Significance
The complexity of biological signaling pathways parallels the design of modern electronic circuits. The AIM2-ASC inflammasome is a filamentous signaling platform that plays an essential role in defense against foreign dsDNA arising from invading pathogens. Currently, the design principles of the AIM2-ASC “signaling circuit” remain poorly understood. Here, we found that the assembly of the AIM2-ASC inflammasome filters short dsDNA as noise at each signaling step. dsDNA induces hysteresis, and the assembly generates multistage, virtually infinite signal amplification. Moreover, an array of positive feedback loops reinforces the assembly. Together with a quantitative model of the assembly pathway, we demonstrate that an ultrasensitive digital circuit drives the assembly of the AIM2-ASC inflammasome on foreign dsDNA.
Keywords: innate immunity, inflammasome, digital circuit, signaling network
Abstract
The AIM2-ASC inflammasome is a filamentous signaling platform essential for mounting host defense against cytoplasmic dsDNA arising not only from invading pathogens but also from damaged organelles. Currently, the design principles of its underlying signaling network remain poorly understood at the molecular level. We show here that longer dsDNA is more effective in inducing AIM2 assembly, its self-propagation, and downstream ASC polymerization. This observation is related to the increased probability of forming the base of AIM2 filaments, and indicates that the assembly discerns small dsDNA as noise at each signaling step. Filaments assembled by receptor AIM2, downstream ASC, and their joint complex all persist regardless of dsDNA, consequently generating sustained signal amplification and hysteresis. Furthermore, multiple positive feedback loops reinforce the assembly, as AIM2 and ASC filaments accelerate the assembly of nascent AIM2 with or without dsDNA. Together with a quantitative model of the assembly, our results indicate that an ultrasensitive digital circuit drives the assembly of the AIM2-ASC inflammasome.
The complexity of biological signaling pathways rivals the design of modern electronic circuitry (1–5). Notably, both biological and artificial pathways share three key design principles in achieving high fidelity: noise filtering, amplification, and feedback regulation (5). Although such circuit-like behaviors of biological pathways at the cellular level have been well documented (3, 6, 7), the design principles of their underpinning signaling networks often remain poorly understood at the molecular level.
In the metazoan innate immune system a major defense mechanism against intracellular pathogens is to assemble supramolecular signaling platforms termed “inflammasomes” (8). Canonical inflammasomes include a cytosolic receptor that recognizes pathogen-associated molecular patterns (PAMPs; e.g., viral nucleic acids), the central ASC hub that converges multiple pathogen signals by partnering with various upstream receptors [ASC: apoptosis associated speck-forming protein containing caspase recruiting domain (CARD)], and the caspase-1 protease that executes the maturation of proinflammatory cytokines (e.g., IL-1β) and pyroptosis (8).
A hallmark of the molecular mechanism by which inflammasomes transduce signals is the sequential assembly of filaments (9). That is, a PAMP triggers the polymerization of a specific receptor, which induces the polymerization of ASC. ASC filaments then promote the polymerization of procaspase-1, activating the latent protease by proximity-induced autocatalysis. At the cellular level, a major outcome of inflammasome pathways is cell death (8), a digital (binary, “on or off”) process that excludes new equilibrium (6). Nevertheless, little is known about the mechanisms by which such filamentous assemblies generate a signaling circuit capable of triggering a digital response. We address this issue in the assembly of the Absent-In-Melanoma 2 (AIM2)-ASC inflammasome.
AIM2 recognizes cytosolic dsDNA and acts as a receptor for the foreign-dsDNA sensing inflammasome (Fig. 1A) (10–12). In humans, the AIM2-ASC inflammasome is essential for defense against virtually all pathogens entailing dsDNA for replication [e.g., human papilloma virus and Francisella tularensis (8, 13, 14)]. Moreover, not only does AIM2 induce cell death upon radiation-induced DNA damage (15), but its dysregulation is also implicated in promoting autoinflammation (8, 13, 14). Thus, AIM2 is a potential therapeutic target for treating various human maladies (8, 13, 14).
