Abstract
Amino acids (AA) are one of the key nutrients that regulate cell proliferation and casein synthesis in cow mammary epithelial cells (CMEC), but the mechanism of this regulation is not yet clear. In this study, the effect of SESN2 on AA-mediated cell proliferation and casein synthesis in CMEC was assessed. After 12 h of AA starvation, CMECs were cultured in the absence of all AA (AA−), in the presences of only essential AA (EAA+), or of all AA (AA+). Cell proliferation, casein expression, and activation of the mammalian target of rapamycin complex 1 (mTORC1) pathway were increased; but SESN2 expression was decreased in response to increased EAA or AA supply. Overexpressing or inhibiting SESN2 demonstrated that cell proliferation, casein expression, and activation of the mTORC1 pathway were all controlled by SESN2 expression. Furthermore, the increase in cell proliferation, casein expression, and activation of the mTORC1 pathway in response to AA supply was inhibited by overexpressing SESN2, and those effects were reversed by inhibiting SESN2. These results indicate that SESN2 is an important inhibitor of mTORC1 in CMEC blocking AA-mediated cell proliferation and casein synthesis.
Introduction
Proliferation of cow mammary epithelial cells (CMEC) and casein synthesis by those cells are regulated by hormones (e.g. prolactin, insulin and glucocorticoids), nutrients (e.g. glucose and amino acids) and environmental stress (e.g. heat stress)1–5. Among the nutrients, amino acids (AA) are the most important as they are not only the building blocks of protein synthesis but also the regulators of cell proliferation and casein synthesis in mammalian epithelial cells6,7.
The main signaling pathway that mediates AA-induced cell proliferation and protein synthesis is the mammalian target of rapamycin complex 1 (mTORC1) pathway8,9. mTORC1 is the main regulatory factor in the pathway, and it is composed of mTOR, G protein β subunit-like protein (GβL), regulatory associated protein of mammalian target of rapamycin (Raptor), proline-rich Akt substrate of 40 kDa (PRAS40), and Deptor10. When AA are sufficient, mTORC1 is stimulated by an unknown signaling pathway and moves to the lysosomal surface from an undefined location, causing mTOR to be phosphorylated. Phosphorylated mTOR activates the downstream molecules, ribosomal protein S6 kinase 1 (S6K1) and eukaryotic translation initiation factor 4E binding protein 1 (4EBP1), which promotes participation in the translation process and protein synthesis4,11,12. The downstream actions of mTORC1 have been well characterized, but the mechanism of AA action on mTORC1 is poorly understood13–15.
Sestrins are a family of highly conserved, stress-inducible, metabolic regulators. In mammals, there are three members of the family: sestrin1 (SESN1), sestrin2 (SESN2) and sestrin3 (SESN3), of which, SESN2 is the most important16–18. Previous reports have shown that SESN2 can suppress reactive oxygen species arising from oxidative stress through its antioxidant function19. In addition to its antioxidant activity, SESN2 can activate adenosine monophosphate-activated protein kinase (AMPK), subsequently inhibiting the activation of mTORC120,21. In human cells (mainly HELA and human embryonic kidney (HEK) 293 cells), SESN2 protein was found to respond to AA depletion (especially leucine) resulting in negative effects on the mTORC1 pathway. It has been reported that sestrins control mTORC1 signaling by inhibiting Rag GTPase22–25. Kimball et al. reported that sestrins/sestrin2 negatively regulated the amino-acid-sensing pathway upstream of mTORC1 by interacting with GTPase-activating proteins toward Rags 2 (GATOR2)26.
In this study, we investigated the function of SESN2 in the process of cell proliferation and casein synthesis associated with AA-mediated stimulation of the mTORC1 pathway in CMEC. We hypothesized that SESN2 mediated AA responses through inhibition of mTORC1 activation.
Results
SESN2 is negatively regulated by AA in CMECs
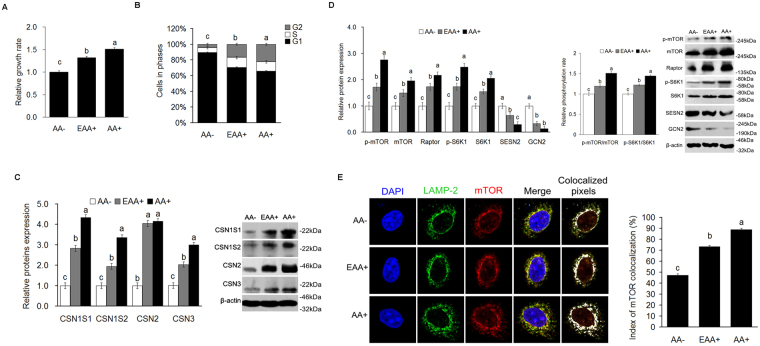
To establish that the lactation cell model was regulated by AA, cell growth and cell cycle responses to AA were analyzed. The results showed that cell growth (Fig. 1A) and the proportion of cells in the G2+S stages (Fig. 1B) were increased (P < 0.05) for essential AA (EAA+) and AA+ groups as compared to the AA- group. Treatment with EAA+ or AA+ also increased (P < 0.05) alpha S1 casein (CSN1S1), alpha S2 casein (CSN1S2), beta casein (CSN2), kappa casein (CSN3), mTOR, p-mTOR, Raptor, S6K1 and p-S6K1 protein expression, and the ratio of p-S6K1 to S6K1, p-mTOR to mTOR (Fig. 1C,D), as well as lysosomal localization of mTOR (Fig. 1E and Supplementary Fig. 1). But the expression of SESN2 and general control nonderepressible 2 (GCN2) protein was decreased (P < 0.05) when cells were treated with EAA+ or AA+ (Fig. 1D). The addition of non EAA (NEAA+) in the absence of EAA showed that cell growth, the expression of CSN2 and the activation of mTORC1 pathway (including expression of mTOR, S6K1, ratios of p-mTOR/mTOR and p-S6K1/S6K1, and lysosomal localization of mTOR) were increased (P < 0.05) compared to the AA- group, but the responses were much smaller than for the EAA+ group (Supplementary Fig. 3A,B).
Figure 1.
SESN2 is negatively regulated by AA in the process of AA-mediated regulation of cell proliferation and casein synthesis in CMEC. (A) Cell proliferation of CMEC treated with EAA or AA was analyzed by MTT assay, the data of “AA− group” was defined as “1”; (B) Cell cycle of CMEC treated with EAA or AA was analyzed by FC; (C) Expression of casein in CMEC treated with EAA or AA was analyzed by WB, the ratio value of “AA− group” was defined as “1”; (D) Expression of proteins related mTORC1 pathway, SESN2 and GCN2, and the phosphorylation ratio of mTOR and S6K1 in CMEC treated with EAA or AA was analyzed by WB, the ratio value of “AA− group” was defined as “1”; (E) Subcellular localization of mTOR in CMEC treated with EAA or AA for 6 h was analyzed by IF. Each bar represents the LSM ± SEM. In the bar charts, different superscript lowercase letters indicate significant difference (p < 0.05). Uncropped images for all Western blots can be found in Supplementary Fig. 4.
