for the analysis of several cell populations simultaneously20–22. It takes advantage of the general characteristics of conventional flow cytometry for cell surface protein expression analysis but incorporates a high-speed sample loading device that allows for fast analysis of a range of antibodies, usually arrayed in 96- or 384-well plates20–22. This tool can also be combined with fluorescent cell barcoding that allows for parallel analysis of distinct populations20–22. To date, no high-throughput flow cytometry analysis has been reported on MSC-PLT, although such analyses have been used to screen surface proteins of MSC-FCS and other cell types21,23–27. This study characterises the overall phenotypic ‘finger print’ of MSC-PLT to identify differentially expressed surface proteins compared to MSC-FCS. This study also provides a searchable database of cell surface proteins expressed by MSCs expanded under different culture conditions that can be used as a resource to appreciate the heterogeneity and functional diversity of the MSC lineage.
Results
Characterisation of MSC-FCS and MSC-PLT
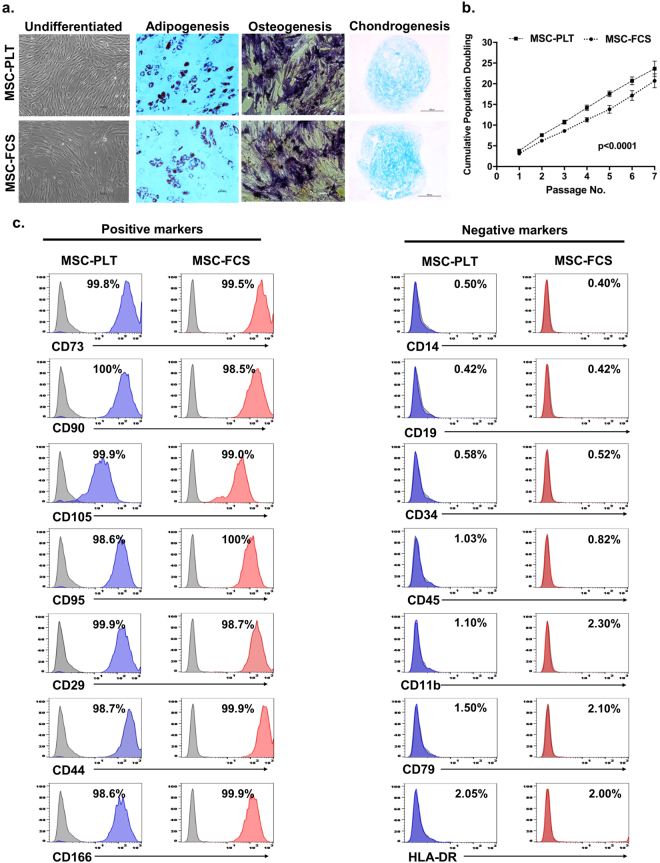
Human bone marrow derived MSCs from healthy donors were expanded in parallel in culture media containing either 5% PLT or 10% FCS. At passage 3, MSCs were characterised based on morphology, tri-lineage differentiation potential and phenotype as described by the ISCT criteria28. MSC-PLT and MSC-FCS exhibited comparable spindle-shaped morphology (Fig. 1a). The ability to differentiate into adipocytes, osteoblasts and chondroblasts was observed in both MSC populations as examined by Oil-Red-O, alkaline phosphatase/Von Kossa and Alcian blue staining, respectively (Fig. 1a). Analysis of cumulative population doublings of paired samples showed that MSC-PLT had higher proliferative capacity than MSC-FCS (p < 0.0001) (Fig. 1b). The paired MSC samples also exhibited the characteristic phenotype, positive for the expression of CD73, CD90, CD105, CD95, CD29, CD44 and CD166 whilst negative for the expression of CD14, CD19, CD34, CD45, CD11b, CD79 and HLA-DR (Fig. 1c).
Figure 1.
Characteristics of MSC-PLT and MSC-FCS. (a) Representative images of MSC-PLT and MSC-FCS showing their morphology and ability to differentiate into adipocytes, osteoblasts and chondroblasts; Scale bars represent 100 or 200 μm; (b) Cumulative population doublings of MSC-PLT and MSC-FCS presented as the mean ± SEM of 13 independent samples; (c) Representative histograms of MSC-PLT and MSC-FCS phenotype showing comparable expression profile of the positive and negative markers.
MSC-PLT show a differential surface protein expression pattern compared to MSC-FCS
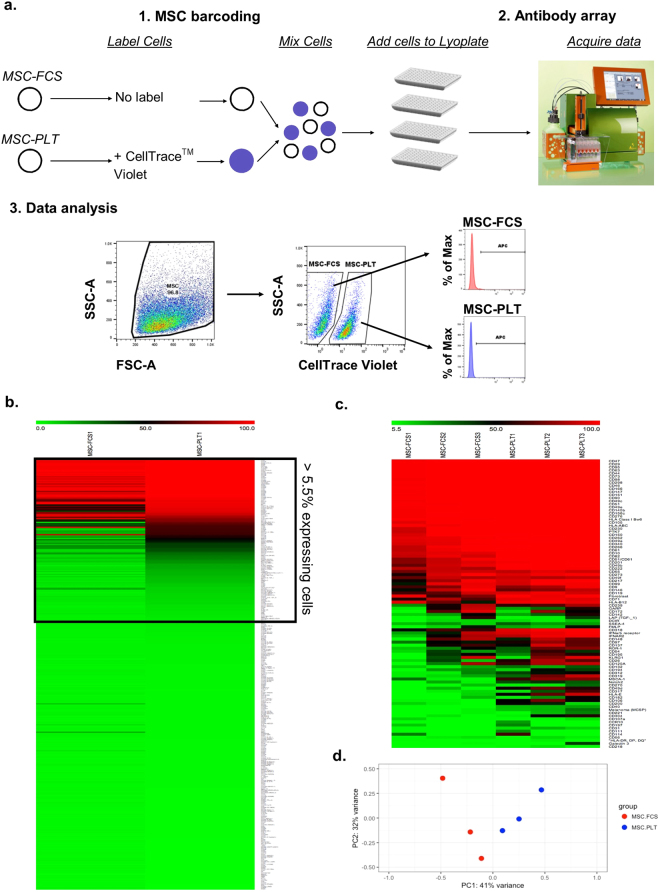
The surface protein expression of both MSC populations was analysed using the Lyoplate system consisting of a panel of monoclonal antibodies specific for 356 human surface proteins. A general overview of the experimental design is illustrated in Fig. 2a. Briefly, paired MSC samples at passage 3 were used for surface protein analysis. To discriminate between MSC-PLT and MSC-FCS, MSC-PLT populations were labelled with CellTraceTM Violet, while MSC-FCS populations were kept unlabelled. MSC-FCS and labelled MSC-PLT cells were then pooled and arrayed on 96 well plates containing APC-conjugated antibodies and isotype controls. APC positivity for each specific antibody in MSC-PLT and MSC-FCS was determined as illustrated in the gating strategy in Fig. 2a.
Figure 2.