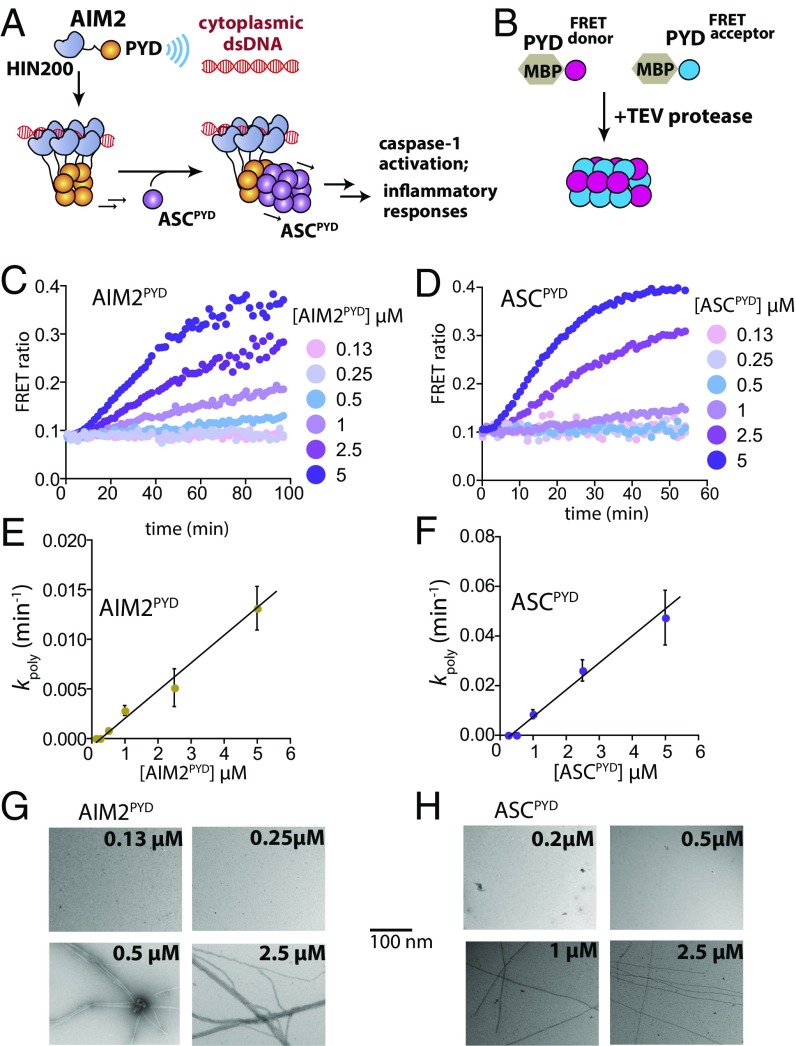
Fig. 1.
The assembly of AIM2PYD and ASCPYD filaments displays threshold behaviors. (A) The assembly of the AIM2-ASC inflammasome. AIM2 assembles into a filament on foreign dsDNA, which then induces the polymerization of ASC. Only ASCPYD is shown for simplicity. (B) FRET-based polymerization assays. FRET donor-labeled species is colored in magenta and FRET acceptor-labeled species is colored in blue. (C) The polymerization of AIM2PYD at each given concentration was monitored over time. The FRET ratio is calculated by (acceptor emission)/(acceptor + donor emission). (D) The polymerization of ASCPYD was monitored as described in C. (E and F) Plots of protein concentration dependence in the polymerization rates (kpoly) of AIM2PYD and ASCPYD; [kpoly = ln(2)/th]. The lines are linear fits (n = 3; ± SD). (G and H) Negative-staining electron micrographs of AIM2PYD and ASCPYD at each indicated concentration after incubating with TEVp for 1 h.
How does the AIM2-ASC inflammasome assemble? Full-length AIM2 (AIM2FL) contains one pyrin domain (PYD; AIM2PYD) that polymerizes into filaments and one HIN200 domain that binds dsDNA without sequence specificity (Fig. 1A; HIN200: hematopoietic IFN inducible nuclear antigen of 200 amino acids). We have recently shown that dsDNA length plays a major role in regulating the polymerization activity of AIM2 (16), which also correlated with its dsDNA length-dependent cell death and IL-1β processing activities (12, 17). This correlation thus prompted us to propose that dsDNA length-dependent formation of the AIM2 filament is critical for regulating its signaling activity (16).
ASC contains one PYD (ASCPYD) and one CARD (ASCCARD) (Fig. 1A) and autopolymerizes in a prion-like manner (9, 18). Moreover, multiple upstream receptors [e.g., AIM2 and NOD-like receptors (NLRs)] induce the polymerization of ASC via PYD–PYD interactions, which in turn allows ASC to converge various PAMPs to the activation of procaspase-1 (9, 18). Of note, we found that the helical symmetry of the AIM2PYD filament is consistent with that of the ASCPYD filament (six-start helix with threefold symmetry; refs. 9 and 16), indicating that upstream AIM2 provides a structural template for the polymerization of downstream ASC.
