Significance
The Notch signaling pathway performs a vital role in biological processes ranging from stem cell maintenance to cell viability. This highly conserved pathway must be tightly controlled, since defects in signaling can promote disease. The E3 ubiquitin ligase DTX1 has emerged as a key negative regulator of Notch signaling, where Notch ubiquitination by DTX1 is thought to control intracellular sorting decisions of the receptor. Here we show that DTX1 can regulate Notch activity independent of directly ubiquitinating the receptor, suggesting that DTX1 targets other factors involved in Notch transport. Using an activity-based screen for DTX1 substrates, we identify PI5P4Kγ, a lipid kinase, and discover that PI5P4Kγ and DTX1 have opposing activities in regulating Notch transit through recycling endosomes.
Keywords: Notch, recycling, endocytosis, PI5P4Kγ
Abstract
Notch signaling is an evolutionarily conserved pathway that is essential for development, where it controls processes ranging from cell differentiation to survival. Transport through endosomes is a critical step in regulating Notch signaling capacity, where the E3 ubiquitin ligase DTX1 is thought to control Notch1 intracellular transport decisions by direct receptor ubiquitination. However, how DTX1 regulates Notch1 transport within endosomes and the consequence of Notch1 ubiquitination by DTX1 remain unresolved. Here we demonstrate that DTX1 colocalizes with Notch1 on tubulovesicular recycling endosomes. We find that DTX1 silencing leads to enhanced Notch1 recycling from this compartment to the cell surface via a rab4a-mediated transport route. This, in turn, increases Notch1 cell-surface levels and enhances signaling. Surprisingly, we discovered that DTX1 depletion also elevates Notch1 activity mediated by a mutant form of the receptor that lacks lysine residues for ubiquitination, suggesting that DTX1 targets additional factors. Using an activity-based screen for ubiquitination targets, we identified multiple DTX1 substrates including PI5P4Kγ, a lipid kinase involved in PI(4,5)P2 production. Immunolocalization analysis reveals that PI5P4Kγ, like DTX1 and Notch1, is present on tubulovesicular recycling endosomes. However, in contrast to DTX1, Notch1 signaling is inhibited by pharmacological inactivation or siRNA depletion of PI5P4Kγ. Moreover, loss of PI5P4Kγ activity decreases Notch1 recycling rates and reduces receptor cell-surface levels. Collectively, these findings argue that PI5P4Kγ positively regulates the Notch pathway by promoting receptor recycling. Additionally, they support a model where DTX1 controls Notch1 endosomal sorting decisions by controlling PI5P4Kγ-mediated production of PI(4,5)P2.
The Notch signaling pathway is central to processes ranging from cell-fate specification to cell viability (1). In mammals, signaling is initiated when Notch binds to one of several ligands (Delta-like 1, 3, and 4 or Jagged 1 and 2) expressed on the surface of neighboring cells (2). After ligand binding, the Notch extracellular region undergoes a conformational change that exposes an ADAM family metalloprotease cleavage site (3–5). This leads to proteolytic release of the ectodomain and leaves behind a membrane-tethered Notch fragment. This fragment is subsequently cleaved by γ-secretase, which liberates the Notch intracellular domain (NICD; a transcription factor) into the cytoplasm (6). NICD subsequently translocates to the nucleus to coordinate gene expression.
Notch activity must be tightly regulated, given that defects in signaling can lead to disease. For example, Notch loss-of-function mutations give rise to aortic valve disease and keratinocyte hyperplasia (7, 8). By contrast, gain-of-function mutations lead to diseases like T-cell acute lymphoblastic leukemia and cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (9, 10). Furthermore, ex vivo and animal model studies indicate that Notch signaling can promote metastasis in a variety of cancers, including those of the prostate, breast, bone, and colon (11–14).
To ensure the appropriate timing, intensity, and duration of Notch signaling, cells must modulate receptor exposure to ligand by regulating the amount of Notch at the cell surface. Accumulating evidence points to a critical role for endosomal transport in regulating Notch activity (15). One such factor is mammalian DTX1 (deltex-1), which encodes a highly conserved RING-domain E3 ubiquitin ligase (16). Originally identified in Drosophila, Deltex was initially thought to positively regulate Notch signaling in a context-dependent manner (17, 18). However, subsequent studies argue that Deltex, in combination with Kurtz, the fly homolog of mammalian nonvisual β-arrestin, down-regulates signaling by promoting Notch degradation (19). This latter finding is consistent with a range of studies demonstrating that DTX1 antagonizes Notch activity in mammals (20–26).
While accumulating evidence argues that DTX1 negatively regulates Notch activity, its mechanism of action remains unclear. Deltex directly binds to and regulates Notch ubiquitination status (19, 27). However, the consequence of Notch ubiquitination by DTX1 has yet to be tested. Here we discovered that mammalian DTX1 acts independent of direct Notch ubiquitination, arguing that DTX1 regulates Notch activity by targeting other factors. We used an activity-based screen to determine DTX1 substrates and, in doing so, identified PI5P4Kγ, a lipid kinase. Here we provide evidence that PI5P4Kγ positively regulates signaling by promoting Notch recycling. Data presented here point to a model where DTX1-regulated lipid modification is critical to controlling Notch signaling capacity.
Results
DTX1 Negatively Regulates Notch1 Signaling.
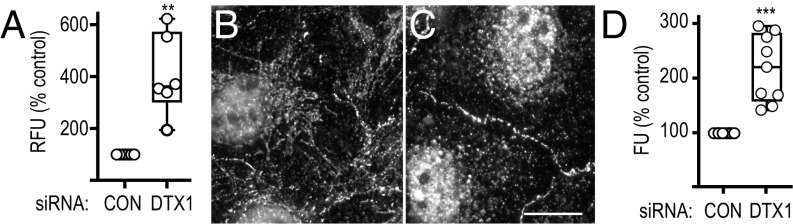
Our published findings using a recombinant Notch1 chimera suggest that DTX1 negatively regulates signaling where DTX1 depletion promotes Notch activity by elevating receptor cell-surface levels (26). To validate this observation, we evaluated endogenous Notch activity in HeLa cells, which express both Notch1 and its ligand JAG1. Following DTX1 depletion by siRNA, Notch signaling is markedly increased relative to control (Fig. 1A), in agreement with our previous findings using recombinant proteins (26). To extend our analysis, we next investigated how DTX1 depletion impacts endogenous Notch1 localization in U2OS cells given that they spread well and are more easily visualized. Immunolocalization analysis in subconfluent cell cultures reveals that Notch1 is present at the cell surface; however, a significant proportion of protein was also observed in a tubulovesicular compartment that radiates from a perinuclear region (Fig. 1B). By contrast, when DTX1 is silenced, Notch1 presence in this compartment is markedly reduced. Instead, a corresponding increase in the nucleus was observed (Fig. 1C), with an apparent increase in Notch1 at cell–cell contact sites. We quantified this later observation using a cloned single-chain variable-fragment antibody against the extracellular domain of Notch1 (28), which we fused in tandem to superfold GFP and Gaussia luciferase (scFv-N1-sfGFP-GLuc), and measured luciferase activity of surface-bound antibody. In doing so, we discovered that DTX1 depletion leads to an approximately twofold increase in receptor at the cell surface relative to control cells (Fig. 1D). Combined, these results validate our published findings and suggest that DTX1 negatively regulates Notch1 activity by preventing receptor delivery to the cell surface.
Fig. 1.
Endogenous Notch signaling and endosomal transport are negatively regulated by DTX1. (A) tTA HeLa cells were treated with control (CON) or DTX1 siRNA for 48 h and then transfected with the dual-luciferase reporter plasmid. Twenty-four hours later, endogenous Notch signaling activity was evaluated with the luciferase reporter. Firefly luciferase activity was normalized to constitutively expressed Renilla luciferase driven by a tTA-driven promoter to calculate a relative fluorescence unit (RFU). (B and C) U2OS cells were treated with control (B) or DTX1 (C) siRNA for 48 h. Cells were then processed for immunolocalization analysis using polyclonal antibodies directed against the Notch1 cytoplasmic tail. (Scale bar, 10 µm.) (D) Gaussia luciferase activity of surface-bound scFv-N1-sfGFP-GLuc on U2OS cells pretreated with the indicated siRNA. Luciferase activity was normalized to cell number. Box and whisker plots (A and D) show each data point from at least three independent experiments performed in triplicate. **P < 0.005, ***P < 0.0005.
DTX1 Inhibits Rab4a-Mediated Recycling of Notch1.