SESN2 negatively regulates AA-mediated cell proliferation in CMEC
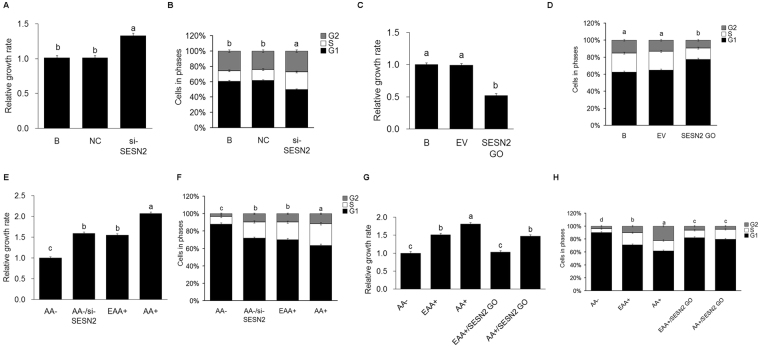
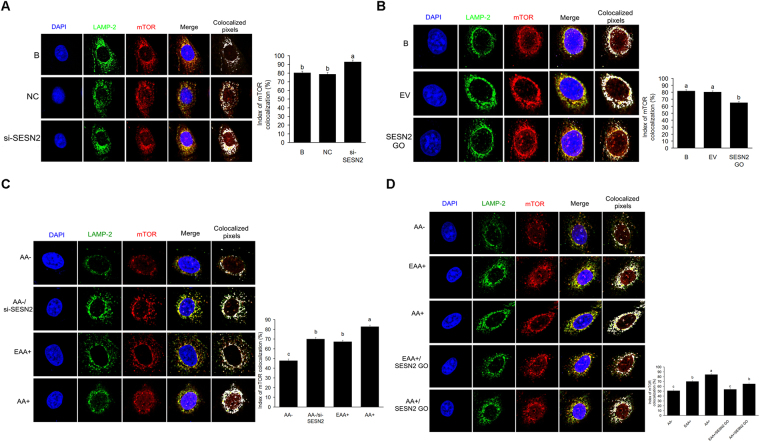
To investigate whether the expression of SESN2 mediates the effects of AA on cell proliferation, SESN2 was inhibited (si-SESN2) or overexpressed (SESN2 GO). The null transfection group (B) was used as blank control; the negative control siRNA transfected group (NC) was used as negative control; and the empty plasmid transfected group (EV) was used as empty plasmid control. Cell growth and cell cycle were analyzed. The results showed that cell growth (Fig. 2A) and the proportion of cells in G2 and S stages (Fig. 2B) were increased (P < 0.05) when SESN2 expression was inhibited, and the reverse when it was overexpressed (Fig. 2C,D).
Figure 2.
SESN2 negatively regulates AA-mediated cell proliferation in CMEC. (A) Cell proliferation of CMEC treated with si-SESN2 was analyzed by MTT assay, the date of the “B group” was defined as “1”; (B) Cell cycle of CMEC treated with si-SESN2 was analyzed by FC; (C) Cell proliferation of CMEC treated with SESN2 GO was analyzed by MTT assay, the date of the “B group” was defined as “1”; (D) Cell cycle of CMEC treated with SESN2 GO was analyzed by FC; (E) Cell proliferation of CMEC treated with AA−, AA−/si-SESN2, EAA+, and AA+ was analyzed by MTT assay, the date of “AA− group” was defined as “1”; (F) Cell cycle of CMEC treated with AA−, AA−/si-SESN2, EAA+, and AA+ was analyzed by FC; (G) Cell proliferation of CMEC treated with AA−, EAA+, AA+, EAA+/SESN2 GO, and AA+/SESN2 GO was analyzed by MTT assay, the date of “AA− group” was defined as “1”; (H) Cell cycle of CMEC treated with AA−, EAA+, AA+, EAA+/SESN2 GO, and AA+/SESN2 GO was analyzed by FC. Each bar represents the LSM±SEM. In the bar charts, different superscript lowercase letters indicate significant differences (p < 0.05). B: Cells were no transfected; NC: Cells were transfected with negative control siRNA; EV: Cells were transfected with empty vector.
To investigate whether AA regulate cell growth and the cell cycle via SESN2, CMEC were treated with AA−, EAA+, AA+, AA−/si-SESN2, EAA+/SESN2 GO and AA+/SESN2 GO; and cell growth and cell cycle were analyzed. The results showed that decreases in cell growth and decreases in the proportion of cells in the G2 and S stages caused by AA starvation were blocked (P < 0.05) when SESN2 was inhibited (Fig. 2E,F) resulting in growth and cell cycle patterns equivalent to those of the EAA+ treatment. Conversely, the stimulatory effects of AA+ on cell growth and proportions of cells in the G2+S phases were inhibited (P < 0.05) by SESN2 overexpression (Fig. 2G,H).
SESN2 negatively regulates AA-mediated casein synthesis in CMEC
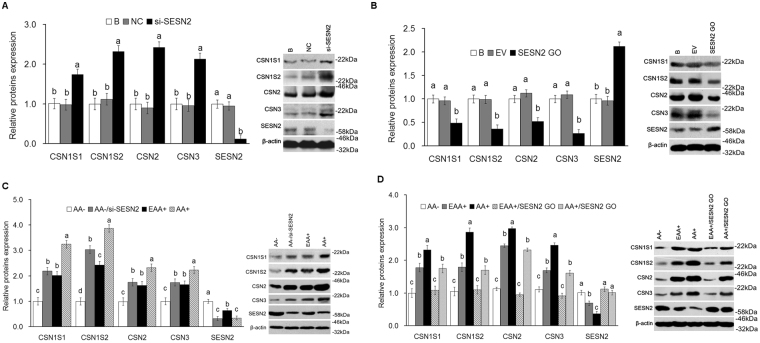
The effect of SESN2 on casein synthesis was analyzed. The results showed that expression of CSN1S1, CSN1S2, CSN2 and CSN3 was increased (P < 0.05) (Fig. 3A) in cells with SESN2 inhibited and decreased (P < 0.05) (Fig. 3B) in cells overexpressing SESN2.
Figure 3.