The assembly of suprastructures such as inflammasomes has emerged as a new paradigm in intracellular signaling (19), and the role of the AIM2-ASC axis in host innate defense is increasingly well established (13, 14). Moreover, multiple high-resolution structures of AIM2 and ASC are now available (9, 17, 20). Nevertheless, little is known about how the interplay among the incoming signal (dsDNA), receptor (AIM2), and downstream hub (ASC) generates a signaling circuit that drives the assembly of suprastructures capable of triggering a digital response (6). Here, using quantitative assays, negative-stain electron microscopy (nsEM), and computation, we set forth the design principles of the biological circuit that drives the assembly of the AIM2-ASC inflammasome.
We found here that the assembly of the AIM2-ASC inflammasome employs a high-pass filter function, which discerns short dsDNA as noise. That is, the length of dsDNA regulates the ability of AIM2 to assemble, self-propagate, and induce ASC polymerization. A Monte Carlo simulation of the filament assembly suggests that the dsDNA length dependence arises because longer fragments provide higher probability of assembling the intact helical base of AIM2 filaments. Moreover, the assembly results in three-stage, sustained amplification. That is, not only AIM2PYD and ASCPYD filaments persist, but the joint AIM2PYD-ASCPYD filaments also persist. Of note, dsDNA induces assembly, but it is not needed for AIM2 filaments to persist, indicating that AIM2 immediately generates a memory of infection (hysteresis). We also found that multiple positive feedback loops reinforce the assembly, as both AIM2 and ASC filaments accelerate the assembly of each other in the presence and absence of dsDNA. Finally, a mathematical model of the assembly suggests that this highly reinforced signaling network positions the inflammasome assembly in “standby,” ready to trigger ultrasensitive digital responses against invading pathogens.
Results
The Assembly of AIM2 and ASC Filaments Displays a Threshold Behavior.
To decipher the signaling network of the AIM2-ASC inflammasome at the molecular level we first optimized our Förster resonance energy transfer (FRET)-based assays (16) for kinetic measurements. Here, because both AIM2PYD and ASCPYD autoassemble without cofactors (9, 16), we fused an N-terminal maltose binding protein (MBP) to suppress autopolymerization as reported by Wu and coworkers (9). Cleaving MBP by the tobacco etch virus protease (TEVp) triggers polymerization (9), which can be monitored by the increase in FRET ratio (Fig. 1B; see also Fig. S1A). These observations indicate that our FRET-based assay can track and recapitulate the polymerization activities of AIM2 and ASC in vitro (9).
It has been speculated that AIM2PYD and ASCPYD filaments assemble in an actin-like nucleation-elongation mechanism (9, 18); however, this model still remains to be directly tested (an alternative is highly cooperative one-step assembly). To address this issue and establish the basic framework for our study we first monitored the relationship between protein concentrations and polymerization kinetics of AIM2PYD and ASCPYD using our FRET assay (Fig. 1 C and D). FRET signals did not change below certain protein concentrations (i.e., threshold); the time-dependent increase in FRET signals then linearly increased with protein concentrations [i.e., the apparent half-time (th) of polymerization and its related rate constant (kpoly); Fig. 1 E and F]. Moreover, filaments only appeared when protein concentrations were high enough to generate FRET signals (Fig. 1 G and H). Overall, these behaviors are consistent with the stepwise mechanism observed from actin filaments (nucleation-elongation) in which a critical concentration must be reached to initiate assembly (21).
AIM2 Discriminates Against dsDNA Length at the Initial Assembly.