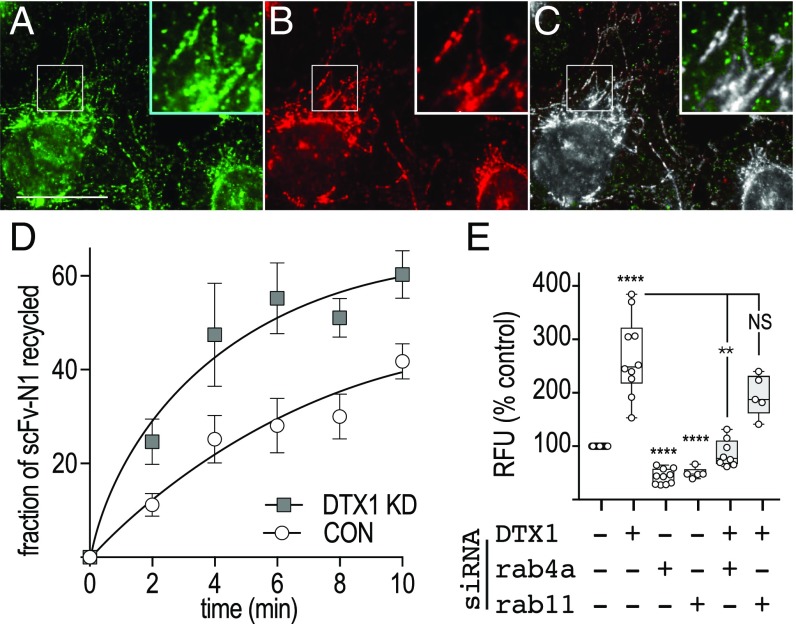
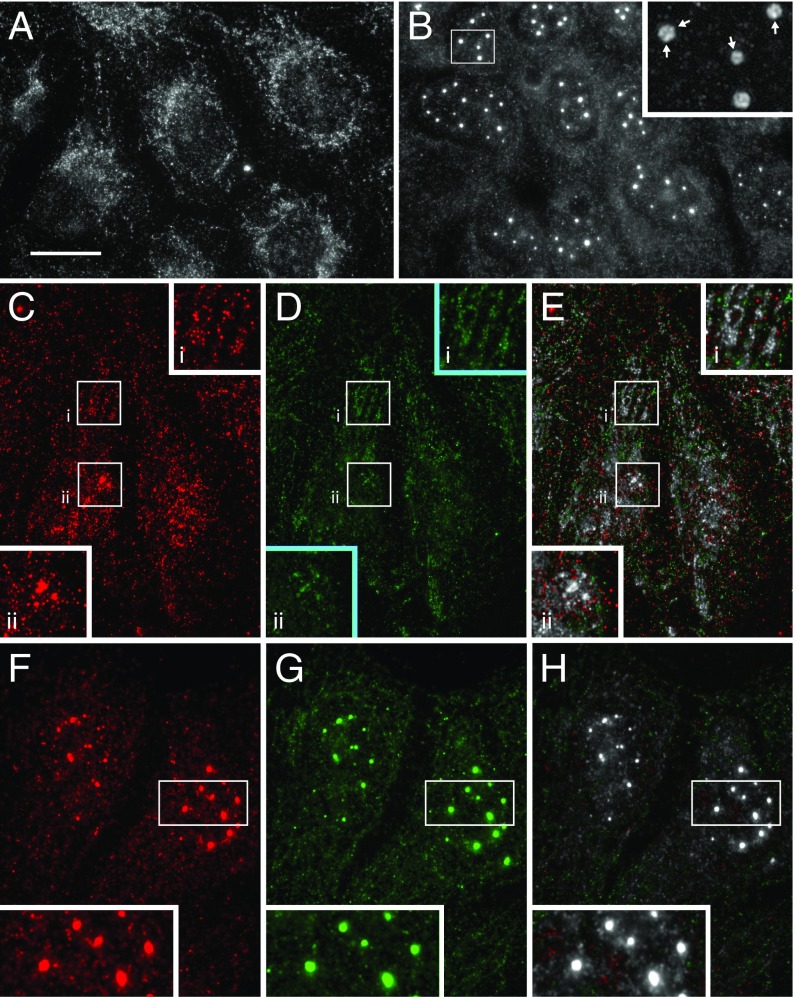
We postulated that DTX1 might limit Notch1 delivery to the plasma membrane by inhibiting receptor recycling. To test this idea, we first visualized endogenous N1 endocytosis using the recombinant antibody against Notch1. After a 15-min uptake, we found that the antibody was readily internalized and accumulated in a tubulovesicular compartment (Fig. 2A), where it colocalized with antibodies that recognize the Notch1 cytoplasmic tail (Fig. 2C). This indicates that full-length, unactivated receptor is internalized and targeted to tubulovesicular endosomes. Given this, we next performed a pulse–chase experiment to evaluate Notch1 recycling and discovered that internalized antibody is recycled at a faster rate in DTX1-depleted cells relative to control cells (Fig. 2D). This result indicates that DTX1 prevents Notch1 recycling following receptor internalization.
Fig. 2.
DTX1 inhibits rab4-mediated recycling. (A and B) U2OS cells were incubated with scFv-N1-sfGFP (A, green) antibody for 15 min at 37 °C and then processed for immunolocalization analysis with polyclonal antibodies against the Notch1 cytoplasmic tail (B, red). Boxed regions are shown at higher (2×) magnification (Insets). (Scale bar, 20 µm.) (C) A colocalized pixel map, obtained using the Colocalization Threshold plugin for ImageJ, shows colocalized pixels in grayscale. (D) U2OS cells were treated with control or DTX1-specific siRNA, and endogenous Notch1 recycling was evaluated by pulse–chase using the recombinant scFv-N1-sfGFP-GLuc antibody (Materials and Methods). (E) tTA HeLa cells were treated with the indicated siRNA combination for 48 h before transfection with plasmids encoding the dual-luciferase reporter and the Notch1 chimera CD8-NΔE. Notch signaling from the Notch chimera was measured after an additional 24-h incubation. Box and whisker plots show each data point from three independent experiments. **P < 0.005, ****P < 0.0001; NS, not significant.
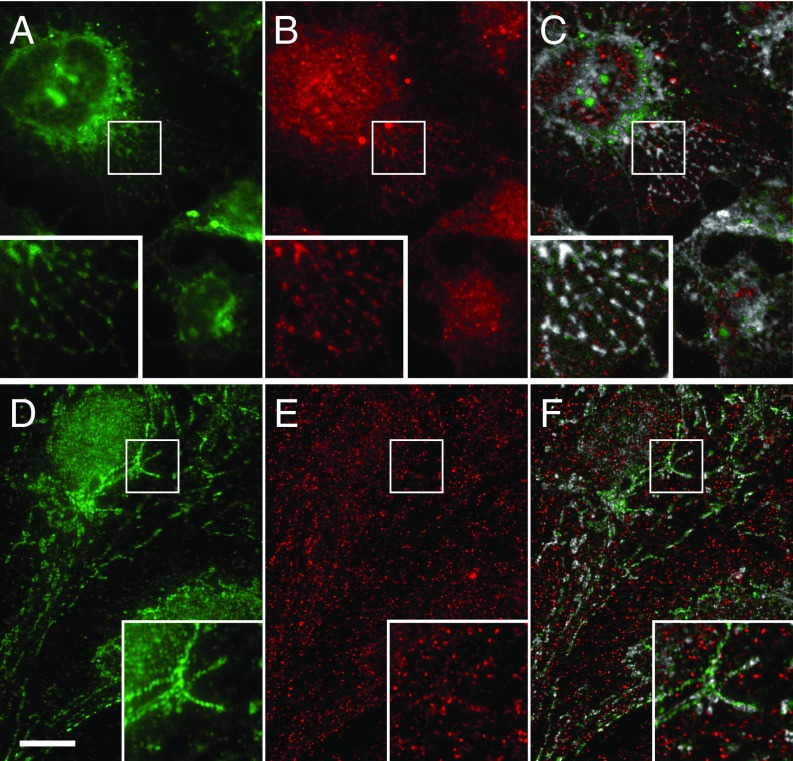
To identify the recycling pathway used by Notch following DTX1 depletion, we disrupted the rapid and slow recycling pathways by silencing rab4a and rab11, respectively, and measured Notch1 signaling activity. We find that silencing either rab4a or rab11 reduced Notch1 activity relative to control (Fig. 2E). These results indicate that receptor recycling is critical to controlling Notch1 activity. We next performed double-siRNA knockdowns to silence DTX1 and either rab4a or rab11. We reasoned that if DTX1 inhibits recycling via either pathway, then the elevated Notch1 signaling that results from DTX1 depletion should be suppressed when the relevant pathway is also impaired. Significantly, elevated Notch1 signaling levels that arise from DTX1 depletion are only suppressed when rab4a is also silenced (Fig. 2E). This finding argues that DTX1 suppresses Notch activity by preventing receptor recycling via a rab4a-dependent transport route. Consistent with this conclusion, coimmunolocalization analysis reveals that internalized scFv-N1-sfGFP antibodies colocalize with endogenous DTX1 on tubulovesicular endosomes (Fig. 3 A–C). Similarly, Notch1 was also found to colocalize with rab4a on tubulovesicular endosomes (Fig. 3 D–F). Taken together, these findings indicate that DTX1 inhibits Notch1 signaling by preventing receptor recycling via a rab4a-dependent pathway.