MSC-PLT show a differential surface protein expression pattern compared to MSC-FCS. (a) Experimental overview of MSC surface protein screening using the Lyoplate technology. MSC-PLT and MSC-FCS were discriminated by labelling MSC-PLT with celltrace violet. Both samples were then mixed and seeded into wells of 96-well plates containing an array of APC-conjugated antibodies. Data was acquired using the automated MACSQuant flow cytometer and analysed by manual gating using MACSQuantify or FlowJo software. MSCs were selected based on SSC/FSC properties. MSC-FCS and MSC-PLT were gated as CelltraceTM violet negative and positive populations, respectively, and the percentage of positive cells for each antibody was determined based on gates drawn for each isotype control. (b) Heatmap showing the expression of all 356 markers on the surface of MSC-FCS and MSC-PLT. (c) Heatmap depicting the expression of the 99 markers detected as positive on the surface of MSC-FCS and MSC-PLT samples. (d) Principle component analysis plot illustrating the differences between MSC-PLT and MSC-FCS sample cohorts.
An initial surface protein screening was performed on one paired MSC-PLT and MSC-FCS samples to identify the positively expressed proteins, which were defined by showing positive staining in at least 5.5% of the cells in one or both cell populations. This threshold has been used as standard criteria in similar studies23–25. Overall, 99 out of 356 markers were positively expressed on both cell populations while the remaining markers were neither expressed on MSC-PLT nor MSC-FCS (Fig. 2b). Both MSC populations expressed the characteristic markers, positive for CD73, CD90, CD105, CD44 and CD29 and negative for CD45, CD34, CD3, CD19, CD14, CD11b and CD79, suggesting the reliability of this analysis platform. Detailed information on the 356 analysed proteins and their expression on MSC-PLT and MSC-FCS is presented in Supplementary Table S1.
The 99 surface proteins positively expressed on both MSC populations were further analysed in three paired MSC samples generated from three independent bone marrow donors. The results showed that 51 out of 99 positively expressed proteins were equally highly expressed on both MSC populations, including those that have been consistently detected in previous studies, such as, CD46, CD47, CD49a, CD9, CD81, CD63, CD47, among others23–25,29. The remaining 48 proteins displayed variable expression levels in MSC-PLT and MSC-FCS (Fig. 2c). Principal component analysis (PCA) was performed to illustrate the differences in global surface protein expression patterns between the two MSC populations. PCA analysis segregated MSC-PLT and MSC-FCS into two distinct clusters. In addition, a lower variability was depicted within the MSC-PLT samples, with all samples closely clustered together, whilst MSC-FCS samples were spread across the spectrum, suggesting that MSC-PLT could be less heterogeneous compared to their MSC-FCS counterpart (Fig. 2d). The percentage of cells expressing the positively detected surface proteins in MSC-PLT and MSC-FCS is presented in Supplementary Table S2, which provides a searchable database for future studies.
Thirteen surface proteins are significantly enriched on MSC-PLT
To identify the differentially expressed surface proteins we focused on the 48 proteins that had a variable expression in MSC-PLT and MSC-FCS. All 48 proteins showed a higher percentage of positive cells in MSC-PLT compared to MSC-FCS, of which 13 were significantly enriched on MSC-PLT with a fold increase in percentage of positive cells of ≥1.5 and a p-value < 0.05 (Fig. 3a,b). The unsupervised hierarchical clustering using percentage value of positive cells of these 13 proteins stratified MSC-PLT and MSC-FCS into two distinct clusters (Fig. 3c). No downregulation of surface proteins was detected in MSC-PLT compared to MSC-FCS in our study (Supplementary Table S2). None was reported in the literature either.
Figure 3.
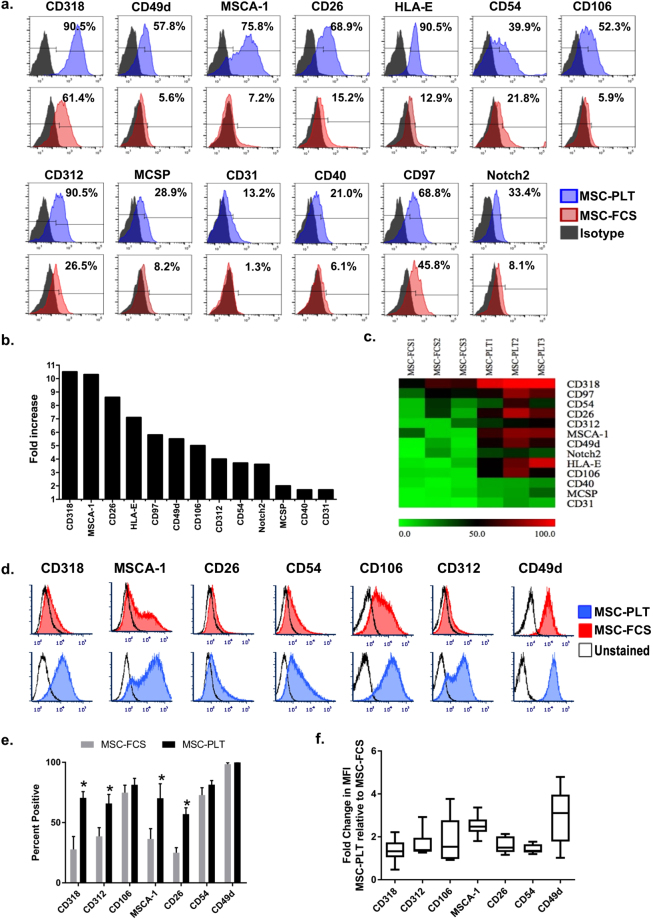
Surface proteins significantly enriched in MSC-PLT. From the analysis of the 48 surface proteins showing variable expression between MSC-PLT and MSC-FCS, 13 of which were significantly enriched on MSC-PLT. (a) Representative histograms of the 13 enriched surface proteins in MSC-PLT (blue) and MSC-FCS (red) as assessed by the Lyoplate analysis. (b) Average fold increase in percentage of positive cells in MSC-PLT (n = 3). (c) heatmap depicting the hierarchical clustering of the 13 markers significantly upregulated in MSC-PLT. (d) Representative histograms of enriched surface proteins in MSC-PLT compared to MSC-FCS in a validation cohort (n = 6), using multiparameter flow cytometry. Surface protein expression was evaluated by (e) the percentage of positive cells in the gated live cell single cell population; data represents mean ± SEM and *p < 0.05 and (f) the fold change of median fluorescence intensity of positively gated cells (MSC-PLT relative to MSC-FCS).
To validate the differentially expressed surface proteins detected using high-throughput screening, an independent validation cohort of paired MSC-PLT and MSC-FCS samples were collected (n = 6) and analysed for the expression of the surface proteins that were highly enriched on MSC-PLT, namely CD318, MSCA-1, CD26, CD54, CD106, CD312 and CD49d, using multiparameter flow cytometry. In comparison with MSC-FCS, higher percentage of positive cells was detected in MSC-PLT for all analysed surface proteins, with the difference in CD318, CD312, MSCA-1 and CD26 reached statistical significance (Fig. 3d,e). Further analysis of MFI for each surface protein confirmed the finding of Lyopate analysis, with MSC-PLT exhibiting higher expression of all analysed antigens across all paired MSC samples (Fig. 3f).