SESN2 negatively regulates AA-mediated casein synthesis in CMEC. (A) Expression of casein in CMEC treated with si-SESN2 was analyzed by WB, the ratio value of “B group” was defined as “1”; (B) Expression of casein in CMEC treated with SESN2 GO was analyzed by WB, the ratio value of “B group” was defined as “1”; (C) Expression of casein in CMEC treated with AA−, AA−/si-SESN2, EAA+, and AA+ was analyzed by WB, the ratio value of “AA− group” was defined as “1”; (D) Expression of casein in CMEC treated with AA−, EAA+, AA+, EAA+/SESN2 GO, and AA+/SESN2 GO was analyzed by WB, the ratio value of “AA− group” was defined as “1”. Each bar represents the LSM ± SEM. In the bar charts, different superscript lowercase letters indicate significant differences (p < 0.05). B: Cells were no transfected; NC: Cells were transfected with negative control siRNA; EV: Cells were transfected with empty vector. Uncropped images for all Western blots can be found in Supplementary Fig. 4.
AA starvation decreased expression of CSN1S1, CSN1S2, CSN2 and CSN3; but this decrease was rescued (P < 0.05) by inhibition of SESN2 (Fig. 3C), and AA stimulating effects were inhibited (P < 0.05) by SESN2 overexpression (Fig. 3D).
SESN2 negatively regulates activation of AA-mediated mTORC1 pathway in CMEC
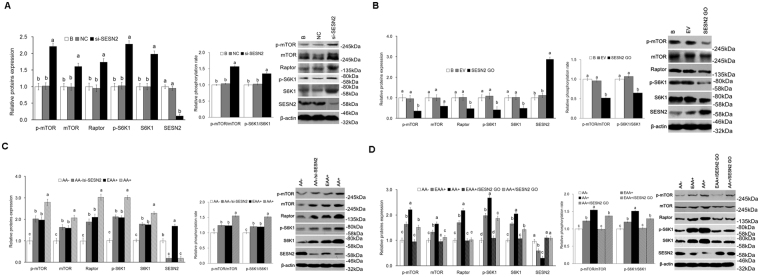
Expression of mTOR, p-mTOR, Raptor, S6K1 and p-S6K1, as well as the ratio of p-S6K1 to S6K1, p-mTOR to mTOR was increased (P < 0.05) (Fig. 4A) in cells with SESN2 inhibited and decreased (P < 0.05) (Fig. 4B) in cells overexpressing SESN2. Additionally, the decrease in expression of mTOR, p-mTOR, Raptor, S6K1 and p-S6K1, as well as the ratio of p-S6K1 to S6K1, p-mTOR to mTOR caused by AA starvation was rescued (P < 0.05) by SESN2 inhibition (Fig. 4C). AA stimulation increased (P < 0.05) expression of p-mTOR, Raptor, S6K1 and p-S6K1, as well as the ratio of p-S6K1 to S6K1, p-mTOR to mTOR; and the increase was inhibited (P < 0.05) by SESN2 overexpression (Fig. 4D). The independent additions of leucine (Leu), isoleucine (Ile), valine (Val), methionine (Met) or glutamine (Gln) to the AA- media down regulated (P < 0.05) the expression of SESN2 and up regulated (P < 0.05) the mTORC1 signaling pathway (Supplementary Fig. 2), however, the effects were not as great as AA + indicating that some additivity of responses must exist. The independent additions of arginine (Arg) or lysine (Lys) to AA− media did not affect (P > 0.05) expression of SESN2 but up regulated (P < 0.05) the mTORC1 signaling pathway (Supplementary Fig. 3B); and the independent additions of proline (Pro) or Alanine (Ala) to the AA− media did not affect (P > 0.05) the expression of SESN2 or the mTORC1 signaling pathway (Supplementary Fig. 3B).
Figure 4.
SESN2 negatively regulates AA-mediated mTORC1 activation in CMEC. (A) Expression of proteins related mTORC1 pathway and the phosphorylation ratio of mTOR and S6K1 in CMEC treated with si-SESN2 was analyzed by WB, the ratio value of “B group” was defined as “1”; (B) Expression of proteins related mTORC1 pathway and the phosphorylation ratio of mTOR and S6K1 in CMEC treated with SESN2 GO was analyzed by WB, the ratio value of “B group” was defined as “1”; (C) Expression of proteins related mTORC1 pathway and the phosphorylation ratio of mTOR and S6K1 in CMEC treated with AA−, AA−/si-SESN2, EAA+, and AA+ was analyzed by WB, the ratio value of “AA− group” was defined as “1”; (D) Expression of proteins related mTORC1 pathway and the phosphorylation ratio of mTOR and S6K1 in CMEC treated with AA−, EAA+, AA+, EAA+/SESN2 GO, and AA+/SESN2 GO was analyzed by WB, the ratio value of “AA− group” was defined as “1”. Each bar represents the LSM ± SEM. In the bar charts, different superscript lowercase letters indicate significant differences (p < 0.05). B: Cells were no transfected; NC: Cells were transfected with negative control siRNA; EV: Cells were transfected with empty vector. Uncropped images for all Western blots can be found in Supplementary Fig. 4.
Lysosomal localization of mTOR was increased (P < 0.05) (Fig. 5A) in cells with SESN2 inhibited, and decreased (P < 0.05) (Fig. 5B) in cells overexpressing SESN2. Additionally, lysosomal localization of mTOR was decreased (P < 0.05) by AA starvation, and this decrease was rescued (P < 0.05) by SESN2 inhibition (Fig. 5C). Lysosomal localization of mTOR in response to increased AA was inhibited (P < 0.05) by SESN2 overexpression (Fig. 5D).
Figure 5.
SESN2 negatively regulates AA-mediated lysosomal localization of mTOR in CMEC. (A) Subcellular localization of mTOR in CMEC treated with si-SESN2 was analyzed by IF; (B) Subcellular localization of mTOR in CMEC treated with SESN2 GO was analyzed by IF; (C) Subcellular localization of mTOR in CMEC treated with AA−, AA−/si-SESN2, EAA+, and AA+ was analyzed by IF; (D) Subcellular localization of mTOR in CMEC treated with AA−, EAA+, AA+, EAA+/SESN2 GO, and AA+/SESN2 GO was analyzed by IF. Each bar represents the LSM ± SEM. In the bar charts, different superscript lowercase letters indicate significant differences (p < 0.05). B: Cells were no transfected; NC: Cells were transfected with negative control siRNA; EV: Cells were transfected with empty vector.