Fig. 3.
Notch1 colocalizes with DTX1 and rab4a on tubulovesicular endosomes. U2OS cells were analyzed by immunolocalization. Endogenous Notch1 was identified using the scFv-N1-sfGFP antibody (A and D, green), while DTX1 (B, red) and rab4a (E, red) were identified with polyclonal and monoclonal antibodies, respectively. A colocalized pixel map shows colocalized pixels in grayscale (C and F). (Scale bar, 10 μm.) Boxed regions are shown at higher (2×) magnification (Insets).
DTX1 Regulates Notch1 Activity Independent of Direct Receptor Ubiquitination.
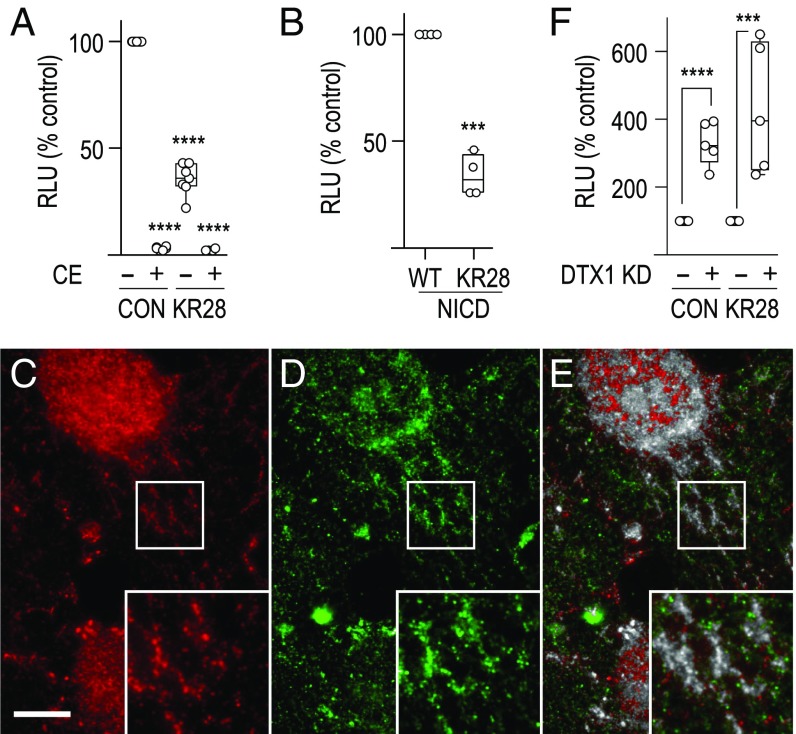
DTX1 directly binds to and regulates Notch1 ubiquitination status (27). However, the impact of direct ubiquitination of Notch1 by DTX1 has not been tested. To determine if DTX1 regulates signaling by directly ubiquitinating Notch1, we mutated all 28 lysine residues within the receptor cytoplasmic tail to arginine (KR28 Notch1) to prevent receptor ubiquitination. We then measured signaling and found that mutated KR28 Notch1 mediates reduced signaling relative to control (Fig. 4A). Although the precise reason for the diminished signaling remains to be elucidated, we note that KR28 Notch1 activity is γ-secretase–dependent, as signaling is reduced by pretreatment with compound E (CE), a γ-secretase inhibitor. Additionally, we observed a similar decrease in signaling for KR28 NICD relative to the WT control (Fig. 4B). This latter observation argues that the lysine mutations impact Notch1 activities downstream of γ-secretase cleavage, possibly by influencing nuclear targeting efficiency or interaction with cofactors. Consistent with this idea, recombinant CD8-KR28 Notch1 localizes to tubulovesicular endosomes, like those observed for endogenous receptor (Fig. 4E), suggesting that the lysine mutations do not significantly impact receptor trafficking. Importantly, however, DTX1 silencing elevates KR28 Notch1 signaling to a similar extent as the control (Fig. 4F). This indicates that DTX1 regulates Notch1 activity without the need to directly ubiquitinate the receptor. Moreover, this result reveals the presence of additional DTX1 substrates that regulate Notch1 activity.
Fig. 4.
DTX1 regulates Notch signaling independent of direct receptor ubiquitination. (A and B) tTA HeLa cells were transfected with (A) plasmid encoding Notch1 chimeras CD8-NΔE (CON) or KR28 CD8-NΔE (KR28) in the presence of 10 µM compound E (CE; +) or DMSO (−) for 16 h or (B) plasmid encoding WT NICD (WT) or KR28 NICD (KR28). Notch signaling was then measured using the dual-luciferase assay. RLU, relative luciferase unit. (C and D) Immunolocalization analysis of CD8-KR28 Notch1 with pAb antibody against the Notch cytoplasmic tail (C, red; which recognizes recombinant and endogenous protein) and mAb 51.1 that recognizes the recombinant extracellular CD8 tag (D, green). (E) A colocalized pixel map shows colocalized pixels in grayscale. (Scale bar, 10 μm.) Boxed regions are shown at higher (2×) magnification (Insets). (F) Notch signaling was measured in tTA HeLa cells treated with control (−) or DTX1 (+) siRNA and expressing the indicated control or KR28 CD8 Notch chimera. Notch signaling was normalized to the siRNA negative control for each chimera to allow a relative comparison. Box and whisker plots show each data point from three independent experiments. KD, knockdown. ***P < 0.0005, ****P < 0.0001.
ProtoArray Screen Identifies DTX1 Substrates.
To identify DTX1 substrates, we first determined the cognate E2 ubiquitin-conjugating enzyme. DTX1 self-ubiquitination is reconstituted by incubation with the ubiquitin-activating enzyme E1 and one of nine E2s [E2C, E2D1 to 4, E2E1, E2L3, E2N, and E2U (29)]. To determine the Notch1-relevant E2, we silenced each using siRNA and monitored Notch1 activity. We reasoned that if a particular E2 partnered with DTX1 to regulate Notch1 signaling, then its depletion should recapitulate the elevated signaling observed following DTX1 knockdown. Indeed, E2C or E2N silencing elevates signaling in a similar manner (Fig. S1A). We next validated DTX1-E2 ubiquitination activity in vitro by incubating DTX1 with E1, ATP, biotinylated ubiquitin, and either E2C, E2N, or E2D1 as a positive control (16). In each case, activity was reconstituted, as evidenced by DTX1 autoubiquitination (Fig. S1B). Using this reconstituted DTX1 activity, we then identified DTX1 substrates with an activity-based screen using commercially available microarray chips that contain over 9,000 recombinant human proteins. In doing so, we identified 165 DTX1 ubiquitination targets that had an average Z score greater than or equal to 3 and a coefficient of variation for two replicate signals of <0.5 (Dataset S1).
Given that DTX1 regulates receptor endosomal transport, we anticipated that substrates relevant to Notch sorting would also function in receptor trafficking. Thus, we cross-referenced the list of DTX1 substrates with genes implicated in receptor trafficking (30). This resulted in an overlap of 47 genes (Dataset S2), from which we tested a subset of 8 for their potential role in the Notch pathway. To do so, we silenced expression of each gene with siRNA and measured Notch signaling using a dual-luciferase assay (26). Not surprisingly, each factor impacted Notch1 signaling capacity when depleted (Fig. S2), where depletion of seven factors (CAMKK1, HCK, PIP5K2C, PLXDC2, PPP2R2C, PPP2R5C, and SCFD1) suppressed Notch activity. By contrast, GSK3β knockdown enhanced Notch activity, consistent with its known role as a regulator of Notch signaling (31–35). Collectively, these findings suggest the possibility that DTX1 controls Notch activity by regulating the activities of multiple substrates.
PI5P4Kγ Is Required for Notch1 Recycling.