To explore the molecular mechanisms underpinning the differential surface protein expression between MSC-PLT and MSC-FCS, qRT-PCR was performed in an independent cohort of paired MSC samples (n = 3) to quantify the mRNA expression levels of CD318 (CDCP1), MSCA-1 (ALPL), and CD26 (DPP4), which are the top three most significantly enriched surface proteins expressed on MSC-PLT with the highest fold increase in the percentage of positive cells (Fig. 3b). The analysis revealed that MSCA-1 (ALPL) and CD26 (DPP4) were significantly overexpressed in MSC-PLT at mRNA levels (p < 0.05). MSC-PLT also exhibited a clear trend of elevated mRNA level for CD318 (CDCP) (Fig. S1).
Quantitative analysis of the enriched surface proteins on MSC-PLT
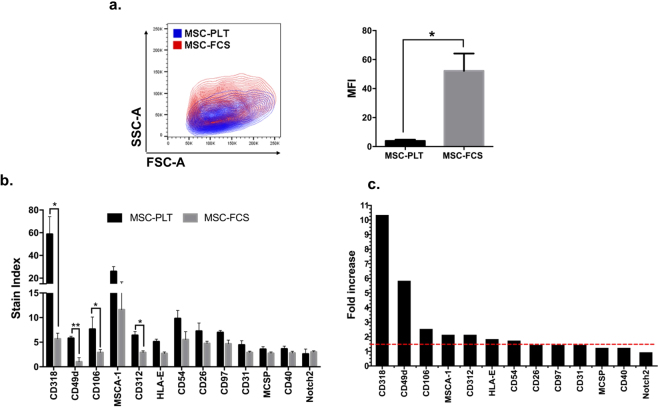
To quantify the expression levels of the enriched surface proteins identified in MSC-PLT, we analysed the differences in staining intensity, herein expressed as stain index (SI), between MSC-PLT and MSC-FCS. The SI is a measure that allows the comparison of the relative brightness of a fluorochrome. The SI takes into consideration of the background fluorescence which can be influenced by autofluorescence, nonspecific staining, electronic noise and optical background from other fluorochromes30. As illustrated in Fig. 4a, MSC-FCS displayed higher complexity than MSC-PLT, as evidenced by increased SSC-A properties. This complexity further influences the background fluorescence of MSC-FCS, which displayed a 13-fold higher background fluorescence intensity than MSC-PLT (p < 0.05). Despite the higher background intensity of MSC-FCS, higher SI was detected in MSC-PLT for 10 out of 13 proteins. The proteins CD318 and CD49d were the most enriched proteins in MSC-PLT exhibiting 10.4-fold and 5.9-fold higher SI than MSC-FCS respectively (p < 0.05 for both). Significantly higher expression of the surface protein CD106 was also detected in MSC-PLT, showing a 2.6-fold higher SI than MSC-FCS (p < 0.05). The surface proteins MSCA-1, CD312, HLA-E, CD54, CD26, CD97 and CD31 showed concomitant ≥1.5-fold higher SI in MSC-PLT (ranging from 2.2-fold to 1.5-fold). However, a statistical significance was not reached in the small sample cohort tested (n = 3). The remaining 3 proteins, MCSP, CD40, and Notch2 showed comparable SI levels between MSC-PLT and MSC-FCS (Fig. 4b,c). A summary of median fluorescence intensity and SI values for all positively expressed markers is presented in Supplementary Table S3, which provides a searchable database for future studies.
Figure 4.
Stain index of enriched surface proteins in MSC-PLT. (a) Representative contour plot of paired MSC samples with MSC-PLT displaying smaller size and lower complexity than MSC-FCS. MSC-FCS showed significantly higher background fluorescence, herein defines as median fluorescence intensity (MFI), in the APC channel, than MSC-PLT. (b) Stain index of 13 surface proteins enriched on MSC-PLT, in comparison with MSC-FCS (n = 3); error bars represent mean ± SEM and *p-value < 0.05. (c) Fold increase in stain index shown as MSC-PLT relative to MSC-FCS). Red line represents a fold increase of 1.5.
Pathway and Network enrichment analysis
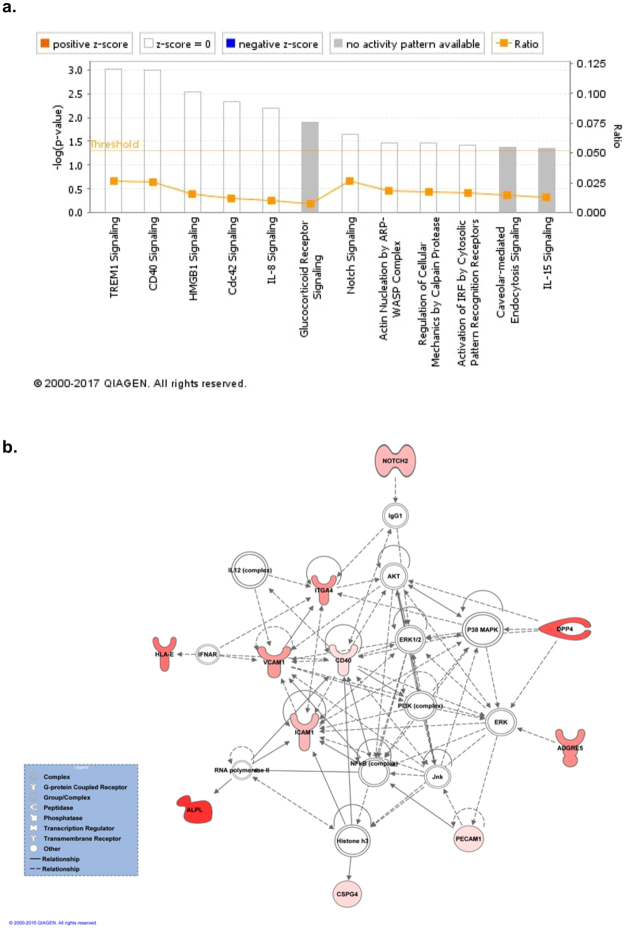
The 13 enriched proteins in MSC-PLT were analysed through the use of Ingenuity Pathway Analysis (IPA, QIAGEN INC., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis) to identify enriched pathways and associated networks based on a calculated probability score of ≥2 and a p-value of <0.05 using IPA software application. The top statistically significant (p < 0.05) canonical pathways influenced by these 13 proteins were shown in Fig. 5a. The pathways associated with “TREM1 signalling”, “CD40 signalling” and “HMGB1 signalling” were the most significantly enriched pathways in MSC-PLT (p < 0.001 for all). These pathways have been previously reported to be involved in the regulation of inflammatory responses and osteogenic differentiation of MSCs31–33.
Figure 5.
Canonical pathway and network analysis by IPA. The 13 enriched proteins in MSC-PLT were analysed through the use of Ingenuity Pathway Analysis (IPA, QIAGEN INC., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis). (a) Significantly altered canonical pathways associated with the enriched surface proteins in MSC-PLT. The canonical pathways included in this analysis are shown in the x-axis of the bar chart. The y-axis indicates the statistical significance calculated using the right-tailed Fisher exact test. (b) Graphical representation of the top ranked network, related to inflammatory responses, with a score of 28, as nodes (molecules) and edges (biological relationship between the different molecules). Red nodes represent the investigated enriched proteins involved in this network. Meanings of node shapes and edges are indicated in the legend within the figure.