All signaling pathways must filter out noise to minimize spurious responses (5). This is particularly critical for the AIM2 pathway because its major outcome is cell death (6, 10–12). One type of noise AIM2 has to filter is length of dsDNA: Prior biochemical cellular studies showed that at least 70-bp dsDNA is required to initiate the AIM2 pathway, with optimal efficiency reached with ∼200-bp dsDNA (12, 16, 17). Nonetheless, it is not known whether dsDNA length regulates the assembly or disassembly dynamics of AIM2 filaments. This is an important question not only for understanding the assembly mechanism of the AIM2 inflammasome but also for decoding the design principles of the underpinning signaling circuit at the molecular level. We thus determined the assembly kinetics of FRET donor- and acceptor-labeled AIM2FL on various dsDNA lengths well below protein concentrations that permit dsDNA-free assembly (ref. 16 and Fig. 2A). We also used a saturating mass concentration for all dsDNA lengths to ensure that the initial encounter is not rate-limiting (16). Here, the observed assembly rates (kassm) nonlinearly increased nearly 700-fold between 24- and 600-bp dsDNA (10-h reduction in th; Fig. 2B). These results correlate with our previous dsDNA length-dependent binding experiments (16), indicating that AIM2 discerns short dsDNA as noise at the first assembly step of the inflammasome in a manner similar to high-pass filters found in electronic circuits (i.e., low frequency signals cannot enter the pathway; Fig. S1B, a).
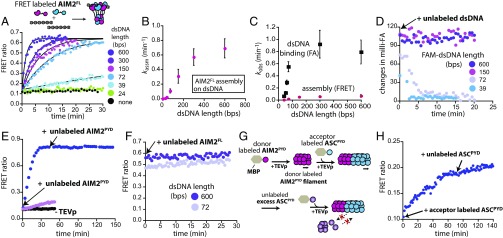
Fig. 2.
dsDNA length regulates the assembly kinetics of AIM2. (A) A scheme for FRET experiments using an equal mix of donor- (magenta) and acceptor- (blue) labeled AIM2FL (72 nM). The time-dependent change in the emission ratio was monitored with 10 µg/mL of each dsDNA (at least 10-fold higher than the dissociation constant for 24-bp dsDNA). Lines are fits to a single-exponential equation. (B) A plot of observed assembly rates determined by single-exponential fitting (kassms) vs. various dsDNA lengths (n = 4; ± SD). (C) A plot of observed rates (kobs) determined by FA and FRET experiments via single exponential fits vs. various dsDNA lengths (n = 3; ± SD). (D) AIM2FL (300 nM) was preincubated on 150 nM FAM-labeled dsDNA for 30 min (binding-site normalized). At time 0, 10 µg/mL linearized plasmid was added and the change in FA of each FAM-labeled dsDNA was monitored over time. (E) Blue trace: MBP-tagged FRET donor- and acceptor-labeled AIM2PYD (5 µM) was allowed to polymerize for 45 min upon adding TEVp; unlabeled MBP-AIM2PYD (25 µM) was subsequently added with additional TEVp and the FRET emission ratio was monitored over time. Violet trace: 5 µM FRET-labeled MBP-AIM2PYD was mixed with 25 µM unlabeled MBP-AIM2PYD then TEVp was added. Black trace: No FRET signal change was detected without TEVp. (F) The 300 nM FRET-labeled AIM2FL was allowed to polymerize on each dsDNA (150 nM, binding-site normalized) by preincubating for 30 min. At time 0, 5 µM unlabeled MBP-AIM2FL was added with TEVp and the change in the emission ratio was monitored over time. (G) A scheme for FRET-based experiments testing the dynamics of the AIM2PYD⋅ASCPYD joint filament. (H) The emission ratio between FRET donor-labeled AIM2PYD and acceptor-labeled ASCPYD was monitored over time before and after adding an excess amount of unlabeled ASCPYD (20 µM).
The IFN-inducible protein 16 (IFI16), which is related to AIM2, one-dimensionally tracks dsDNA then assembles into filaments; this stepwise mechanism allows IFI16 to assemble faster on longer dsDNA (22). It is not known whether AIM2 assembles in a similar stepwise mechanism, especially considering that IFI16 has one additional dsDNA-binding HIN200 domain. For instance, we observed here that AIM2 assembles faster on longer dsDNA (Fig. 2A) as seen from IFI16 (22); however, the faster assembly can either be achieved by the IFI16-like stepwise assembly (i.e., tracking then assembly, ref. 22) or from a highly cooperative assembly (i.e., concerted, one step). This is another important issue to be resolved not only for understanding the overall operating principles of AIM2-family proteins but also for quantitatively modeling the AIM2 signaling pathway. We thus conducted the following experiments. For instance, an experimental prediction from a stepwise model is that protein–dsDNA association rates determined by monitoring the changes in fluorescence anisotropy (FA) of fluorescein (FAM)-labeled dsDNA would be faster than those determined by FRET between AIM2 molecules at the same protein and dsDNA concentrations; an alternative concerted assembly predicts similar rates from these two experiments. Here, we found that the observed rates obtained by monitoring the FA of FAM-dsDNA were significantly faster than the protein oligomerization rates determined by FRET of AIM2 for all dsDNA lengths (Fig. 2C and Fig. S1 C and D). We also observed a linear relationship between AIM2FL concentrations and dsDNA-dependent assembly rates (Fig. S1E), indicating that cooperativity is not the main driver. Further supporting the tracking-assisted assembly mechanism (23), the kassms eventually decreased with increasing dsDNA concentrations (Fig. S1F). Overall, our results suggest that AIM2 assembles in an IFI16-like stepwise manner (22) and also indicate that different AIM2 family proteins share a similar kinetic noise filtering strategy.