SESN2 negatively regulates cell proliferation and casein synthesis via mTORC1 pathway in CMEC
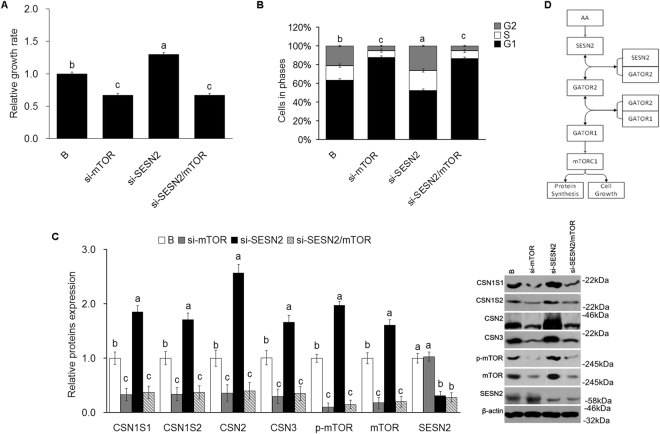
Cell growth was decreased (P < 0.05) and the proportion of cells in the G2 and S stages decreased (P < 0.05) when mTOR gene expression was inhibited. mTOR inhibition also prevented (P < 0.05) cell growth and G2 and S stage changes caused by SESN2 inhibition (Fig. 6A,B). Inhibition of mTOR mRNA expression inhibited (P < 0.05) expression of CSN1S1, CSN1S2, CSN2 and CSN3, and inhibition of SESN2 increased each; however, inhibition of mTOR blocked (P < 0.05) the SESN2 responses (Fig. 6C).
Figure 6.
SESN2 negatively regulates cell proliferation and casein synthesis via mTORC1 pathway in CMEC. (A) Cell proliferation of CMEC treated with si-SESN2, si-mTOR, and si-SESN2/si-mTOR was analyzed by MTT assay, the date of “B group” was defined as “1”; (B) Cell cycle of CMEC treated with si-SESN2, si-mTOR, and si-SESN2/si-mTOR was analyzed by FC; (C) Expression of casein in CMEC treated with si-SESN2, si-mTOR, and si-SESN2/si-mTOR was analyzed by WB, the ratio value of “B group” was defined as “1”. (D) The model of SESN2 negative regulates the AA-mediated cell proliferation and casein synthesis via inhibiting mTORC1 pathway. Each bar represents the LSM ± SEM. In the bar charts, different superscript lowercase letters indicate significant differences (p < 0.05). B: Cells were no transfected. Uncropped images for all Western blots can be found in Supplementary Fig. 4.
Discussion
AA regulate cell proliferation and milk protein synthesis primarily through their regulation of the mTORC1 pathway and subsequent effects on translation27–29. The observed increases in mTORC1 phosphorylation, cell growth, and casein synthesis in response to AA supplementation (Fig. 1 and Supplementary Fig. 1) herein were all consistent with previous research2,30,31 despite the lack of inclusion of lactogenic hormones in the media. Although Osorio et al.1 indicated that lactogenic hormones are required for regulation of casein expression, there is some dichotomy between requirements to initiate lactation from a nonlactating state and maintenance of an existing lactational state, particularly in ruminants. Clearly ruminants are much less sensitive to prolactin than rodents, and the role of prolactin may be more associated with long-term maintenance of the cell numbers32. Ruminants also appear to have little to no sensitivity to cortisol after lactation has been initiated33. In our study, the CMEC were isolated and cultured from the udder of a mid-lactation healthy Holstein dairy cow, i.e. expression of caseins had been initiated. Additionally, CMEC were cultured prior to experiments in DMEM/F12 media containing 10% FBS. As FBS contains insulin, cortisol, and prolactin34, maintenance of expression of caseins by the cells would have been supported by the FBS during cell growth35–37. The FBS was removed during the experiments with no hormone replacement to avoid the potential of saturating the mechanisms utilized by AA through the inclusion of hormones at supraphysiological concentrations. While one cannot rule out any effects of hormone removal, the CMEC continued to transcribe and translate caseins for the 18 h period required to complete the casein measurements. As identical hormone conditions were used for all treatments, any potential effects of hormone removal would not have affected treatment comparisons. Additional work will be required to assess interactions among these hormones and AA.
The activation of mTORC1 is important for cell growth and protein synthesis, but how AA affect mTORC1 is still uncertain. A number of independent reports have identified several potential pieces of the mechanism, but a clear picture has not yet emerged. They include AA activation of mTORC1 via protein phosphatase 2 oligomer containing the PR61ε-targeting subunit (PP2AT61ε), Ste20 family kinase MAP4K338; an inside-out mechanism39; a G protein coupled receptor (GPCR) T1R1/T1R340; PB1-containing kinase MEEK3/p38δ/p62/E3-ubiquitin ligase TRAF641; and Sestrins/GATOR2/GATOR122,25. We have shown herein that in CMEC, the expression of SESN2 was significantly decreased in response to EAA or AA supply (Fig. 1D), which is consistent with the results of Chantranupong et al.22 and Parmigiani et al. 23 in HEK 293 T and Kimball et al.26 in mouse embryonic fibroblasts (MEF) cells. These results support SESN2 as the mechanism of regulation of AA effects on the mTORC1 pathway in CMEC.
SESN2, the product of hypoxia-inducible gene 95 (Hi95), is a member of the sestrins family42. Previous studies have shown that the expression of SESN2 was induced by DNA damage, oxidative stress and glucose starvation43,44. Expression of SESN2 was stimulated by genotoxic stress via tumor suppressor p5342,45 in HEK293 and MCF-7 cells; by glucose starvation through activation of transcription factor 4 (ATF4) and nuclear factor (erythroid-derived 2)-like 2 (NRF2) transcription factors43,46 in human lung adenocarcinoma H1299 and H460 cell lines and MEF; and by AA deprivation acting through GCN2, ATF4 in MEF cells47. In this study, we showed that the expression of SESN2 and GCN2 was significantly decreased in response to increased EAA or AA supply (Fig. 1D), suggesting that AA starvation stimulates SESN2 and GCN2 expression. The AA response was elicited by individual AA including Leu, Ile, Val, Met and Gln (Supplementary Fig. 2), which is consistent with the findings of Ye et al. in MEF44.
Sestrins (in particular, SESN2) play an important metabolic regulation role in mammals. Early studies concluded that SESN2 inhibited the mTORC1 pathway through AMPK-mTORC1 signaling48,49. However, recent studies demonstrated that SESN2 regulates mTORC1 in mouse embryonic fibroblast (MEF), HEK-293T, and Hela cells22,26,50 by binding to GATOR2, which disrupts the GATOR1/GATOR2 complex thus releasing GATOR1 to inhibit mTORC124,25. Kimball et al. reported that SESN2 regulates the mTORC1 pathway depending on its phosphorylation state which is driven by Leu deprivation, and no other EAA26. Control can also be exerted through changes in SESN2 gene expression. Ye et al. and Wolfson et al. showed that Ile, lysine, Val, Gln and Met regulate the expression of SESN2 which, in turn, inhibits the mTORC1 pathway25,47. In this study, we found that Leu, Ile, Val, Met and Gln independently regulated the mTORC1 pathway through changes in expression of SESN2 (Fig. 1D and Supplementary Fig. 2), but Arg and Lys regulation of the mTORC1 pathway was not through changes in expression of SESN2 (Supplementary Fig. 3B). These findings are consistent with the results of Ye et al.47, and collectively indicate SESN2 can respond to many, but not all, individual AA in CMEC.