From the data for canonical pathways and biological functions, IPA assessed networks which revealed the direct or indirect association of the 13 enriched markers in MSC-PLT with each other, and other molecules identified in the canonical pathways. The IPA software identified 3 different networks. The first and most relevant network is depicted in Fig. 5b. This network has a score of 28, included 11 of 13 the analysed markers and is associated with inflammatory responses. This network’s key components included p38 mitogen-activated protein kinases (MAPK), extracellular signal-regulated kinase (ERK) 1/2 and protein kinase B/phosphatidylinositol 3-kinase (AKT/PI3K) family of kinases. The scores and number of markers associated to the second and third networks are depicted in Table 1. The second network, scoring 9, is associated with the carbohydrate metabolism, and contains 4 of the 13 analysed proteins. This network has as key component the G-protein-coupled receptor (Gpcr) family, which mediates the energy metabolism and lead to the activation of important pathways such as the vascular endothelial growth factor (VEGF)34 and the insulin growth factor (IGF) pathway35. The third network, with a score of 4, is associated to the cellular mobility and migration and includes 2 of the 13 enriched markers. This network has as central component the GTPase family which regulate the actin cytoskeleton and the mobility of various cells36. Details of the components of each network are represented in Supplementary Table S4.
Table 1.
Summary of biological networks derived using IPA network analysis, with respective scores, number of analysed proteins and key components to each specific network.
| Networks | Score | Number analysed proteins in network | Key component (s) |
|---|---|---|---|
| Inflammatory response | 28 | 11 | ERK1/2; P38 MAPK; AKP/PI3K complex |
| Carbohydrate metabolism | 9 | 4 | Gpcr |
| Cellular movement | 4 | 2 | GTPase |
Discussion
Human PLT has been shown, as a xenogeneic-free supplement for MSC expansion, to promote the proliferation and genomic stability of MSCs while maintaining their multipotent properties. MSCs expanded in PLT-containing media have been increasingly used in clinical trials. Yet the global surface protein expression of MSC-PLT remains unreported. Here, we present novel data demonstrating the expression pattern of 356 surface proteins on MSC-PLT screened using discovery-orientated high-throughput flow cytometry. This screening platform has been successfully used for the identification of surface markers in several alternative cell types21,24,37,38.
Among 99 surface proteins that were positively expressed on both MSC-PLT and MSC-FCS, 48 were enriched on MSC-PLT compared to MSC-FCS. Further principal component analysis of the overall expression profiles revealed a lower variability within the MSC-PLT sample cohort compared to the MSC-FCS cohort. Previous reports have shown that PLT expanded MSCs exhibited smaller and narrower morphology than FCS-expanded MSCs17,39. This morphology was further associated to lower contamination with other resting mature cell types and higher uniformity of the cell surface protein expression, suggesting that PLT expanded MSCs consist of a more homogeneous population than their FCS-expanded counterparts40. Consistent with the previous findings, the enriched expression of the 48 variably expressed proteins on MSC-PLT detected in our study suggests an increased homogeneity of PLT expanded MSCs.
Beyond serving as markers for cell characterisation, surface proteins are critical mediators of important biological functions including cell-to-cell contact, extracellular matrix interactions, cell migration and signal transduction. The main objective of our study was to address whether there are differences in the surface protein expression profiles between MSC-PLT and MSC-FCS. We identified 13 surface proteins (CD40, HLA-E, MSCA-1, CD97, CD106, CD54, Notch2, CD318, CD26, CD312, MCSP, CD31 and CD49d) that were significantly enriched on MSC-PLT. Several of the overexpressed markers have been shown to be heterogeneously expressed by FCS-expanded MSCs41–46. In comparison to FCS, human PLT contains high concentration of growth factors including fibroblast growth factor (FGF), epidermal growth factor (EGF), transforming growth factor (TGF-β1), platelet derived growth factor (PDGF), insulin growth factor-1 (IGF-1), and vascular endothelial growth factor (VEGF)12. Collectively, these growth factors are involved in repair mechanisms by the activation of signalling pathways involved in cell proliferation, recruitment and motility thus promoting wound healing and neovascularisation47–49, therefore, they may have a direct effect on the increased expression of the 13 identified surface proteins.
In our study, the proteins CD318, CD26 and MSCA-1 showed the highest enrichment in MSC-PLT, which may have contributed to the enhanced proliferative capacity of PLT expanded cells. CD318, also known as CUB-domain-containing protein 1 is a type I transmembrane protein involved in cell adhesion and extracellular matrix interaction43. CD318 is expressed mainly by cultured MSCs50 and confers higher proliferative capacity to adherent progenitors43. CD26 is a surface glycoprotein with peptidase activity that has been detected on the surface of various cells51. It has been demonstrated that the expression of CD26 on MSCs was associated with higher CFU-F properties52. MSCA-1, also known as alkaline phosphatase liver/bone/kidney, has been associated with a subpopulation of MSCs with enhanced chondrogenic and osteogenic differentiation properties41,50,53.
Other enriched proteins, including CD97, CD312, MCSP and Notch2, have been reported to be expressed on the surface of MSCs but their relevance is still being investigated54–56. These proteins may have an important role in MSC proliferation and adhesion. CD97 and CD312, two closely related members of the EGF-TM7 transmembrane receptors, are important mediators of stemness and invasiveness in several carcinomas57. Similarly, MCSP, a transmembrane receptor with high affinity to a range of integrins and growth factors, including, bFGF and PDGF-AA, has been shown to promote cellular migration and metastasis58, while Notch2 signalling regulates cell proliferation56. Some of the 13 proteins enriched on MSC-PLT belong to the family of cell adhesion molecules, including CD49d, CD31, CD54 and CD106. The expression of CD49d has been detected in MSCs in a previous study59; however, the biological relevance of CD49d in MSCs has not yet been elucidated. CD31 is commonly regarded as a negative marker in MSCs. In our study, a small percentage of MSC-PLT (13.8% ± 2.3 SEM) had positive CD31 expression whilst MSC-FCS showed negativity for this marker (2.0% ± 0.6 SEM). The expression of CD31 has been shown to be up-regulated by PDGF, FGF and other growth factors60,61, therefore its enrichment on MSC-PLT may be a result of the high concentration of these growth factors in the PLT. Expression of the adhesion molecules CD54 and CD106 has been reported to identify a subpopulation of MSCs with enhanced immunosuppressive properties42,62.
Lack of expression in co-stimulatory molecules CD80, CD86 and CD40 is generally considered as a feature of MSCs that is associated to their low immunogenic properties63. In our study, there was a small proportion of MSC-PLT that expressed very low levels of CD40 (18.3% ± 1.4 SEM) while MSC-FCS were detected negative (4.9% ± 2.0 SEM) for this antigen. This marker binds to its ligand CD40L and contributes to the pro-inflammatory signalling cascade32. This up-regulation of pro-inflammatory markers may indicate an impaired immunogenic potential of MSC-PLT in comparison to their MSC-FCS counterparts. However, further investigation is necessary to evaluate whether this enrichment is biologically relevant in MSC-PLT immunogenicity. MSC-PLT also showed enhanced expression of HLA-E. This protein is a non-classical major histocompatibility complex (MHC) which binds to CD94/NKG2A receptor in NK cells and activates a signalling cascade which inhibits NK cell cytotoxicity64. Increased expression of HLA-E by MSC-PLT indicates these cells may be less susceptible to NK cell mediated rejection. In fact, one previous report illustrated that PLT-expanded MSCs show increased resistance to NK cell mediated lysis29.