Persisting AIM2 and ASC Filaments Would Generate Three-Stage Signal Amplification.
Increasing the lifetime of signaling complexes at each stage of a cascade is key to amplification (5), which is also mathematically equivalent to multistage amplifiers found in electronic circuits (4). Currently, the disassembly dynamics of AIM2 filaments remain unknown, and we asked whether the length of bound dsDNA regulates the stability of AIM2 filaments. To test this idea, we assembled AIM2FL⋅FAM-dsDNA complexes at saturating concentrations (16) then added excess linearized plasmid as a competitor (Fig. 2D). The FA of bound FAM-dsDNA rapidly decreased for dsDNA ≤72 bp, while the FA remained constant for the larger dsDNA (Fig. 2D). These results suggest that dsDNA length regulates not only the assembly kinetics but also the lifetime of bound duplexes.
PYD⋅PYD interactions, but not HIN200⋅dsDNA interactions, drive the assembly of AIM2 nucleoprotein filaments (16). Thus, we suspected that filament disassembly could be independent of dsDNA dissociation. To test this idea, we first determined whether AIM2PYD filaments are intrinsically dynamic or static by adding excess unlabeled AIM2PYD to preassembled FRET-labeled AIM2PYD filaments (Fig. 2E). Here, the FRET did not change for more than 1 h, indicating that subunits do not readily exchange (Fig. 2E, blue). In contrast, when we mixed excess unlabeled AIM2PYD with FRET-labeled AIM2PYD before removing MBP, we observed markedly decreased FRET efficiency (Fig. 2E, violet). Next, to test whether dsDNA-bound AIM2FL filaments are dynamic, we assembled AIM2FL⋅dsDNA complexes under the same conditions as described in Fig. 2E, but using FRET-labeled AIM2FL proteins and unlabeled dsDNA instead. Adding excess unlabeled AIM2FL did not change FRET signals even for 72-bp dsDNA, suggesting that the nucleoprotein filaments are static (Fig. 2F).
We then asked whether ASCPYD and the joint AIM2PYD⋅ASCPYD filaments also persist. Using the same strategy described in Fig. 2E we first found that the subunits of ASCPYD filaments do not exchange for more than 1 h (Fig. S1G). Next, we first assembled FRET donor-labeled AIM2PYD filaments then added FRET acceptor-labeled ASCPYD (Fig. 2G); the increase in FRET signals indicated that ASCPYD assembled on the AIM2PYD filament (Fig. 2H). Adding excess unlabeled ASCPYD after 90 min did not reduce the FRET signal for nearly 1 h, indicating that the joint complex is also static (Fig. 2 G and H). Together, our results indicate that AIM2, ASC, and AIM2⋅ASC filaments are all persistent and thus capable of generating three-stage, virtually infinite signal amplification. Moreover, our results also indicate that the assembly of the AIM2-ASC inflammasome is an intrinsically irreversible, nonequilibrium process.
dsDNA Induces Hysteresis: AIM2 Switches into the Filamentous State via dsDNA.
Host nucleases eventually destroy foreign dsDNA [e.g., TREX-1 (14)]. Thus, we asked whether AIM2-bound dsDNA could be accessed and destroyed by nucleases, triggering filament disassembly. To test this idea, we challenged AIM2FL⋅FAM-dsDNA complexes with micrococcal nuclease and found that FA signals rapidly decrease even for large dsDNA (Fig. 3A). Because the long dsDNA fragments do not dissociate within the time frame of the experiment (Fig. 2D), our observation suggests that degradation occurs within the filament; visualizing AIM2⋅dsDNA with agarose gel followed by SYBR-gold staining before and after nuclease treatment also indicated that dsDNA is essentially obliterated (Fig. S2A). In contrast, when we monitored the stability of AIM2FL filaments under the same conditions by FRET between labeled proteins upon adding nuclease the signal did not change over 1 h even for 39-bp dsDNA (Fig. 3B), suggesting that once assembled on dsDNA AIM2FL oligomers persist even if dsDNA is degraded.