The current work cannot differentiate between the above alternatively proposed upstream mechanisms; however, it clearly demonstrates that SESN2 is involved in mediating AA effects on mTORC1 and that SESN2 lies upstream of mTORC1. It also extends prior observations in other species and cell types to CMEC. Cell proliferation, casein synthesis and activation of the mTORC1 pathway were all significantly increased with SENS2 inhibition and significantly decreased with SESN2 overexpression (Figs 2A,B, 3A,B, 4A,B and 5B). Additionally, responses to AA (EAA+ and AA+) were significantly blocked when SESN2 was overexpressed (Figs 2D, 3C,D, 4A,B and 5C,D). Finally, the effects of SESN2 over or under expression were significantly blocked when mTORC1 protein expression was inhibited (Fig. 6). The potential system is schematically summarized in Fig. 6D.
Because AA act as both a substrate and a regulator of protein synthesis, it is of interest to discern the effects of each mechanism. From Figs 3 and 4, it is clear that essentially all of the mTORC1 and casein responses to EAA (as compared to AA−) and total AA (as compared to EAA+) were significantly blocked by overexpression of SESN2, supporting the conclusion that cell signaling is the primary controller of rates of protein synthesis. Had AA exerted any control as a substrate, one would expect to see a response to the addition of AA independent of the expression levels of SESN2, which was not the case.
The results also demonstrate that NEAA are impacting SESN2 with subsequent effects on mTORC1 signaling, cellular growth rates, and casein synthesis. Appuhamy et al. and Arriola et al. observed independent responses to multiple EAA6,51; however, preliminary screenings of NEAA, using a bovine tissue slice model, did not show any responses to the NEAA52,53. The current results provide evidence that the NEAA should be screened for regulatory activity and possibly included in models of AA effects on mammary function54. This is an important finding as it potentially adds considerable complexity to the system where the response surface may have as many as 20 dimensions, thus making parameterization of the system exceedingly difficult.
In summary, the present work shows that SESN2 is an important negative regulatory factor which was induced by EAA and NEAA starvation, and it negatively regulates AA-mediated cell proliferation and casein synthesis via inhibition of the mTORC1 pathway in CMEC.
Materials and Methods
All experiments involving dairy cattle were conducted according to the principles of the Chinese Academy of Agricultural Sciences Animal Care and Use Committee (Beijing, China), which approved study protocols.
Cell preparation and treatments
Mammary gland tissue was dissected from the udder of mid-lactation healthy Holstein dairy cow and cut into 1 mm3 pieces using sterile techniques. The explants were plated into 25 cm2 flasks (Corning, USA) coated with 5 mg/ml sterile rat tail collagen type II (diluted with 0.006 M acetic acid) (Shengyou, Hangzhou, China) and cultured with DMEM/F12 media (11320033, Gibco, USA) containing 10% fetal bovine serum (FBS), 10,000U of penicillin, 10,000U of streptomycin, and 25 mg/mL of amphotericin B in an incubator at 37 °C in and atmosphere of 5% CO2. At 80% confluence, the explants were removed from the culture. Epithelial and fibroblast cells were segregated using 0.25% trypsin and 0.15% trypsin plus 0.02% ethylenediaminetetraacetic acid (EDTA) (C0203, Beyotime, China) as previously described55 yielding primary cow mammary epithelial cells (CMEC).
For the experiments, CMEC were plated into 6 well plates at a density of 1.0 × 105 cells per well and cultured with DMEM/12 containing 10% FBS. When they reached 80% confluence, cells were starved of AA (cells cultured with DMEM/F12 media without FBS and devoid of all AA) for 12 h, and then treated with DMEM/F12 media without FBS and devoid of all AA (AA−), containing only essential AA (EAA, containing lysine, methionine, valine, leucine, isoleucine, tryptophan, phenylalanine and threonine in this manuscript) at normal DMEM/F12 concentrations (EAA+), or complete DMEM/F12 (AA+) for 6 h. For cell growth tests, treatment duration was 24 h. The concentrations of each EAA in the EAA+ treatment were those of normal DMEM/F12 medium. All AA used in the study were obtained from Sigma (San Francisco, USA). The main effects of AA were also assessed with overexpression (SESN2 GO) or inhibition of SESN2 (si-SESN2) yielding the additional treatments of AA−/si-SESN2, EAA+/SESN2 GO, AA+/SESN2GO, si-mTOR and si-SESN2/si-mTOR.
Cell growth
Cell growth was analyzed with 3-(4,5)-dimethylthiahiazo (-z-y1)-3, 5-di-phenytetrazoliumromide (MTT) (M5655, Sigma, San Francisco, USA). CMEC were plated in 96 well plates at a density of 5.0 × 104 cells/ml and cultured with DMEM/12 containing 10% FBS. After 12 h, media were switched to an AA and FBS free form followed by treatment media (DMEM/F12 media devoid of all AA, containing only EAA or complete DMEM/F12, Leagene Biotechnology, Beijing, China) for 24 h. At the end of the treatment period, MTT (5 mg/mL diluted in PBS) was added to the media at a final concentration of 0. 5 mg/mL and the cells were cultured for an additional 4 h. The media were then discarded, and 150 μl of DMSO (D2650, Sigma, USA) was added to each well. The plates were shaken for 10 minutes at room temperature, and the absorbance at 450 nm was determined using an automated enzyme-linked immunosorbent assay (ELISA) reader (MD, Spectramax M3, USA)56. The relative growth rate of the treated cells as compared to the control cells (AA− group) was reported. This experiment was repeated with three different batches of cells.
Cell cycle
The cell cycle was analyzed by flow cytometry (FC) as previously reported57. The CMEC were plated into 6 well plates and treated as described for cell growth analysis, but the density of plated cells was 1.0 × 105 cells per well. Cells were digested with 0.25% trypsin-0.02% EDTA, harvested, washed 3 times with PBS (135 mM NaCl, 4.7 mM KCl, 10 mM Na2HPO4, 2 mM NaH2PO4, pH = 7.4), fixed overnight with ice-cold 70% ethanol at 4 °C, and washed 3 times with PBS followed by incubation with 1 mL RNase A (0.1 mg/mL, diluted with PBS, Sigma, USA) for 0.5 h at 37 °C, and 3 additional washes with PBS. The cells were then incubated with 1 mL propidium iodide (PI, 50 μg/mL, diluted in PBS, Sigma, USA) for 0.5 h at 37 °C in a darkened environment, washed 3 times with PBS, and re-suspended in PBS. The cell cycle was analyzed using FACS Calibur (Becton-Dickinson, USA). The resulting data were analyzed using Modfit software58.