In silico network analysis with the 13 upregulated markers, showed a significant enrichment of inflammatory responses, carbohydrate metabolism and cell movement pathways. The most significant network, related to inflammatory responses, includes the protein complexes, ERK, PI3K and p38 MAPK65,66, which are essential signalling cascades involved in cellular growth and differentiation. This pathway is an important regulator of both pro- and anti-inflammatory responses67. Activation of these pathways is mediated by a variety of growth factors, including FGF, VEGF and TGF-β168, all of which are detected at high concentration in platelet lysate product. The carbohydrate metabolism pathway, having as key components the Gpcr family, is involved in regulation of cell metabolism69 and implicated in their proliferative and migratory capacity70. The cell movement pathway has as key components the Rho family of guanosine triphosphatases (GTPases), which play important roles in cytoskeletal rearrangements that are crucial in a set of specialised functions, including, self-renewal, adhesion and migration36. In MSCs, the Rho-GTPase signalling regulates commitment into osteogenic and adipogenic differentiation71.
In conclusion, this is the first study reporting a differential cell surface protein expression between MSC-PLT and MSC-FCS. The identified cell surface proteins enriched in MSC-PLT are potentially associated with increased proliferation and migration capacity, as well as enhanced chondrogenic and osteogenic differentiation properties. Further studies are required to further investigate into the activation status of the identified pathways in MSC-PLT and uncover the impact of those enriched proteins on biological functions of MSC-PLT. This study also provides a searchable database of cell surface proteins expressed by MSCs expanded in different culture media, which can inform further studies to characterise specific MSC subsets.
Materials and Methods
Human bone marrow-derived MSCs
Each MSC sample was derived from independent bone marrow aspirates collected from different healthy donors (surplus of haematopoietic stem cell transplant) with informed consent and Local Research Ethical Committee approval (NRES Committee North East – Newcastle & North Tyneside 2). All methods were performed in accordance with the relevant guidelines and regulations. All MSC samples were derived from bone marrow mononuclear cells using standard plastic adherent method. Each paired MSC-PLT and MSC-FCS sample was generated from the same bone marrow sample by splitting the bone marrow mononuclear cells into two sub-fractions, one was cultured in DMEM based non-haematopoietic media (Miltenyibiotec, Germany) supplemented with 10% FCS while the other in DMEM (Sigma-Aldrich, USA) supplemented with 5% human platelet lysate (PLTmax, Mill Creek, USA) and 2 IU/ml Heparin. The same FCS and PLT products were used across all cultures throughout the study. The platelet lysate utilised in this work was commercially available and manufactured under good manufacturing practice (GMP) conditions. The specifications about the composition and manufacturing process of the PLT used in this study are described in details by Crespo-Diaz R et al.12 Both media were also supplemented with 2 mM L-glutamine, 100 IU/ml penicillin and 100 µg/ml streptomycin (Sigma-Aldrich, USA). Paired MSC samples were then incubated at 37 °C, 5% CO2 humidified atmosphere. Non-adherent cells were removed after 48 h by media changes. The adherent cells were maintained in culture, with media changed twice weekly. Cells were then passaged when reaching 70–80% confluence using 1x Trypsin-EDTA (Sigma-Aldrich, USA) and plated at a density of 4 × 103 cells/cm2. Cells harvested from passage 3 were used for all subsequent analysis. Phenotype, morphology, differentiation capacity and proliferation kinetics was monitored on paired samples as described in supplementary methods.
Cell barcoding for discrimination of MSC populations
To discriminate between MSC-FCS and MSC-PLT, MSC-PLT were labelled with CellTraceTM Violet (Life Technologies, USA), and MSC-FCS were kept unlabelled. MSC-FCS were suspended in PEB buffer, consisting of phosphate buffered saline (PBS) with 2 mM of EDTA (Sigma-Aldrich, USA), and 0.5% of bovine serum albumin (Sigma-Aldrich, USA), and used directly for subsequent analyses. MSC-PLTs were washed in PBS and re-suspended in a 0.2 µM solution of CellTraceTM violet at a density of 1 × 104 cells/µl. Cells were incubated for 5 minutes at room temperature in the dark, washed twice with PEB and re-suspended in the same buffer for subsequent analyses.
Surface protein screening using Lyoplate technology
Non-labelled MSC-FCS and CelltraceTM violet labelled MSC-PLT were pooled at a 1:1 ratio and analysed using the Miltenyi 96 well human antibody panel (Miltenyibiotec, Germany), containing APC-conjugated antibodies with specificity for 356 cell surface markers and 9 isotype controls, arrayed on four round bottomed 96-well plates. A detailed list of the antibodies can be found in Supplementary Table S5. Staining and plate preparation was performed as described by the manufacturer’s protocol. Briefly, the lyophilized antibodies were reconstituted with 25 µl of deionized water. The previously prepared MSCs were added to each well of the 96 well plates at a density of 5 × 104 cells/well. Cell incubation with the antibodies was carried out for 10 minutes in the dark at 4 °C. The cells were then washed twice with PEB and centrifuged at 300 × g for 5 minutes. The cell pellet was re-suspended in 50 µl PEB and surface protein expression data was acquired by flow cytometry using an automated MACSQuant flow cytometer (Miltenyibiotec, Germany), where 35 µl of sample were analysed and at least 10,000 events per well were collected. The workflow is shown in Fig. 2a.
Surface protein screening data analysis
Data acquisition was accomplished using the MACSQuant analyser 10 software (Miltenyibiotec, Germany). Data analysis was performed using either MACSQuant analyser software or FlowJo v10 software (FlowJo LLC, Treestar, USA). An initial analysis template was created to discriminate between MSC-PLT and MSC-FCS and to analyse each population for its APC positivity. Analytical data (event count, percentage of APC positive clean events, median fluorescence intensity and standard deviation) were exported to Excel and associated to sample ID, plate number, row and column. The gating strategy for the flow cytometry data analyses is depicted in Fig. 2a.