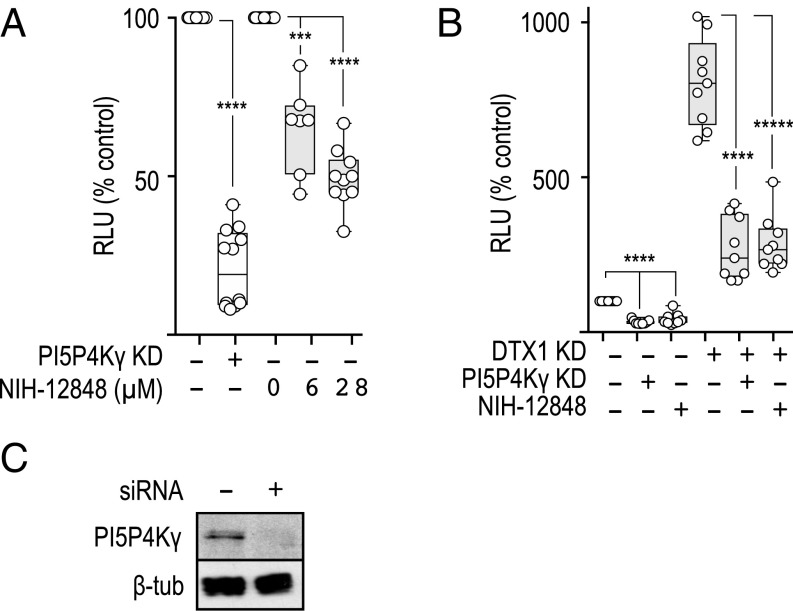
To begin unraveling the mode by which DTX1 regulates the Notch pathway, we pursued a single gene for additional investigation. Of particular interest was PIP5K2C, of which little is known regarding the biological context in which it functions (36). PIP5K2C encodes a lipid kinase (PI5P4Kγ) that phosphorylates PI5P to generate PI(4,5)P2 (37). PI5P4Kγ can be inhibited with NIH-12848, an isoform-specific drug that impairs lipid kinase activity by preventing its recruitment to PI5P (38). Therefore, we initially used the drug to validate the role of PI5P4Kγ in Notch signaling. Consistent with siRNA depletion studies, Notch signaling is reduced in a concentration-dependent manner when cells are pretreated with NIH-12848 relative to the DMSO control (Fig. 5). Given that PI5P4Kγ inactivation with either siRNA or NIH-12848 reduces Notch activity, we conclude that the lipid kinase positively regulates Notch signaling.
Fig. 5.
PI5P4Kγ opposes DTX1 activity and positively regulates Notch signaling. (A) Notch signaling was measured with the dual-luciferase assay in tTA HeLa cells expressing the CD8-NΔE Notch1 chimera that were treated with control (−) or PI5P4Kγ (+) siRNA or treated with increasing concentrations of NIH-12848 to inhibit PI5P4Kγ or DMSO as a control. (B) CD8-NΔE Notch1 chimera-expressing tTA HeLa cells were treated with the indicated siRNA and drug combinations. Notch signaling was then measured as stated above. Box and whisker plots show each data point from three independent experiments. (C) Immunoblot analysis of tTA HeLa cell lysates that were treated with control (−) or PI5P4Kγ (+) siRNA. Polyclonal antibodies were used to detect PI5P4Kγ, and the monoclonal antibody E7 was used to detect beta tubulin (β-tub), used as a loading control. ***P < 0.0005, ****P < 0.0001, *****P < 0.00001.
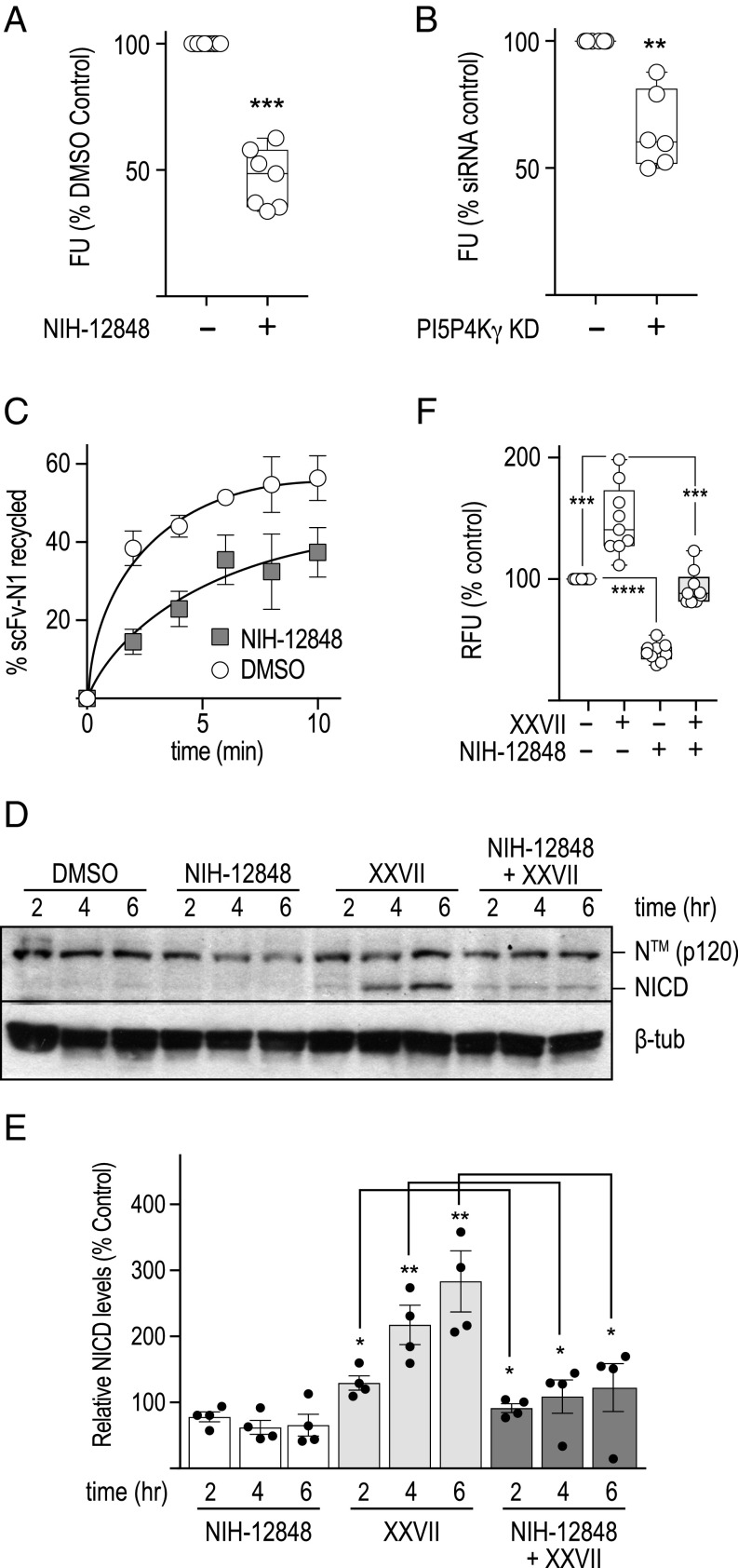
Our results demonstrate that DTX1 and PI5P4Kγ have opposing activities in regulating Notch. Given that loss of DTX1 promotes Notch recycling via a rab4a-dependent pathway, we postulated that PI5P4Kγ might up-regulate Notch activity by promoting recycling. To test this idea, we first measured the impact of PI5P4Kγ inactivation on Notch signaling when DTX1 expression is also silenced. We reasoned that if PI5P4Kγ promotes Notch1 recycling, inactivation of the kinase should suppress increases in Notch activity when DTX1 expression is reduced. Indeed, PI5P4Kγ depletion or pharmacological inactivation suppresses increases in Notch1 signaling that arise when DTX1 activity is also reduced (Fig. 5B). To further investigate the potential role for PI5P4Kγ in regulating Notch1 activity, we evaluated both Notch1 cell-surface levels and receptor recycling rates following inactivation of the lipid kinase. Pharmacological inhibition or siRNA depletion of PI5P4Kγ reduced Notch1 cell-surface levels relative to control (Fig. 6 A and B). Correspondingly, we found that acute PI5P4Kγ inhibition also reduces the Notch1 recycling rate using the pulse–chase assay (Fig. 6C). Collectively, these observations indicate that PI5P4Kγ positively regulates Notch1 by promoting receptor recycling.
Fig. 6.
PI5P4Kγ regulates Notch1 recycling. (A and B) Notch1 cell-surface levels were measured in U2OS cells (Materials and Methods) following a 3-h pretreatment with (A) DMSO (−) or the PI5P4Kγ inhibitor NIH-12848 (+; 28 µM), or (B) control or PI5P4Kγ-specific siRNA. (C) Notch1 recycling kinetics were measured in U2OS cells treated identically as those in A. (D) Representative immunoblot analyses for Notch1 from HeLa cell lysates after the cells were incubated for the specified times with the indicated compounds. For samples treated with both inhibitors, cells were incubated with NIH-12848 (28 µM) for 1 h before addition of the GSK3β inhibitor XXVII (32 µM). (E) Immunoblot quantification from four individual blots where each data point is shown with error bars representing ± SEM. (F) Endogenous Notch signaling using the dual-luciferase assay following HeLa cell treatment with the indicated drug combination corresponding to conditions outlined in D. *P < 0.05, **P < 0.005, ***P < 0.0005, ****P < 0.0001.
We recently discovered that GSK3β inhibition enhances Notch1 activity by promoting receptor recycling via a rab4a-mediated transport route (39). Given that GSK3β, like PI5P4Kγ, was identified as a DTX1 substrate (Dataset S2) and that DTX1 inactivation also promotes rab4a-dependent recycling (Fig. 2), we postulated that PI5P4Kγ and GSK3β might act along the same pathway. Acute GSK3β inhibition with the drug XXVII (40) increases NICD production and elevates Notch signaling (39). Thus, we reasoned that if PI5P4Kγ promotes recycling via a rab4a-dependent transport route, acute inhibition of PI5P4Kγ with NIH-12848 should suppress increases in NICD production and Notch1 signaling that result when GSK3β is also inactivated. Indeed, acute inactivation of PI5P4Kγ activity suppresses the enhanced production of NICD and downstream signaling that arises when GSK3β is also acutely inhibited (Fig. 6 D–F). These findings suggest that the competing activities of PI5P4Kγ and GSK3β modulate Notch1 access to a rab4a-mediated recycling route.