Protein expression
Protein expression was analyzed by Western blotting (WB) analysis. Treated cells were harvested with a lysis buffer (P0013, Beyotime, China), and total protein concentrations were analyzed using a BCA Protein Assay Kit (P0011, Beyotime, China). About 30 μg of total protein was separated on a 10% SDS-PAGE gel and transferred onto a polyvinylidene fluoride (PVDF) membrane (FFP39, Beyotime, China) using 200 mA of constant electrical current. The membrane was blocked with TBST buffer (1.21 g of Tris, 5.84 g of NaCl and 1 mL of Tween-20 in 1 L ddH2O, pH 7.5) containing 5% skim milk (M/V) for 1.5 h at room temperature (about 25 °C), and then incubated with primary antibody (diluted with TBST containing 5% skim milk) for 1.5 h at room temperature or overnight at 4 °C. The membrane was washed 3 times (5 min/time) with TBST and then incubated with HRP-conjugated secondary antibody (diluted with TBST containing 5% skim milk, 1:2000, Bioss, China) for 1 h at room temperature. The membrane was washed 3 times (10 min/time) with TBST, and the chemiluminescence of HRP was visualized with Super ECL Plus (32106, Thermo, USA). Protein band intensity was analyzed with ImageJ2X software. The β-actin was use as the reference protein. The primary antibodies used were the following: anti-CSN1S1 (1:1000, SAB1401093, Sigma, USA), anti-CSN1S2 (1:500, bs-10034R, Bioss, China), anti-CSN2 (1:500, orb18512, Biorbyt, UK), anti-CSN3 (1:1000, SAB1401094, Sigma, USA), anti-β-actin (1:1000, 4967, Cell Signaling Technology, USA), anti-mTOR (1:1000, 2972, Cell Signaling Technology, USA), anti-p-mTOR (Ser2481) (1:1000, 2974, Cell Signaling Technology, USA), anti-Raptor (1:500, sc-27744, Santa Cruz, USA), anti-S6K1 (1:500, sc-230,Santa Cruz, USA), anti-p-S6K1 (Thr389) (1:500, sc-11759, Santa Cruz, USA), anti-GCN2 (1:500, sc-374609, Santa Cruz, USA) and anti-SESN2 (1:500, sc-292558, Santa Cruz, USA).
Plasmid construction and transfection
Plasmid construction was performed as previously reported59. Total RNA of CMEC was extracted using a Trizol reagent (15596-026, Invitrogen, USA), and the cDNA was synthesized with M-MLV reverse transcriptase (2641 A, Takara, China). The CDS area of SESN2 was amplified with PCR. The SESN2 primers (GenBank: BT021909.1) were the following: Forward: 5′-CGGAATTCCACACCATGATCGTTGCGG-3′, the EcoR I site is underlined; and Reverse: 5′-GAAGATCTCAGGTGAGTAAATGGGCTTCC-3′, the Bgl II site is underlined. The PCR product was sequenced (BGI, China) and then subcloned into the MCS of eukaryotic expression vector pCMV-C-Flag (D2632, Beyotime, China). The plasmid will be subsequently refered to as SESN2-Flag.
For the SESN2 GO, the transfection of SESN2-Flag was performed. The CMEC were plated into 6 well plates a density of 1.0 × 105 cells per well, and at about 70% confluence, the medium was changed with OPTI-MEM I medium (31985-070, Invitrogen, USA). SESN2-Flag vector and pCMV-C-Flag empty vector were transfected with Lipofectamine 2000 transfection reagent (11668019, Invitrogen, USA) according to the manufacturer’s instructions. Briefly, for cells of each well to be transfected, 5 ìg DNA plasmid and 10 μl Lipofectamine 2000 transfection reagent were diluted into 250 ìl OPTI-MEM I medium, respectively. After incubating for 5 min at room temperature, the diluted DNA plasmid and Lipofectamine 2000 transfection reagent were mixed, and incubated for 20 min at room temperature. Then the mixture was added to well containing cells. After 6 h, the OPTI-MEM I media were switched to DMEM/12 media containing 10% FBS.
Small interfering RNA transfection
The specific siRNA of genes indicated in this experiment and the negative control siRNA were synthesized (GenePharma, Shanghai, China). The si-SESN2 was transfected using Lipofectamine 2000 transfection reagent according to the manufacturer’s instructions. The operation process was the as same as that of SESN2-Flag DNA plasmid transfection, but the amount of siRNA and Lipofectamine 2000 transfection reagent were 100 pM and 10 μl per well, respectively. The siRNA sequences used in this study are shown in Table 1.
Table 1.
List of siRNA sequences.
| Gene name | Sequences of siRNA (5′-3′) | |
|---|---|---|
| SESN2 | sense | CCUUUGCAAACCCAGAUAUTT |
| antisense | AUAUCUGGGUUUGCAAAGGTT | |
| mTOR | sense | CCACUCGAAUUGGAAGAUUTT |
| antisense | AAUCUUCCAAUUCGAGUGGTT | |
| Negative control | sense | UUCUCCGAACGUGUCACGUTT |
| antisense | ACGUGACACGUUCGGAGAATT | |
Immunofluorescence
The CMEC were plated on cover slips in 6 well plates with the concentration of 1.0 × 105 cells per well and cultured with DMEM/12 containing 10% FBS for 12 h. The cells were treated for 6 h and used for lysosomal localization analysis of mTOR in the following manner. Media were removed. Cells were washed 3 times with PBS, fixed with ice-cold methanol at 4 °C for 10 min, and washed 3 times (5 min/time) with TBST. Cells were blocked with a blocking buffer (P0102, Beyotime, China) for 1 h at 37 °C and then incubated with an anti-mTOR rabbit polyclonal primary antibody [1:200, Cell Signaling Technology, USA, diluted with primary antibody dilution buffer (P0103, Beyotime, China)] and an anti-LAMP2 goat polyclonal primary antibody (1: 100, Santa Cruz, USA, diluted with primary antibody dilution buffer) for 1 h at 37 °C. Cells were subsequently washed 3 times (5 min/time) with TBST and incubated with mouse-anti-rabbit alexa fluor 488-conjugated secondary antibody [Bioss, China, diluted 1:100 with secondary antibody dilution buffer (P0108, Beyotime, China)] and mouse-anti-goat alexa fluor 647-conjugated secondary antibody (Bioss, China, diluted 1: 100 with secondary antibody dilution buffer) for 45 min at 37 °C in the dark. The cells were washed 3 times (5 min/time) with TBST, and incubated with 4′,6-diamidino-2-phenylindole (DAPI) (0.5–10 μg/ml, C1002, Beyotime, China) for 10 min at 37 °C in a dark environment, and then washed 3 times (10 min/time) with TBST. The cells were fixed on the slides with antifade mounting medium (P0126, Beyotime, China) and observed with laser scanning confocal microscopy (LEICA, Germany). Co-localization was analyzed with ImageJ2X software, and at least 10 treated cells in each co-localization were used to analyze the index of co-localization.