Surface protein validation using multiparameter flow cytometry analysis
To validate the findings of the high throughput screen, paired MSC-PLT and MSC-FCS samples were incubated with an optimally titrated master-mix of antibodies (CD318-FITC, CD312-APC, CD26 PE, CD54 PE-Vio770, MSCA-1 BUV395, CD106 APC-Vio770) or singularly with CD49d-APC. Samples were washed and incubated with DAPI for dead cell exclusion and acquired on a BD FACSymphony A5 flow cytometer (Becton Dickinson, USA) using PMT voltages that were optimised to achieve best resolution without saturating signals in any channel. Detailed Ab information and the detector channels used, including optical filters, are shown in Supplemental Table S6. Spectral compensation was performed using the BD FACSDIVA auto-compensation wizard in conjunction with BD CompBeads (Becton Dickinson) stained with each individual antibody-fluorochrome in the panel. Data was then analysed offline with FCSExpress V6 (DeNovo software, USA) using a classical sequential gating strategy. Briefly, live cells were identified as having an intermediate to high FSC_A value in conjunction with DAPI negativity. Doublets and aggregates were then eliminated using a combination of FSC_A, FSC_H and SSC-W. The percentage of cells (MSC-PLT or MSC-FCS) positive for a given marker was determined relative to their own respective unstained control by setting a gate on the latter to a 99.9% confidence. Median fluorescent intensities were then derived for cells deemed positive for each respective marker only
Reverse transcription and quantitative Real Time-Polymerase Chain Reaction
RNA from paired MSC samples was extracted using the RNeasy mini kit (Qiagen, Germany) as per the manufacturer’s protocol. RNA purity and quantity was analysed using Nanodrop 2000 (Thermo Fisher Scientific, USA) and reverse transcribed using High-Capacity cDNA reverse transcription reagents (Applied Biosystems, USA). Taqman® (Thermo Fisher Scientific, USA) gene expression assays were used to measure the expression levels of the genes CDCP1 (CD318, Hs01080405_m1), ALPL (MSCA-1, Hs01029144_m1) and DPP4 (CD26, Hs00897391_m1) all purchased from Thermo Fisher Scientific, USA. Reactions were carried out in triplicate using 7900HT Fast Real Time PCR system (Applied Biosystems, USA) and standard thermal cycling conditions. Ct values were normalised to the reference gene Glyceraldehyde 3-phosphate dehydrogenase (GAPDH, Hs9999905_m1, Thermo Fisher Scientific, USA).
Pathway and network analysis
For further understanding the relevance of the proteins significantly enriched on the surface of MSC-PLT, the 13 enriched markers were analysed through the use of Ingenuity Pathway Analysis (IPA, QIAGEN INC., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis) Canonical pathway analysis identified the pathways from the IPA library of canonical pathway that were most significant to the data set. The markers from the data set that met the fold-change cut-off of 2 in association with a canonical pathway in Ingenuity knowledge base were considered for analysis. The significance of the association between the data set and the canonical pathways was analysed by IPA software algorithms in 2 ways: a ratio of the number of markers from the data set that map to the pathway, divided by the total number of molecules mapping to the canonical pathway, a Fisher exact test was used to calculate a p-value determining the association probability between the molecules in the data set and the canonical pathway. Network analysis was used to exhibit relationships between enriched genes and the canonical pathways identified by the IPA software. Networks were algorithmically constructed by IPA software on the bases of the functional and biological connectivity of the 13 enriched surface proteins on MSC-PLT and by limiting the number of molecules in a network to 35 and displaying only the most significant interactions.
Statistical analyses
Statistical analyses were performed using GraphPad prism v5.0 (GraphPad software, USA). Unless otherwise stated, data are presented as mean and standard error of the mean (SEM). One-way ANOVAs were used for statistical analysis and p-values of <0.05 were considered significant.
Electronic supplementary material
Acknowledgements
This work was supported by FP7-People-2012-ITN, (315963), the Newcastle Healthcare Charities, and Arthritis Research UK Tissue Engineering Centre (Awards 19429 and 21156). Part of this work was carried out at Miltenyi Biotec, Bergisch Gladbach, Germany.
Author Contributions
M.R., X.W.: Conceived the research idea. M.R., K.G., S.K., A.F.: Designed the Lyoplate and multiparameter validation flow cytometry experiments. M.R., X.W., D.M.: Performed the experiments and analysed the data. M.R., X.W., L.N., D.M., A.F.: Wrote the paper and contributed to the analysis and interpretation of data. A.M.D.: Critical review of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-22326-5.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
4/16/2018
The version of this Article previously published quoted an incorrect email address for Monica Reis. Correspondence and requests for materials should also be addressed to Monica.Reis@childrens.harvard.edu. This has now been corrected in the HTML and PDF versions of the Article.
Contributor Information
Monica Reis, Email: Monica.Reis@childrens.harvard.edu.
Xiao-nong Wang, Email: x.n.wang@newcastle.ac.uk.
References
- 1.Gao F, et al. Mesenchymal stem cells and immunomodulation: current status and future prospects. Cell Death Dis. 2016;7:e2062. doi: 10.1038/cddis.2015.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le Blanc K, et al. Mesenchymal stem cells for treatment of steroid-resistant, severe, acute graft-versus-host disease: a phase II study. Lancet (London, England) 2008;371:1579–1586. doi: 10.1016/S0140-6736(08)60690-X. [DOI] [PubMed] [Google Scholar]
- 3.Sensebe L, Krampera M, Schrezenmeier H, Bourin P, Giordano R. Mesenchymal stem cells for clinical application. Vox sanguinis. 2010;98:93–107. doi: 10.1111/j.1423-0410.2009.01227.x. [DOI] [PubMed] [Google Scholar]
- 4.EMA, E. M. A. Note for guidance on minimizing the risk of transmitting animal spongiform encephalopathy agents via human and veterinary medicinal products. EMEA410/01 rev. 2 (2008).