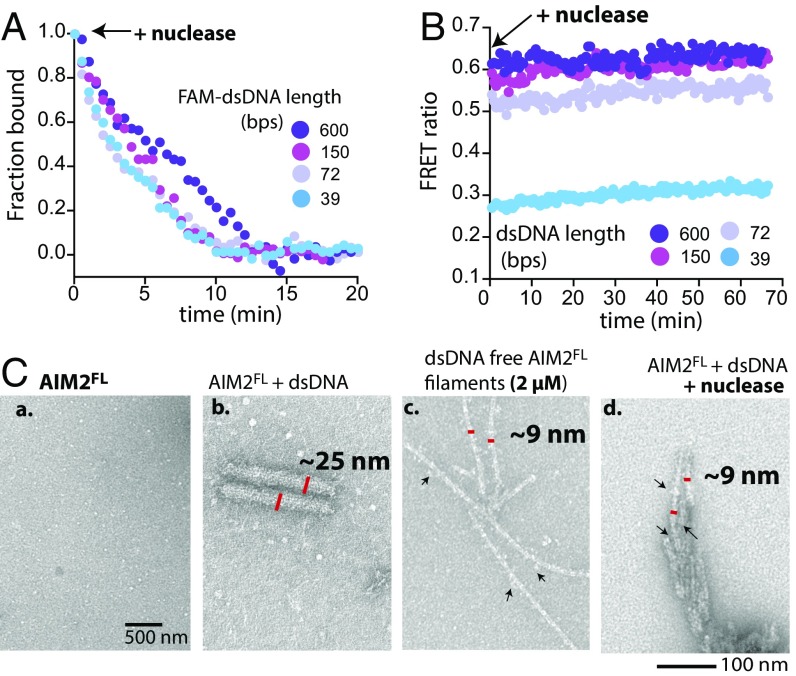
Fig. 3.
dsDNA induces hysteresis. (A) The 300 nM unlabeled AIM2FL was preincubated with each FAM-labeled dsDNA for 30 min (150 nM, binding-site normalized). At time 0, 1 mg/mL micrococcal nuclease was added and the change in FA of each FAM-dsDNA was monitored over time. The bound fractions calculated for each dsDNA based on its bound and unbound FA value. (B) The 300 nM FRET-labeled AIM2FL was preincubated with each unlabeled dsDNA for 30 min (150 nM, binding-site normalized). At time 0, 1 mg/mL micrococcal nuclease was added and the FRET emission ratio was monitored over time. (C) Negative-stain electron micrographs of (a) untagged 300 nM AIM2FL, (b) with 150 nM (binding-site normalized) 600-bp dsDNA, (c) apo-AIM2FL filament auto-assembled at 2 µM, and (d) after adding 1 mg/mL micrococcal nuclease to b and incubating it for 30 min. Arrows indicate HIN200 domains around the periphery of the PYD filament (“sprouts”).
We also imaged AIM2FL⋅dsDNA samples before and after nuclease treatment by EM (Fig. 3C; see also Fig. S2A, c). Of note, dsDNA-bound AIM2FL filaments resemble cylinders (∼25 nm wide; Fig. 3C, b), while dsDNA-free AIM2FL forms ∼9-nm-wide “Brussels sprouts”-like filaments in high concentrations [≥500 nM, Fig. 3C, c; the PYD filament constitutes the thin stem and AIM2Hin randomly clusters at periphery like sprouts (16)]. Upon nuclease treatment AIM2 filaments were still detected (Fig. 3C, d). Moreover, the filaments were also thinner and resembled Brussels sprouts rather than wide cylinders, indicating the absence of dsDNA (Fig. 3C; see also Fig. S2A, c). To further test that nuclease-treated filaments are not bound to shorter incompletely digested dsDNA we imaged AIM2FL in complex with various dsDNA lengths using nsEM (Fig. S1H). Here, we found that short dsDNA (≤72 bp) produced mostly unstructured oligomeric complexes (very short filaments are possible for 72-bp dsDNA, but it is difficult to distinguish due to the intrinsic resolution limit of nsEM). However, filaments were observed with longer dsDNA (≥150 bp) and they consistently resembled those observed before nuclease treatment (Fig. 3C, b), but not the thin filaments (Fig. 3C, c and d). These observations consistently suggest that the lingering filaments are not bound to dsDNA. Overall, we concluded that dsDNA induces hysteresis, allowing AIM2 to switch into the filamentous state.