PI5P4Kγ Inactivation Enhances Notch1 Targeting to LAMP1-Positive Endolysosomes.
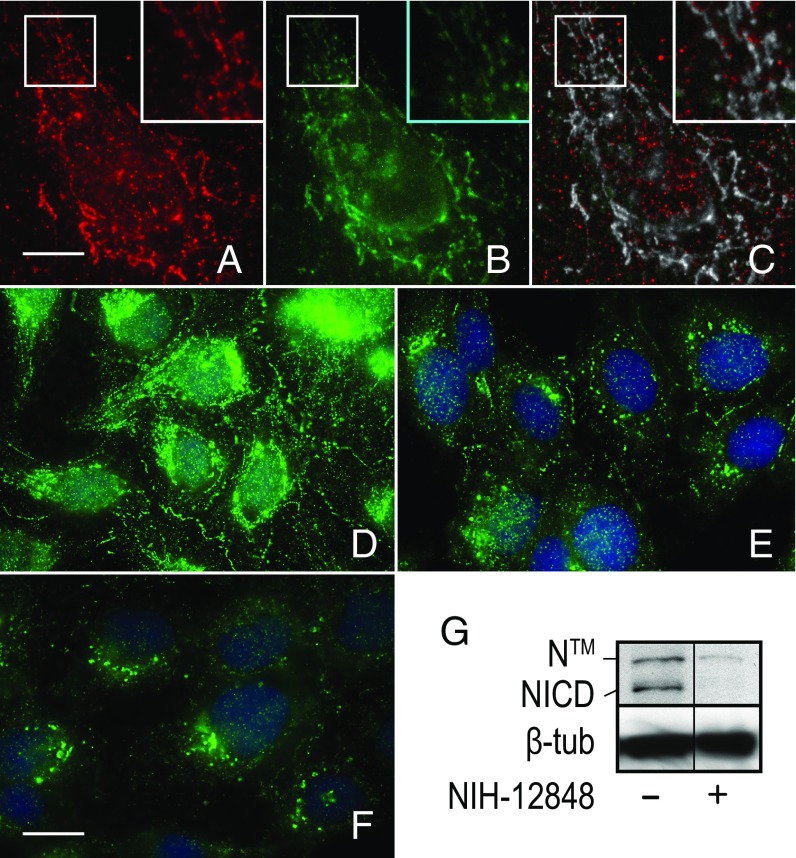
To gain insight into how PI5P4Kγ regulates Notch1 activity, we next examined PI5P4Kγ subcellular distribution by immunolocalization. Consistent with its role in Notch recycling, we found that PI5P4Kγ colocalizes with the receptor on tubulovesicular endosomes (Fig. 7C). By contrast, inactivation of the lipid kinase by siRNA or overnight incubation with NIH-12848 drug diminished the levels of Notch1 at cell–cell interfaces and reduced its association with tubulovesicular endosomes relative to the control (Fig. 7 E and F). Moreover, Notch1 protein levels appeared to be reduced in PI5P4Kγ-inactivated cells. Correspondingly, immunoblot analysis revealed a significant reduction in both membrane-tethered Notch1 (NTM) and activated receptor (NICD) protein levels in NIH-12848–treated cells (Fig. 7G). This reduction in receptor protein levels suggested to us that loss of PI5P4Kγ activity might alter Notch1 transport following internalization and redirect the receptor toward lysosomes for degradation. We tested this idea by tracking scFv-N1-sfGFP antibody uptake following PI5P4Kγ depletion or acute inactivation of the kinase by pretreating cells with NIH-12848 for 3 h. As expected, scFv-N1-sfGFP was readily internalized and targeted to tubulovesicular endosomes after a 12-min incubation in control cells (Fig. 8 A and C). By contrast, when PI5P4Kγ was depleted (Fig. 8B) or pharmacologically inhibited (Fig. 8F), internalized antibody was observed in vesiculated endosomes distributed throughout the cytoplasm, which coimmunolocalization analysis revealed were positive for the endolysosomal marker LAMP1 (Fig. 8H). By comparison, internalized antibody was also found to colocalize with LAMP1 in control cells (Fig. 8E), although the extent of colocalization was half that observed in cells following PI5P4Kγ inhibition (Fig. S3). Collectively, these results argue that PI5P4Kγ counters the inhibitory effects of DTX1 by promoting Notch1 targeting to or recycling from tubulovesicular endosomes. Moreover, they reveal that Notch1 is more efficiently targeted to endolysosomes for degradation following loss of PI5P4Kγ activity.
Fig. 7.
PI5P4Kγ inactivation alters Notch1 trafficking and leads to receptor degradation. (A) Coimmunolocalization analysis of endogenous Notch1 (A, red) and PI5P4Kγ (B, green) using recombinant scFv-N1-sfGFP and pAb against PI5P4Kγ, respectively. (C) A colocalized pixel map shows colocalized pixels in grayscale. Boxed regions are shown at higher (2×) magnification (Insets). (D–F) Endogenous Notch1 subcellular localization was evaluated in U2OS cells treated with control siRNA (D) or PI5P4Kγ siRNA (E) for 72 h or with 28 µM NIH-12848 (F) for 16 h. DAPI stain was used to mark nuclei. (G) Immunoblot analysis of endogenous Notch1 of U2OS cells treated with DMSO (−) or 28 µM NIH-12848. The immunoblot reflects two nonadjacent lanes from the same blot. Polyclonal antibodies against the cytoplasmic tail were used to detect Notch1. Beta tubulin was used as a loading control and detected with the mAb E7. [Scale bars, 10 µm (A–C) and 20 µm (D–F)].
Fig. 8.
PI5P4Kγ inactivation enhances Notch1 targeting to LAMP1-positive endolysosomes. (A and B) Immunolocalization analysis of Notch1 in U2OS cells treated with control (A) or PI5P4Kγ-specific siRNA (B). (C–H) Coimmunolocalization analysis of scFv-N1-sfGFP-GLuc (C and F, green) with LAMP1 (D and G, red) following recombinant antibody uptake for 15 min in U2OS cells pretreated with DMSO (C–E) or 28 µM NIH-12848 for 3 h. Colocalized pixel maps show colocalized pixels in grayscale (E and H). Boxed regions are shown at higher (2×) magnification (Insets). (Scale bars, 20 µm.)
DTX1 Depletion Alters PI5P4Kγ Distribution Within Cells.
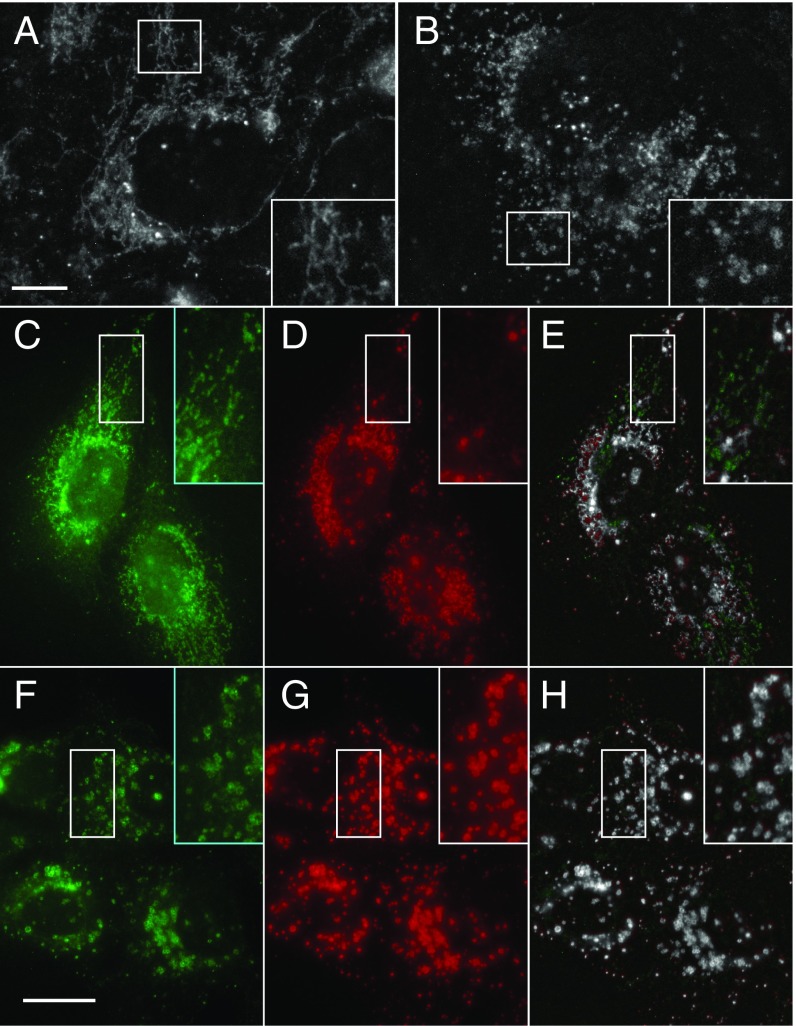
Given that PI5P4Kγ and DTX1 have opposing activities in regulating Notch1 transport and signaling activity, we postulated that DTX1 depletion might impact PI5P4Kγ distribution within cells. Consistent with our previous immunolocalization analyses (Fig. 7), PI5P4Kγ is present on tubulovesicular endosomes in control siRNA-treated cells (Fig. 9A). In contrast, DTX1 silencing leads to a redistribution of PI5P4Kγ from tubulovesicular endosomes to punctate structures that appear to consist of smaller, coalesced vesicles which are enriched near or possibly within the nucleus (Fig. 9B). We next attempted to resolve the identity of these PI5P4Kγ-positive structures. Since DTX1 depletion enhances Notch1 recycling via a rab4a-mediated pathway, we first evaluated PI5P4Kγ colocalization with rab4a. In controls, PI5P4Kγ colocalizes with rab4a throughout cells, including tubulovesicular endosomes and sporadic perinuclear endosomes (Fig. 9E). By comparison, in DTX1-silenced cells, rab4a remains associated with tubulovesicular endosomes; however, it also becomes enriched on the enlarged PI5P4Kγ-positive compartment (Fig. 9H). Given that there are no known reports of nuclear rab4a, we conclude that DTX1 depletion redistributes PI5P4Kγ to rab4a-positive endosomes adjacent to the nucleus. Moreover, these observations suggest that DTX1 modulates formation of these rab4a/PI5P4Kγ-positive endosomes, a compartment from which Notch1 might be directly recycled.