- 5.Horwitz EM, et al. Isolated allogeneic bone marrow-derived mesenchymal cells engraft and stimulate growth in children with osteogenesis imperfecta: Implications for cell therapy of bone. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:8932–8937. doi: 10.1073/pnas.132252399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reinhardt, J., Stuhler, A. & Blumel, J. Safety of bovine sera for production of mesenchymal stem cells for therapeutic use. Human gene therapy22, 775, author reply 776 (2011). [DOI] [PubMed]
- 7.Spees JL, et al. Internalized antigens must be removed to prepare hypoimmunogenic mesenchymal stem cells for cell and gene therapy. Molecular therapy: the journal of the American Society of Gene Therapy. 2004;9:747–756. doi: 10.1016/j.ymthe.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 8.Poloni A, et al. Selection of CD271+ cells and human AB serum allows a Large expansion of mesenchymal stromal cells from human bone marrow. Cytotherapy. 2009;11:153–162. doi: 10.1080/14653240802582125. [DOI] [PubMed] [Google Scholar]
- 9.Díez JM, Bauman E, Gajardo R, Jorquera JI. Culture of human mesenchymal stem cells using a candidate pharmaceutical grade xeno-free cell culture supplement derived from industrial human plasma pools. Stem Cell Res Ther. 2015;6:1–10. doi: 10.1186/s13287-015-0016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stute N, et al. Autologous serum for isolation and expansion of human mesenchymal stem cells for clinical use. Experimental hematology. 2004;32:1212–1225. doi: 10.1016/j.exphem.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Doucet C, et al. Platelet lysates promote mesenchymal stem cell expansion: a safety substitute for animal serum in cell-based therapy applications. Journal of cellular physiology. 2005;205:228–236. doi: 10.1002/jcp.20391. [DOI] [PubMed] [Google Scholar]
- 12.Crespo-Diaz R, et al. Platelet lysate consisting of a natural repair proteome supports human mesenchymal stem cell proliferation and chromosomal stability. Cell transplantation. 2011;20:797–811. doi: 10.3727/096368910X543376. [DOI] [PubMed] [Google Scholar]
- 13.Bieback K. Platelet lysate as replacement for fetal bovine serum in mesenchymal stromal cell cultures. Transfus Med Hemother. 2013;40:326–335. doi: 10.1159/000354061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wuchter P, et al. Evaluation of GMP-compliant culture media for in vitro expansion of human bone marrow mesenchymal stromal cells. Experimental hematology. 2016;44:508–518. doi: 10.1016/j.exphem.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Fekete N, et al. Platelet lysate from whole blood-derived pooled platelet concentrates and apheresis-derived platelet concentrates for the isolation and expansion of human bone marrow mesenchymal stromal cells: production process, content and identification of active components. Cytotherapy. 2012;14:540–554. doi: 10.3109/14653249.2012.655420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hemeda H, Giebel B, Wagner W. Evaluation of human platelet lysate versus fetal bovine serum for culture of mesenchymal stromal cells. Cytotherapy. 2014;16:170–180. doi: 10.1016/j.jcyt.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Griffiths S, Baraniak PR, Copland IB, Nerem RM, McDevitt TC. Human platelet lysate stimulates high-passage and senescent human multipotent mesenchymal stromal cell growth and rejuvenation in vitro. Cytotherapy. 2013;15:1469–1483. doi: 10.1016/j.jcyt.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 18.Lucchini G, et al. Platelet-lysate-Expanded Mesenchymal Stromal Cells as a Salvage Therapy for Severe Resistant Graft-versus-Host Disease in a Pediatric Population. Biology of Blood and Marrow Transplantation. 2010;16:1293–1301. doi: 10.1016/j.bbmt.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 19.Introna M, et al. Treatment of Graft versus Host Disease with Mesenchymal Stromal Cells: A Phase I Study on 40 Adult and Pediatric Patients. Biology of Blood and Marrow Transplantation. 2014;20:375–381. doi: 10.1016/j.bbmt.2013.11.033. [DOI] [PubMed] [Google Scholar]
- 20.Black CB, Duensing TD, Trinkle LS, Dunlay RT. Cell-based screening using high-throughput flow cytometry. Assay and drug development technologies. 2011;9:13–20. doi: 10.1089/adt.2010.0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gedye CA, et al. Cell Surface Profiling Using High-Throughput Flow Cytometry: A Platform for Biomarker Discovery and Analysis of Cellular Heterogeneity. PLOS ONE. 2014;9:e105602. doi: 10.1371/journal.pone.0105602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramirez S, Aiken CT, Andrzejewski B, Sklar LA, Edwards BS. High-throughput flow cytometry: validation in microvolume bioassays. Cytometry. Part A: the journal of the International Society for Analytical Cytology. 2003;53:55–65. doi: 10.1002/cyto.a.10035. [DOI] [PubMed] [Google Scholar]
- 23.Donnenberg AD, Meyer EM, Rubin JP, Donnenberg VS. The cell-surface proteome of cultured adipose stromal cells. Cytometry. Part A: the journal of the International Society for Analytical Cytology. 2015;87:665–674. doi: 10.1002/cyto.a.22682. [DOI] [PubMed] [Google Scholar]
- 24.Baer PC, et al. Comprehensive phenotypic characterization of human adipose-derived stromal/stem cells and their subsets by a high throughput technology. Stem cells and development. 2013;22:330–339. doi: 10.1089/scd.2012.0346. [DOI] [PubMed] [Google Scholar]
- 25.Walmsley GG, et al. High-Throughput Screening of Surface Marker Expression on Undifferentiated and Differentiated Human Adipose-Derived Stromal Cells. Tissue engineering. Part A. 2015;21:2281–2291. doi: 10.1089/ten.tea.2015.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rostovskaya M, Anastassiadis K. Differential expression of surface markers in mouse bone marrow mesenchymal stromal cell subpopulations with distinct lineage commitment. PLOS ONE. 2012;7:7. doi: 10.1371/journal.pone.0051221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ong WK, et al. Identification of specific cell-surface markers of adipose-derived stem cells from subcutaneous and visceral fat depots. Stem Cell Reports. 2014;2:171–179. doi: 10.1016/j.stemcr.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dominici M, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 29.Abdelrazik H, Spaggiari GM, Chiossone L, Moretta L. Mesenchymal stem cells expanded in human platelet lysate display a decreased inhibitory capacity on T- and NK-cell proliferation and function. European journal of immunology. 2011;41:3281–3290. doi: 10.1002/eji.201141542. [DOI] [PubMed] [Google Scholar]
- 30.Frings, K. R. S. V. V. d. C. P. Comparison between antibodies from different providers: determination of median fluorescent intensities and stain indices. MACS & More 14 (2012).
- 31.Li W, et al. Leukemia Decreased TREM-1 Expression In Hematopoiesis Stem/Progenitor Cells. Blood. 2013;122:5166–5166. [Google Scholar]
- 32.Elgueta R, et al. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunological reviews. 2009;229:10.1111. doi: 10.1111/j.1600-065X.2009.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng L, et al. HMGB1 promotes the secretion of multiple cytokines and potentiates the osteogenic differentiation of mesenchymal stem cells through the Ras/MAPK signaling pathway. Exp Ther Med. 2016;12:3941–3947. doi: 10.3892/etm.2016.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richard DE, Vouret-Craviari V, Pouyssegur J. Angiogenesis and G-protein-coupled receptors: signals that bridge the gap. Oncogene. 2001;20:1556–1562. doi: 10.1038/sj.onc.1204193. [DOI] [PubMed] [Google Scholar]