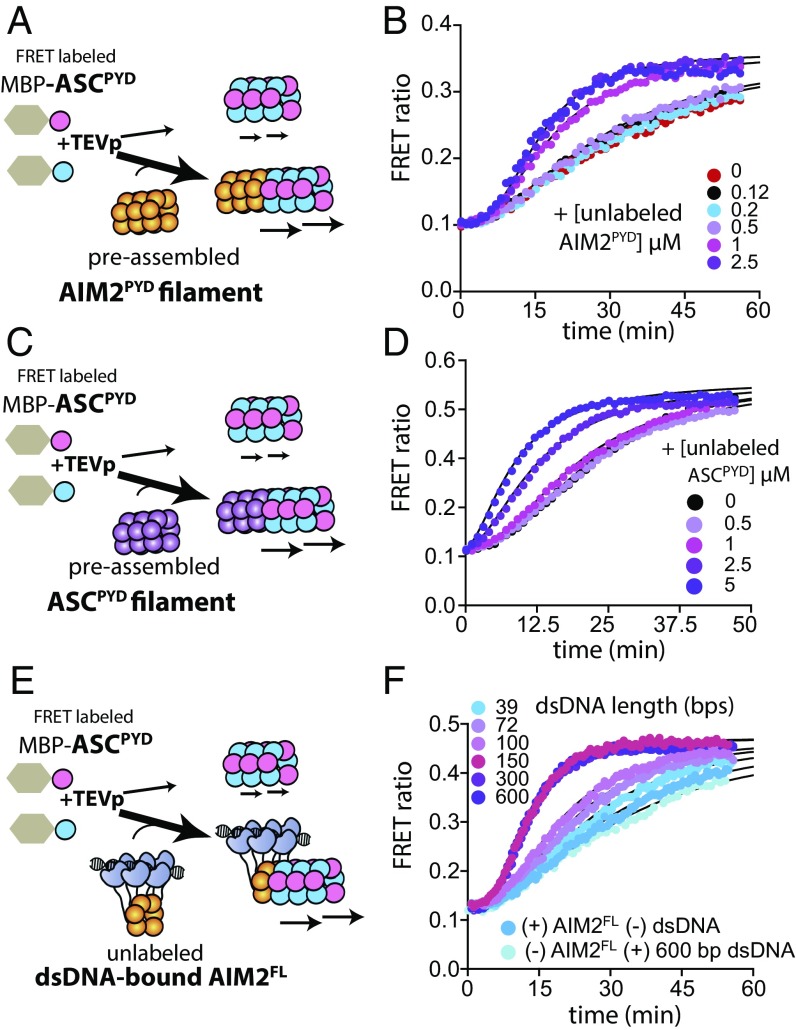
AIM2 Filaments Self-Propagate via Multiple Positive Feedback Loops.
Positive and negative feedback loops play important roles in steering the overall outcome of signaling pathways (5, 24, 25): Negative feedback induces new equilibria [e.g., metabolic pathways (5, 24, 25)], while positive feedback promotes binary responses [e.g., apoptosis (5, 24, 25)]. Whether either type of feedback is in place for the AIM2-ASC pathway remains poorly understood. For instance, although ASCPYD filaments provide a positive feedback to its own assembly in prion-like fashion (18), it is not known whether upstream AIM2 can also facilitate its own polymerization in trans. To address this issue we tested whether preassembled, unlabeled AIM2PYD filaments could enhance the polymerization of FRET-labeled AIM2PYD (Fig. 4A). We found here that the presence of unlabeled AIM2PYD accelerated the assembly of the FRET-labeled species sixfold (100 min). Of note, significant enhancements were observed only when the concentration of the unlabeled AIM2PYD was high enough to be filamentous (≥500 nM; Fig. 4B and Fig. S2B), indicating that polymerization is a prerequisite for trans activation. Currently, it is thought that dsDNA length dependence is limited to the initial binding/assembly step (16); however, we do not yet understand whether the resulting AIM2 complexes on different dsDNA lengths are equally effective in downstream or this newly observed self-propagation activity. We thus asked whether dsDNA lengths that support AIM2FL oligomers could regulate the trans-polymerization activity. To ensure that signals only arise from trans-PYD–PYD interactions, we used FRET-labeled AIM2PYD and unlabeled AIM2FL in these experiments (Fig. 4C). Here, a catalytic amount of AIM2FL accelerated the polymerization of AIM2PYD only when bound to dsDNA, again indicating that polymeric AIM2 is required to promote self-propagation (Fig. 4D). Moreover, AIM2FL bound to 52-bp dsDNA or shorter failed to accelerate the polymerization of AIM2PYD, while the maximal acceleration (sixfold, 100 min) was reached at ∼200 bp (Fig. 4D and Fig. S2C), suggesting that the high-pass filter against short dsDNA also regulates its self-propagation.