Fig. 9.
DTX1 depletion alters PI5P4Kγ subcellular distribution. Immunolocalization analysis of PI5P4Kγ in U2OS cells treated with control (A) or DTX1-specific siRNA (B). Coimmunolocalization analysis of PI5P4Kγ (C and F, red) and rab4a (D and G, green) in cells treated with control (C–E) or DTX1-specific siRNA (F–H). Colocalized pixel maps show colocalized pixels in grayscale (E and H). Boxed regions are shown at higher (2×) magnification (Insets). Arrows indicate smaller punctate structures (B). [Scale bar, 10 µm (A and B) and 20 µm (C–H).]
Discussion
Regulation of Notch Signaling by DTX1.
Results from this study support a model where DTX1 negatively regulates Notch signaling in mammals. This idea is reinforced by results from multiple published studies. Increased signaling is observed in progenitor T cells of mice lacking DTX1/2 (22). By contrast, DTX1 overexpression antagonizes Notch activity during hematopoiesis (20, 24, 25) and suppresses Notch-induced neurite outgrowth in mouse models (21). Collectively, these studies clearly indicate a role for DTX1 in down-regulating the Notch pathway, although the detailed mechanism by which it does so remains unclear.
Based on the results presented here, we propose that DTX1 prevents Notch delivery to the cell surface by inhibiting receptor recycling. In turn, this prevents receptor access to ligand—the result of which is a down-regulation of Notch1 signaling capacity. This is supported by several lines of evidence where DTX1 depletion (i) increases Notch1 recycling rates, (ii) elevates receptor cell-surface levels, and (iii) increases Notch1 signaling activity. Although biochemical evidence is currently lacking, studies in flies suggest that Drosophila deltex (Dx) regulates Notch activity by directly ubiquitinating the receptor (19, 27). Our data do not exclude this possibility; however, our mutagenesis studies clearly demonstrate that DTX1 can control signaling without directly ubiquitinating Notch1. This argues that DTX1 controls Notch1 by regulating the activities of other factors. Consistent with this notion, our in vitro ubiquitination screen for DTX1 substrates revealed multiple targets that impact Notch signaling when depleted by siRNA. Whether these in vitro substrates are bona fide DTX1 targets in vivo remains to be resolved. However, the range of putative targets suggests that DTX1 activity may be significantly more complex than previously thought.
PI5P4Kγ Promotes Notch Recycling.
The ubiquitination screen for DTX1 targets led to the identification of PI5P4Kγ. PI5P4Kγ inactivation by either siRNA or pharmacological inhibition demonstrates that the lipid kinase positively regulates Notch signaling. Our findings also indicate that PI5P4Kγ up-regulates Notch activity by enhancing receptor recycling from rab4a-positive endosomes. This conclusion is underpinned by the fact that Notch recycling rates and receptor cell-surface levels are reduced when PI5P4Kγ activity is inhibited. As mentioned above, it currently remains unclear whether DTX1 directly ubiquitinates PI5P4Kγ in vivo. However, a mechanistic relationship between DTX1 and PI5P4Kγ is supported by the finding that DTX1 depletion redistributes the lipid kinase from peripheral, tubulovesicular endosomes to a rab4a-positive perinuclear compartment. Thus, we speculate that PI5P4Kγ promotes recycling from this perinuclear compartment and not directly from tubulovesicular endosomes. Therefore, DTX1-dependent ubiquitination of PI5P4Kγ might suppress recycling by limiting access of the lipid kinase to this perinuclear compartment either by stabilizing PI5P4Kγ association with tubulovesicular endosomes or preventing its recruitment to the rab4a-positive perinuclear compartment.
Alternatively, DTX1 might directly control PI5P4Kγ enzymatic activity. In this case, ubiquitination of PI5P4Kγ by DTX1 might maintain the kinase in an inactive state. As mentioned, PI5P4Kγ belongs to a family of lipid kinases that use PI5P as a substrate to generate PI(4,5)P2 (37). However, this family is thought to generate highly localized pools of PI(4,5)P2 because PI5P abundance in cells is comparatively low relative to PI4P, which is the substrate for PI4P5Ks that synthesize the bulk of PI(4,5)P2 in cells (36). Thus, it is possible that when PI5P4Kγ is active, it facilitates localized production of PI(4,5)P2 on tubulovesicular membranes that mature into the perinuclear endosomes that mediate Notch1 recycling.
In either case, we predict that PI5P4Kγ lipid kinase activity is essential for Notch recycling. This implicates a critical role for PI(4,5)P2-binding proteins that facilitate Notch transport. Potential candidates include members of the eps15 homology domain-containing proteins (EHD1, 3, and 4)—each of which is known to bind PI(4,5)P2 and facilitate receptor transport through recycling endosomes (41–45). Resolving the precise role of PI(4,5)P2 production by PI5P4Kγ and determining the mechanistic consequence of PI5P4Kγ ubiquitination by DTX1 on Notch1 recycling will be the focus of future work.
Materials and Methods
Cells and Culture Conditions.
Tetracycline transactivator (tTA) HeLa and U2OS cells were obtained from Sandra Schmid, UT Southwestern, Dallas, TX and Wendy Gordon, University of Minnesota, Minneapolis, respectively. Cells were cultured in Dulbecco’s modified Eagle’s medium containing 10% FBS, 4.5 g/L glucose, and 100 U/mL penicillin/streptomycin at 37 °C with 5% CO2. tTA HeLa cells were maintained in the presence of 400 mg/mL G418 to maintain expression of the tTA. Each culture was maintained at a subconfluent density for no more than 20 passages.
Antibodies and Other Reagents.
A polyclonal antibody against mammalian Notch1 was generated in rabbit against the Notch1 cytoplasmic tail (mouse amino acids 1,759–2,306) fused to GST. PI5P4Kγ rabbit polyclonal antisera were a generous gift of Jonathan H. Clarke, University of Cambridge, Cambridge, United Kingdom. The mAb E7 was used to identify β-tubulin. Polyclonal antisera against DTX1 (OAGA00965) were purchased from Aviva Systems Biology. mAbs targeting rab4a (4E11; sc-51723) and LAMP1 (H5G11; sc-18821) were purchased from Santa Cruz Biotechnology. Recombinant proteins for UBE1 (E-305), UBE2C (E2-654), UBE2D1 (E2-616), UBE2N (E2-666), human ubiquitin biotin (U-570), and the energy-regenerating system (B-10) were purchased from Boston Biochem. Streptavidin-HRP (PI21126) and secondary antibodies conjugated to Alexa 488 (A11034, A32723) and Alexa 555 (A32727, A32732) were purchased from Thermo Fisher. NIH-12848 (534333), compound E (530509), and XXVII (361570) were purchased from EMD Millipore. ProtoArray Human protein microarray chips (PAH0525101) were purchased from Thermo Fisher.
Constructs.