- 35.Rozengurt E, Sinnett-Smith J, Kisfalvi K. Crosstalk between Insulin/IGF-1 and GPCR Signaling Systems: A Novel Target for the Anti-diabetic Drug Metformin in Pancreatic Cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16:2505–2511. doi: 10.1158/1078-0432.CCR-09-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murali A, Rajalingam K. Small Rho GTPases in the control of cell shape and mobility. Cellular and molecular life sciences: CMLS. 2014;71:1703–1721. doi: 10.1007/s00018-013-1519-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D’Antonio M, et al. High-Throughput and Cost-Effective Characterization of Induced Pluripotent Stem Cells. Stem Cell Reports. 2017;8:1101–1111. doi: 10.1016/j.stemcr.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lakschevitz FS, et al. Identification of neutrophil surface marker changes in health and inflammation using high-throughput screening flow cytometry. Exp Cell Res. 2016;342:200–209. doi: 10.1016/j.yexcr.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 39.Schallmoser K, et al. Human platelet lysate can replace fetal bovine serum for clinical-scale expansion of functional mesenchymal stromal cells. Transfusion. 2007;47:1436–1446. doi: 10.1111/j.1537-2995.2007.01220.x. [DOI] [PubMed] [Google Scholar]
- 40.Li C-y, et al. Comparative analysis of human mesenchymal stem cells from bone marrow and adipose tissue under xeno-free conditions for cell therapy. Stem Cell Research & Therapy. 2015;6:55. doi: 10.1186/s13287-015-0066-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alexander D, et al. MSCA-1/TNAP selection of human jaw periosteal cells improves their mineralization capacity. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2010;26:1073–1080. doi: 10.1159/000323985. [DOI] [PubMed] [Google Scholar]
- 42.Yang ZX, et al. CD106 identifies a subpopulation of mesenchymal stem cells with unique immunomodulatory properties. PLoS One. 2013;8:e59354. doi: 10.1371/journal.pone.0059354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeda H, Fujimori Y, Kai S, Ogawa H, Nakano T. CD318/CUB-domain-containing protein 1 expression on cord blood hematopoietic progenitors. Exp Ther Med. 2010;1:497–501. doi: 10.3892/etm_00000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Battula, V. L. et al. Human placenta and bone marrow derived MSC cultured in serum-free, b-FGF-containing medium express cell surface frizzled-9 and SSEA-4 and give rise to multilineage differentiation. Differentiation75 (2007). [DOI] [PubMed]
- 45.Mutyaba PL, et al. Notch signaling in mesenchymal stem cells harvested from geriatric mice. Journal of orthopaedic trauma. 2014;28(Suppl 1):S20–23. doi: 10.1097/BOT.0000000000000064. [DOI] [PubMed] [Google Scholar]
- 46.Leemans JC, et al. The epidermal growth factor-seven transmembrane (EGF-TM7) receptor CD97 is required for neutrophil migration and host defense. Journal of immunology (Baltimore, Md.: 1950) 2004;172:1125–1131. doi: 10.4049/jimmunol.172.2.1125. [DOI] [PubMed] [Google Scholar]
- 47.Ng F, et al. PDGF, TGF-beta, and FGF signaling is important for differentiation and growth of mesenchymal stem cells (MSCs): transcriptional profiling can identify markers and signaling pathways important in differentiation of MSCs into adipogenic, chondrogenic, and osteogenic lineages. Blood. 2008;112:295–307. doi: 10.1182/blood-2007-07-103697. [DOI] [PubMed] [Google Scholar]
- 48.Ball SG, Shuttleworth CA, Kielty CM. Vascular endothelial growth factor can signal through platelet-derived growth factor receptors. The Journal of cell biology. 2007;177:489–500. doi: 10.1083/jcb.200608093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chiara Barsotti M, et al. Effect of Platelet Lysate on Human Cells Involved in Different Phases of Wound Healing. PLOS ONE. 2013;8:e84753. doi: 10.1371/journal.pone.0084753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Battula, V. L. et al. Isolation of functionally distinct mesenchymal stem cell subsets using antibodies against CD56, CD271, and mesenchymal stem cell antigen-1. Haematologica94 (2009). [DOI] [PMC free article] [PubMed]
- 51.Mentlein R. Dipeptidyl-peptidase IV (CD26)-role in the inactivation of regulatory peptides. Regulatory Peptides. 1999;85:9–24. doi: 10.1016/S0167-0115(99)00089-0. [DOI] [PubMed] [Google Scholar]
- 52.Battula VL, Treml S, Abele H, Buhring HJ. Prospective isolation and characterization of mesenchymal stem cells from human placenta using a frizzled-9-specific monoclonal antibody. Differentiation. 2008;76:326–336. doi: 10.1111/j.1432-0436.2007.00225.x. [DOI] [PubMed] [Google Scholar]
- 53.Esteve D, et al. Multiple Functions of MSCA-1/TNAP in Adult Mesenchymal Progenitor/Stromal Cells. Stem Cells Int. 2016;1815982:29. doi: 10.1155/2016/1815982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Niehage C, et al. The Cell Surface Proteome of Human Mesenchymal Stromal Cells. PLOS ONE. 2011;6:e20399. doi: 10.1371/journal.pone.0020399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wegmeyer H, et al. Mesenchymal stromal cell characteristics vary depending on their origin. Stem cells and development. 2013;22:2606–2618. doi: 10.1089/scd.2013.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sato, Y. et al. Notch2 Signaling Regulates the Proliferation of Murine Bone Marrow-Derived Mesenchymal Stem/Stromal Cells via c-Myc Expression. PLOS ONE11 (2016). [DOI] [PMC free article] [PubMed]
- 57.Aust G, et al. CD97, but not its closely related EGF-TM7 family member EMR2, is expressed on gastric, pancreatic, and esophageal carcinomas. Am J Clin Pathol. 2002;118:699–707. doi: 10.1309/A6AB-VF3F-7M88-C0EJ. [DOI] [PubMed] [Google Scholar]
- 58.Price MA, et al. CSPG4, a potential therapeutic target, facilitates malignant progression of melanoma. Pigment cell & melanoma research. 2011;24:1148–1157. doi: 10.1111/j.1755-148X.2011.00929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maerz JK, et al. Bone marrow-derived mesenchymal stromal cells differ in their attachment to fibronectin-derived peptides from term placenta-derived mesenchymal stromal cells. Stem Cell Research & Therapy. 2016;7:29. doi: 10.1186/s13287-015-0243-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xie H, et al. PDGF-BB secreted by preosteoclasts induces CD31(hi)Emcn(hi) vessel subtype in coupling osteogenesis. Nature medicine. 2014;20:1270–1278. doi: 10.1038/nm.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu, Y. T. et al. Defining minimum essential factors to derive highly pure human endothelial cells from iPS/ES cells in an animal substance-free system. Sci Rep5 (2015). [DOI] [PMC free article] [PubMed]
- 62.Ren G, et al. Inflammatory Cytokine-Induced Intercellular Adhesion Molecule-1 and Vascular Cell Adhesion Molecule-1 in Mesenchymal Stem Cells Are Critical for Immunosuppression. Journal of immunology (Baltimore, Md.: 1950) 2010;184:2321–2328. doi: 10.4049/jimmunol.0902023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Najar M, et al. Immune-related antigens, surface molecules and regulatory factors in human-derived mesenchymal stromal cells: the expression and impact of inflammatory priming. Stem cell reviews. 2012;8:1188–1198. doi: 10.1007/s12015-012-9408-1. [DOI] [PubMed] [Google Scholar]
- 64.Braud VM, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 65.Herlaar E, Brown Z. p38 MAPK signalling cascades in inflammatory disease. Molecular Medicine Today. 1999;5:439–447. doi: 10.1016/S1357-4310(99)01544-0. [DOI] [PubMed] [Google Scholar]
- 66.Cantaert T, Baeten D, Tak PP, van Baarsen LGM. Type I IFN and TNFα cross-regulation in immune-mediated inflammatory disease: basic concepts and clinical relevance. Arthritis Research & Therapy. 2010;12:1–10. doi: 10.1186/ar3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arthur JSC, Ley SC. Mitogen-activated protein kinases in innate immunity. Nat Rev Immunol. 2013;13:679–692. doi: 10.1038/nri3495. [DOI] [PubMed] [Google Scholar]
- 68.Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002;12:9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- 69.Choi HY, et al. G protein-coupled receptors in stem cell maintenance and somatic reprogramming to pluripotent or cancer stem cells. BMB Reports. 2015;48:68–80. doi: 10.5483/BMBRep.2015.48.2.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Doze VA, Perez DM. GPCRs in Stem Cell Function. Progress in molecular biology and translational science. 2013;115:175–216. doi: 10.1016/B978-0-12-394587-7.00005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sherwin, S. Y. et al. In Novel Signaling Mechanisms and Bone Cell Biology Meeting Abstracts OR31-33-OR31-33 (Endocrine Society, 2014).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.