Statistical analysis
Date were analyzed using the Proc Mixed procedure of SAS (version 9.1, SAS Institute, Cary, NC). The fixed effect was treatment for all models. For growth rate measurements which were conducted across 3 cell preparations, cell batch was included as a random effect. For all other measurements, plate was included as a random effect. Least squares means (LSM) of treatments were compared using Tukey’s multiple comparisons procedure, and the effects were considered significant at a probability value of P < 0.05. Data were presented as LSM ± standard error of the means (SEM). All LSM represented at least three independent experiments.
Electronic supplementary material
Acknowledgements
This study was financially supported by the Special Fund for Agro-scientific Research in the Public Interest (201403071), the Agricultural Science and Technology Innovation Program (ASTIP-ISA12), and the Project of Risk Assessment on Raw Milk (GJFP2016008).
Author Contributions
Chaochao Luo, developed the concept and design of the experiments, performed experimental work, analyzed data and drafted the manuscript; Shengguo Zhao helped with design of the experiment, data analysis, and drafting of the manuscript; Muchen Zhang and Yanan Gao assisted in the performance of the experimental work; Mark D Hanigan helped with drafting the manuscript and the statistical analyses; Nan Zheng and Jiaqi Wang helped with development of the concept and experimental design, supervised the project, and helped write the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Chaochao Luo and Shengguo Zhao contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-22208-w.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Osorio JS, Lohakare J, Bionaz M. Biosynthesis of milk fat, protein, and lactose: roles of transcriptional and posttranscriptional regulation. Physiol Genomics. 2016;48:231–256. doi: 10.1152/physiolgenomics.00016.2015. [DOI] [PubMed] [Google Scholar]
- 2.Bionaz M, Loor JJ. Gene networks driving bovine mammary protein synthesis during the lactation cycle. Bioinform Biol Insights. 2011;5:83–98. doi: 10.4137/BBI.S7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bionaz, M., Hurley, W. L. & Loor, J. J. Milk Protein Synthesis in the Lactating Mammary Gland: Insights from Transcriptomics Analyses. In: Milk Protein. 285-324 (Hurley WLInTech, 2012).
- 4.Rhoads R, Grudzien-Nogalska E. Translational regulation of milk protein synthesis at secretory activation. J Mammary Gland Biol Neoplasia. 2007;12:283–292. doi: 10.1007/s10911-007-9058-0. [DOI] [PubMed] [Google Scholar]
- 5.Hu H, Zhang YD, Zheng N, Cheng JB, Wang JQ. The effect of heat stress on gene expression and synthesis of heat-shock and milk proteins in bovine mammary epithelial cells. Anim. Sci. J. 2016;87:84–91. doi: 10.1111/asj.12375. [DOI] [PubMed] [Google Scholar]
- 6.Arriola Apelo SI, et al. Isoleucine, leucine, methionine, and threonine effects on mammalian target of rapamycin signaling in mammarytissue. J Dairy Sci. 2014;97:1047–1056. doi: 10.3168/jds.2013-7348. [DOI] [PubMed] [Google Scholar]
- 7.Lu LM, et al. Comparative phosphoproteomics analysis of the effects of L-methionine on dairy cow mammary epithelial cells. Can. J. Anim. Sci. 2012;92:433–442. doi: 10.4141/cjas2012-063. [DOI] [Google Scholar]
- 8.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by AAs. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han JM, et al. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149:410–424. doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- 11.So L, et al. The 4E-BP-eIF4E axis promotes rapamycin-sensitive growth and proliferation in lymphocytes. Sci Signal. 2016;9:ra57. doi: 10.1126/scisignal.aad8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fingar DC, et al. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol Cell Biol. 2004;24:200–216. doi: 10.1128/MCB.24.1.200-216.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor PM. Role of AA transporters in AA sensing. Am J Clin Nutr. 2014;99:223S–230S. doi: 10.3945/ajcn.113.070086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 15.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 16.Budanov AV, et al. Identification of a novel stress-responsive gene Hi95 involved in regulation of cell viability. Oncogene. 2002;21:6017–6031. doi: 10.1038/sj.onc.1205877. [DOI] [PubMed] [Google Scholar]
- 17.Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 18.Chen YS, Chen SD, Wu CL, Huang SS, Yang. DI. Induction of sestrin2 as an endogenous protective mechanism against amyloid beta-peptide neurotoxicity in primary cortical culture. Exp Neurol. 2014;253:63–71. doi: 10.1016/j.expneurol.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 19.Rhee SG, Bae SH. The antioxidant function of sestrins is mediated by promotion of autophagic degradation of Keap1 and Nrf2 activation and by inhibition of mTORC1. Free Radic Biol Med. 2015;88:205–211. doi: 10.1016/j.freeradbiomed.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 20.Lee JH, et al. Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science. 2010;327:1223–1228. doi: 10.1126/science.1182228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchis-Gomar F. Sestrins: novel antioxidant and AMPK-modulating functions regulated by exercise? J Cell Physiol. 2013;228:1647–1650. doi: 10.1002/jcp.24338. [DOI] [PubMed] [Google Scholar]
- 22.Chantranupong L, et al. The Sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep. 2014;9:1–8. doi: 10.1016/j.celrep.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parmigiani A, et al. Sestrins inhibit mTORC1 kinase activation through the GATOR complex. Cell Rep. 2014;9:1281–1291. doi: 10.1016/j.celrep.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saxton RA, et al. Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science. 2016;351:53–58. doi: 10.1126/science.aad2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolfson RL, et al. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science. 2016;351:43–48. doi: 10.1126/science.aab2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimball SR, Gordon BS, Moyer JE, Dennis MD, Jefferson LS. Leucine induced dephosphorylation of Sestrin2 promotes mTORC1 activation. Cell Signal. 2016;28:896–906. doi: 10.1016/j.cellsig.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doepel L, et al. Milk protein synthesis as a function of AA supply. J Dairy Sci. 2004;87:1279–1297. doi: 10.3168/jds.S0022-0302(04)73278-6. [DOI] [PubMed] [Google Scholar]
- 28.Bruhat A, et al. AAs as regulators of gene expression in mammals: molecular mechanisms. Biofactors. 2009;35:249–257. doi: 10.1002/biof.40. [DOI] [PubMed] [Google Scholar]