The activated Notch1 mimic CD8-NΔE was previously described (46). NICD-KR14-V5, which encodes 14 lysine mutations within the Notch1 cytoplasmic tail, was a gift from Michael Potente, Goethe University, Frankfurt, Germany (47) and was used as a template to generate KR28 CD8-NΔE in pAD-CMV by PCR mutagenesis. Primer sequences used for mutagenesis can be found in Table S1. Anti–Notch1-E6 in pBIOCAM5 was a gift from John McCafferty, University of Cambridge, Cambridge, UK, (Addgene; plasmid 39344) and was used to generate scFv-N1-sfGFP-Gluc in pBIOCAM5. Human DTX1 in pOTB7 was purchased from Open Biosystems and subcloned into pET28a(+) for bacterial expression. The Notch dual-luciferase reporter construct was generated by cloning firefly luciferase behind 10 repeating RBP-Jκ transcription factor binding sites for Notch-induced expression in pAD-Tet. In the same plasmid backbone, Renilla luciferase was cloned in the opposite direction behind two repeating tTA binding sites to provide low-level, constitutive expression in tTA HeLa cells. All constructs were sequence-verified.
scFv-N1-sfGFP-GLuc Antibody Production.
Recombinant N1 antibody was produced by transfecting HEK293 cells with scFv-N1-sfGFP-GLuc pAD-CMV(+) plasmid for 48 h. Culture media containing secreted antibody were then concentrated 50× with a Centricon concentrator (UFC710008; EMD Millipore) with a 10-kDa cutoff by centrifugation at 5,000 × g. Concentrated antibody was stored at 4 °C and used at a 1:200 dilution for immunolocalization analysis and at 1:100 for antibody uptake experiments.
Immunolocalization and Image Quantification.
For imaging studies, U2OS cells were grown on glass coverslips, transferred to ice, and washed with PBS. Cells were then fixed with ice-cold acetone for 2 min followed by methanol for 2 min. Coverslips were then washed with PBS containing 0.1% Tween (PBST) before antibody addition. Cells were incubated with primary antibody for 1 h at room temperature (RT), washed with PBST, and incubated for 1 h at RT with the appropriate secondary antibody conjugated to either Alexa 488 or Alexa 555. Samples were then visualized by epifluorescence using a Zeiss Axio Imager M1 and captured with a 12-bit monochrome Jenoptik CCD camera. Images were imported, cropped, and assembled into panels using Photoshop CS6 and Illustrator CS6 (Adobe Systems). ImageJ (v1.51q; NIH) was used to quantify colocalization between internalized scFv-N1-sfGFP-GLuc and LAMP1. In short, each image was background-subtracted using a 50-pixel rolling-ball radius. The extent of colocalization was then determined using the Colocalization Threshold plugin, which automatically determines the Manders overlap coefficient (48), where data are presented as a fraction of the total.
Notch Signaling Assay and Statistical Analysis.
Signaling was evaluated with the dual-reporter plasmid (Constructs) using a dual-luciferase RBP-Jκ reporter assay (SABiosciences) and assessed according to the manufacturer’s published protocols (Promega). In each case, RBP-Jκ–promoted firefly luciferase activity was normalized to constitutively expressed Renilla luciferase under the control of a tetracycline-regulatable (TRE) promoter driven by the tTA. Relative luciferase units represent signaling expressed as a ratio of Notch-promoted firefly luciferase activity over Renilla luciferase. For all other experiments, Notch signaling was measured using the Notch chimera CD8-NΔE (46), which mimics the activated receptor. This chimera was expressed under the control of a TRE promoter that limits protein expression to near-endogenous levels, as previously described (26). All data were statistically analyzed using a paired t test (two-tailed) to calculate a P value with Prism (v7.0b; GraphPad). P values are represented in each figure with asterisks, where P < 0.05, 0.005, 0.0005, and 0.0001 are shown as *, **, ***, and ****, respectively. NS (not significant) indicates a P > 0.5.
siRNA Silencing.
To silence protein expression, cells were transfected twice with siRNA using RNAiMAX (Thermo Fisher) on the first day and 24 h later following manufacturer protocols. Cells were incubated an additional 18 to 24 h before functional analysis. Validated siRNAs were purchased from Qiagen or GenePharma, and target-specific sequences are listed in Table S2. Where indicated, protein expression silencing was validated by immunoblot analysis.
DTX1 Ubiquitination Assay and ProtoArray Screen.
For in vitro ubiquitination assays, a master mix of ubiquitin-conjugating enzymes was assembled on ice and included 100 nM UBE1, 250 nM UBE2C, UBE2D1, or UBE2N, 0.1 µg/µL biotinylated ubiquitin, and 1× energy-regenerating system. The master mix was aliquoted and 0.25 µg bacterially expressed DTX1-6His was added. Samples were incubated at 30 °C for 30 min and the reaction was stopped by addition of 2× Laemmli protein sample buffer and boiling for 2 min. Samples were then analyzed by SDS/PAGE and immunoblot analysis using DTX1 antisera and HRP conjugated to streptavidin. ProtoArray screens were performed according to manufacturer protocols. The negative control included UBE1, UBE2C, and UBE2N, biotinylated ubiquitin, and the energy-regenerating system, while the experimental array also included DTX1-6His. Results were analyzed using ProtoArray Prospector v5.2 from Thermo Fisher.
Notch1 Cell Surface-Level Quantification.
Following experimental treatment, U2OS cells were counted and equally split between two 35-mm dishes. Cells were then incubated with 1.0 mL 1:100 dilution scFv-N1-sfGFP-GLuc antibody in dPBS on ice for 60 min to allow antibody binding. Cells were washed three times with 1.0 mL ice-cold dPBS and then lysed with 100 µL dPBS containing 1% Triton X-100. Five microliters of lysate was analyzed using the luciferase assay, and signal was normalized by dividing by the cell number.
scFv-N1-sfGFP-GLuc Recycling Assay.
Cells were grown in 35-mm dishes and pretreated with the indicated siRNA or drug. Media were replaced with ice-cold growth media containing scFv-N1-sfGFP-GLuc antibody (1:100 dilution). Cells were then incubated at 4 °C for 60 min to allow antibody binding. Cells were washed three times with dPBS and transferred to 37 °C for 12 min to allow antibody internalization. Antibody uptake was terminated by returning the dishes to ice. Dishes were then washed with ice-cold dPBS. Cells were incubated on ice for 10 min in dPBS containing 5 mM EDTA and 20 mM TCEP [Tris(2-carboxyethyl)phosphine hydrochloride] to resuspend cells and irreversibly inactivate surface-bound scFv-N1-sfGFP-GLuc—Gaussia luciferase activity relies on disulfide bonds and is inactivated with reducing agents (49). Cells were then pelleted and resuspended in 0.5 mL ice-cold DMEM/10% FBS containing DMSO or XXVII. Forty-microliter aliquots were transferred into 1.5-mL tubes for each time point. All tubes, except for the 0 time point (total), were then transferred to 37 °C for the indicated time period before returning to ice to stop recycling. scFv-N1-sfGFP-GLuc antibody, which returned to the cell surface, was then inactivated by addition of 1 µL 0.5 M TCEP. Cells were washed with 1.0 mL dPBS and gently pelleted by centrifugation (180 × g) at 4 °C, the supernatant was aspirated, and cells were lysed with 20 µL dPBS containing 1% Triton X-100. Five microliters of lysate was then used to measure the luciferase activity retained within cells. Luciferase activity at each time point was divided by the total and subtracted from 1 to determine the fraction of recycled scFv-N1-sfGFP-GLuc.
Supplementary Material
Acknowledgments
We thank Drs. Lihsia Chen and Tom Hays for helpful discussions during the course of these studies.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712142115/-/DCSupplemental.
References
- 1.Bray SJ. Notch signalling in context. Nat Rev Mol Cell Biol. 2016;17:722–735. doi: 10.1038/nrm.2016.94. [DOI] [PubMed] [Google Scholar]
- 2.Kopan R, Ilagan MX. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brou C, et al. A novel proteolytic cleavage involved in Notch signaling: The role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 4.van Tetering G, et al. Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J Biol Chem. 2009;284:31018–31027. doi: 10.1074/jbc.M109.006775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gordon WR, et al. Mechanical allostery: Evidence for a force requirement in the proteolytic activation of Notch. Dev Cell. 2015;33:729–736. doi: 10.1016/j.devcel.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Strooper B, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 7.Garg V, et al. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 8.Rangarajan A, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001;20:3427–3436. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 10.Joutel A, et al. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383:707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Imanaka N, Chen J, Griffin JD. Hypoxia potentiates Notch signaling in breast cancer leading to decreased E-cadherin expression and increased cell migration and invasion. Br J Cancer. 2010;102:351–360. doi: 10.1038/sj.bjc.6605486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, et al. miR-598 inhibits metastasis in colorectal cancer by suppressing JAG1/Notch2 pathway stimulating EMT. Exp Cell Res. 2017;352:104–112. doi: 10.1016/j.yexcr.2017.01.022. [DOI] [PubMed] [Google Scholar]
- 13.Kwon OJ, et al. Notch promotes tumor metastasis in a prostate-specific Pten-null mouse model. J Clin Invest. 2016;126:2626–2641. doi: 10.1172/JCI84637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang P, Yang Y, Nolo R, Zweidler-McKay PA, Hughes DP. Regulation of NOTCH signaling by reciprocal inhibition of HES1 and Deltex 1 and its role in osteosarcoma invasiveness. Oncogene. 2010;29:2916–2926. doi: 10.1038/onc.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baron M. Endocytic routes to Notch activation. Semin Cell Dev Biol. 2012;23:437–442. doi: 10.1016/j.semcdb.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 16.Takeyama K, et al. The BAL-binding protein BBAP and related Deltex family members exhibit ubiquitin-protein isopeptide ligase activity. J Biol Chem. 2003;278:21930–21937. doi: 10.1074/jbc.M301157200. [DOI] [PubMed] [Google Scholar]
- 17.Matsuno K, Diederich RJ, Go MJ, Blaumueller CM, Artavanis-Tsakonas S. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development. 1995;121:2633–2644. doi: 10.1242/dev.121.8.2633. [DOI] [PubMed] [Google Scholar]
- 18.Matsuno K, et al. Involvement of a proline-rich motif and RING-H2 finger of Deltex in the regulation of Notch signaling. Development. 2002;129:1049–1059. doi: 10.1242/dev.129.4.1049. [DOI] [PubMed] [Google Scholar]
- 19.Mukherjee A, et al. Regulation of Notch signalling by non-visual beta-arrestin. Nat Cell Biol. 2005;7:1191–1201. doi: 10.1038/ncb1327. [DOI] [PubMed] [Google Scholar]
- 20.Izon DJ, et al. Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity. 2002;16:231–243. doi: 10.1016/s1074-7613(02)00271-6. [DOI] [PubMed] [Google Scholar]
- 21.Sestan N, Artavanis-Tsakonas S, Rakic P. Contact-dependent inhibition of cortical neurite growth mediated by Notch signaling. Science. 1999;286:741–746. doi: 10.1126/science.286.5440.741. [DOI] [PubMed] [Google Scholar]
- 22.Lehar SM, Bevan MJ. T cells develop normally in the absence of both Deltex1 and Deltex2. Mol Cell Biol. 2006;26:7358–7371. doi: 10.1128/MCB.00149-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiaris H, et al. Modulation of Notch signaling elicits signature tumors and inhibits hras1-induced oncogenesis in the mouse mammary epithelium. Am J Pathol. 2004;165:695–705. doi: 10.1016/S0002-9440(10)63333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yun TJ, Bevan MJ. Notch-regulated ankyrin-repeat protein inhibits Notch1 signaling: Multiple Notch1 signaling pathways involved in T cell development. J Immunol. 2003;170:5834–5841. doi: 10.4049/jimmunol.170.12.5834. [DOI] [PubMed] [Google Scholar]
- 25.Hsiao HW, et al. Deltex1 is a target of the transcription factor NFAT that promotes T cell anergy. Immunity. 2009;31:72–83. doi: 10.1016/j.immuni.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 26.Zheng L, Saunders CA, Sorensen EB, Waxmonsky NC, Conner SD. Notch signaling from the endosome requires a conserved dileucine motif. Mol Biol Cell. 2013;24:297–307. doi: 10.1091/mbc.E12-02-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hori K, Sen A, Kirchhausen T, Artavanis-Tsakonas S. Synergy between the ESCRT-III complex and Deltex defines a ligand-independent Notch signal. J Cell Biol. 2011;195:1005–1015. doi: 10.1083/jcb.201104146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falk R, et al. Generation of anti-Notch antibodies and their application in blocking Notch signalling in neural stem cells. Methods. 2012;58:69–78. doi: 10.1016/j.ymeth.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Wijk SJ, et al. A comprehensive framework of E2-RING E3 interactions of the human ubiquitin-proteasome system. Mol Syst Biol. 2009;5:295. doi: 10.1038/msb.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collinet C, et al. Systems survey of endocytosis by multiparametric image analysis. Nature. 2010;464:243–249. doi: 10.1038/nature08779. [DOI] [PubMed] [Google Scholar]
- 31.Kim WY, et al. GSK-3 is a master regulator of neural progenitor homeostasis. Nat Neurosci. 2009;12:1390–1397. doi: 10.1038/nn.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin YH, Kim H, Oh M, Ki H, Kim K. Regulation of Notch1/NICD and Hes1 expressions by GSK-3alpha/beta. Mol Cells. 2009;27:15–19. doi: 10.1007/s10059-009-0001-7. [DOI] [PubMed] [Google Scholar]
- 33.Espinosa L, Inglés-Esteve J, Aguilera C, Bigas A. Phosphorylation by glycogen synthase kinase-3 beta down-regulates Notch activity, a link for Notch and Wnt pathways. J Biol Chem. 2003;278:32227–32235. doi: 10.1074/jbc.M304001200. [DOI] [PubMed] [Google Scholar]
- 34.Foltz DR, Santiago MC, Berechid BE, Nye JS. Glycogen synthase kinase-3beta modulates Notch signaling and stability. Curr Biol. 2002;12:1006–1011. doi: 10.1016/s0960-9822(02)00888-6. [DOI] [PubMed] [Google Scholar]
- 35.Guha S, et al. Glycogen synthase kinase 3 beta positively regulates Notch signaling in vascular smooth muscle cells: Role in cell proliferation and survival. Basic Res Cardiol. 2011;106:773–785. doi: 10.1007/s00395-011-0189-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giudici ML, Clarke JH, Irvine RF. Phosphatidylinositol 5-phosphate 4-kinase γ (PI5P4Kγ), a lipid signalling enigma. Adv Biol Regul. 2016;61:47–50. doi: 10.1016/j.jbior.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 37.Rameh LE, Tolias KF, Duckworth BC, Cantley LC. A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature. 1997;390:192–196. doi: 10.1038/36621. [DOI] [PubMed] [Google Scholar]
- 38.Clarke JH, et al. The function of phosphatidylinositol 5-phosphate 4-kinase γ (PI5P4Kγ) explored using a specific inhibitor that targets the PI5P-binding site. Biochem J. 2015;466:359–367. doi: 10.1042/BJ20141333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng L, Conner SD. Glycogen synthase kinase 3β inhibition enhances Notch1 recycling. Mol Biol Cell. December 13, 2017 doi: 10.1091/mbc.E17-07-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berg S, et al. Discovery of novel potent and highly selective glycogen synthase kinase-3β (GSK3β) inhibitors for Alzheimer’s disease: Design, synthesis, and characterization of pyrazines. J Med Chem. 2012;55:9107–9119. doi: 10.1021/jm201724m. [DOI] [PubMed] [Google Scholar]
- 41.Blume JJ, Halbach A, Behrendt D, Paulsson M, Plomann M. EHD proteins are associated with tubular and vesicular compartments and interact with specific phospholipids. Exp Cell Res. 2007;313:219–231. doi: 10.1016/j.yexcr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 42.Sharma M, Giridharan SS, Rahajeng J, Naslavsky N, Caplan S. MICAL-L1 links EHD1 to tubular recycling endosomes and regulates receptor recycling. Mol Biol Cell. 2009;20:5181–5194. doi: 10.1091/mbc.E09-06-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naslavsky N, Boehm M, Backlund PSJ, Jr, Caplan S. Rabenosyn-5 and EHD1 interact and sequentially regulate protein recycling to the plasma membrane. Mol Biol Cell. 2004;15:2410–2422. doi: 10.1091/mbc.E03-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naslavsky N, Rahajeng J, Chenavas S, Sorgen PL, Caplan S. EHD1 and Eps15 interact with phosphatidylinositols via their Eps15 homology domains. J Biol Chem. 2007;282:16612–16622. doi: 10.1074/jbc.M609493200. [DOI] [PubMed] [Google Scholar]
- 45.Waxmonsky NC, Conner SD. Αvβ3-integrin-mediated adhesion is regulated through an AAK1L- and EHD3-dependent rapid-recycling pathway. J Cell Sci. 2013;126:3593–3601. doi: 10.1242/jcs.122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sorensen EB, Conner SD. γ-Secretase-dependent cleavage initiates Notch signaling from the plasma membrane. Traffic. 2010;11:1234–1245. doi: 10.1111/j.1600-0854.2010.01090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guarani V, et al. Acetylation-dependent regulation of endothelial Notch signalling by the SIRT1 deacetylase. Nature. 2011;473:234–238. doi: 10.1038/nature09917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manders EMM, Verbeek FJ, Aten JA. Measurement of co‐localization of objects in dual‐colour confocal images. J Microsc. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 49.Inouye S, Sahara Y. Identification of two catalytic domains in a luciferase secreted by the copepod Gaussia princeps. Biochem Biophys Res Commun. 2008;365:96–101. doi: 10.1016/j.bbrc.2007.10.152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.