- 29.Rezaei R, Wu Z, Hou Y, Bazer FW, Wu G. AAs and mammary gland development: nutritional implications for milk production and neonatal growth. J Anim Sci Biotechnology. 2016;7:20. doi: 10.1186/s40104-016-0078-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toerien CA, Trout DR, Cant JP. Nutritional stimulation of milk protein yield of cows is associated with changes in phosphorylation of mammary eukaryotic initiation factor 2 and ribosomal s6 kinase 1. J Nutr. 2010;140:285–292. doi: 10.3945/jn.109.114033. [DOI] [PubMed] [Google Scholar]
- 31.Arriola Apelo SI, et al. Casein synthesis is independently and additively related to individual essential AA supply. J. Dairy Sci. 2014;97:2998–3005. doi: 10.3168/jds.2013-7422. [DOI] [PubMed] [Google Scholar]
- 32.Lacasse P, et al. New developments on the galactopoietic role of prolactin in dairy ruminants. J. Dairy Sci. 2012;43:154–160. doi: 10.1016/j.domaniend.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 33.Ely LO, Baldwin RL. Effects of adrenalectomy upon ruminant liver and mammary function during lactation. J. Dairy Sci. 1976;59:491–503. doi: 10.3168/jds.S0022-0302(76)84231-2. [DOI] [PubMed] [Google Scholar]
- 34.Biswas R, Vonderhaar BK. Role of serum in the prolactin responsiveness of MCF-7 human breast cancer cells in long-term tissue culture. Cancer Res. 1987;47:3509–3514. [PubMed] [Google Scholar]
- 35.Hu H, et al. In vitro culture and characterization of a mammary epithelial cell line from Chinese Holstein dairy cow. PLoS One. 2009;4:e7636. doi: 10.1371/journal.pone.0007636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ao J, et al. Tudor-SN Regulates Milk Synthesis and Proliferation of Bovine Mammary Epithelial Cells. Int J Mol Sci. 2015;16:29936–29947. doi: 10.3390/ijms161226212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, et al. MicroRNA-152 regulates DNA methyltransferase 1 and is involved in the development and lactation of mammary glands in dairy cows. PLoS One. 2014;9:e101358. doi: 10.1371/journal.pone.0101358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan L, et al. PP2A T61 epsilon is an inhibitor of MAP4K3 in nutrient signaling to mTOR. Mol Cell. 2010;37:633–642. doi: 10.1016/j.molcel.2010.01.031. [DOI] [PubMed] [Google Scholar]
- 39.Zoncu R, et al. mTORC1 senses lysosomal AAs through an inside-out mechanism that requires the vacuolar H (+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wauson EM, et al. The G protein-coupled taste receptor T1R1/T1R3 regulates mTORC1 and autophagy. Mol Cell. 2012;47:851–862. doi: 10.1016/j.molcel.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Linares JF, et al. AA Activation of mTORC1 by a PB1-Domain-Driven Kinase Complex Cascade. Cell Rep. 2015;12:1339–1352. doi: 10.1016/j.celrep.2015.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jegal KH, et al. Eupatilin induces Sestrin2-dependent autophagy to prevent oxidative stress. Apoptosis. 2016;21:642–656. doi: 10.1007/s10495-016-1233-6. [DOI] [PubMed] [Google Scholar]
- 44.Ding B, et al. Sestrin2 is induced by glucose starvation via the unfolded protein response and protects cells from non-canonical necroptotic cell death. Sci. Rep. 2016;6:22538. doi: 10.1038/srep22538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones RG, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 46.Seo K, Ki SH, Shin SM. Sestrin2-AMPK activation protects mitochondrial function against glucose deprivation-induced cytotoxicity. Cell Signal. 2015;27:1533–43. doi: 10.1016/j.cellsig.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 47.Ye J, et al. GCN2 sustains mTORC1 suppression upon AA deprivation by inducing Sestrin2. Genes Dev. 2015;29:2331–2336. doi: 10.1101/gad.269324.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim H, et al. Janus-faced Sestrin2 controls ROS and mTOR signaling through two separate functional domains. Nat Commun. 2015;6:10025. doi: 10.1038/ncomms10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X, Niu Y, Yuan H, Huang J, Fu L. AMPK binds to Sestrins and mediates the effect of exercise to increase insulin-sensitivity through autophagy. Metabolism. 2015;64:658–665. doi: 10.1016/j.metabol.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 50.Peng M, Yin N, Li MO. Sestrins function as guanine nucleotide dissociation inhibitors for Rag GTPases to control mTORC1 signaling. Cell. 2014;159:122–133. doi: 10.1016/j.cell.2014.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Appuhamy JA, Knoebel NA, Nayananjalie WA, Escobar J, Hanigan MD. Isoleucine and leucine independently regulate mTOR signaling and protein synthesis in MAC-T cells and bovine mammary tissue slices. J Nutr. 2012;142:484–491. doi: 10.3945/jn.111.152595. [DOI] [PubMed] [Google Scholar]
- 52.Felock, A., Arriola, S. I. A., Garnett, R. L. & Hanigan, M. D. Effects of AICAR, Rapamycin, and Non-essential AAs on Cell Signaling in Bovine Mammary Tissue. J. Dairy Sci. 97, E–Suppl. 1 (2014).
- 53.Garnett, R. L. et al. Inhibitory Factors of Casein Synthesis in Mammary Tissue of Lactating Dairy Cows. J. Dairy Sci. 97, E–Suppl. 1 (2014).
- 54.Castro Marquez JJ, Arriola Apelo SI, Appuhamy JA, Hanigan MD. Development of a model describing regulation of casein synthesis by the mammalian target of rapamycin (mTOR) signaling pathway in response to insulin, AAs, and acetate. J. Dairy Sci. 2016;99:6714–6736. doi: 10.3168/jds.2015-10591. [DOI] [PubMed] [Google Scholar]
- 55.Zhao K, Liu HY, Zhou MM, Liu JX. Establishment and characterization of a lactating bovine mammary epithelial cell model for the study of milk synthesis. Cell Biol Int. 2010;34:717–721. doi: 10.1042/CBI20100023. [DOI] [PubMed] [Google Scholar]
- 56.Liu J, Zhang W, Jing H, Popovich DG. Bog bilberry (Vaccinium uliginosum L.) extract reduces cultured Hep-G2, Caco-2, and 3T3-L1 cell viability, affects cell cycle progression, and has variable effects on membrane permeability. J Food Sci. 2010;75:H103–107. doi: 10.1111/j.1750-3841.2010.01546.x. [DOI] [PubMed] [Google Scholar]
- 57.Wang LN, et al. Leucyl-tRNA synthetase regulates lactation and cell proliferation via mTOR signaling in dairy cow mammary epithelial cells. Int J Mol Sci. 2014;15:5952–5969. doi: 10.3390/ijms15045952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kampa-Schittenhelm KM, et al. Cell cycle-dependent activity of the novel dual PI3K-MTORC1/2 inhibitor NVP-BGT226 in acute leukemia. Mol. Cancer. 2013;12:46. doi: 10.1186/1476-4598-12-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luo CC, Yin DY, Gao XJ, Li QZ, Zhang L. Goat mammary gland expression of Cecropin B to inhibit bacterial pathogens causing mastitis. Anim Biotechnol. 2013;24:66–78. doi: 10.1080/10495398.2012.745417. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.