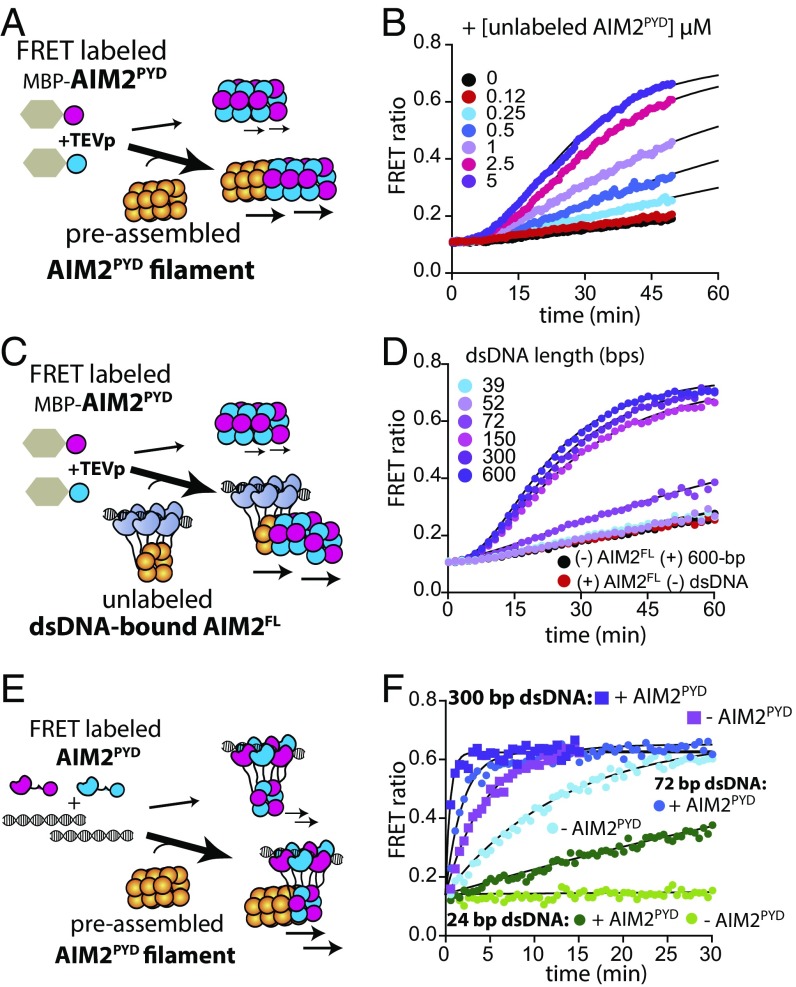
Fig. 4.
AIM2 propagates autonomously. (A) A scheme describing FRET-based experiments testing whether AIM2PYD can self-propagate. (B) The time-dependent increase in FRET emission ratio of donor- and acceptor-labeled AIM2PYD (2.5 µM) was monitored with increasing concentrations of unlabeled AIM2PYD that had been allowed to polymerize by preincubating with TEVp for 1 h. (C) A scheme for FRET-based experiments testing whether the length of AIM2FL-bound dsDNA regulates its ability to induce the polymerization of AIM2PYD in trans. (D) The time-dependent increase in FRET emission ratio of donor- and acceptor-labeled AIM2PYD (2.5 µM) was monitored in the presence of preassembled AIM2FL oligomers (72 nM) on 10 µg/mL of indicated dsDNA lengths (preincubated for 1 h). (E) A scheme for FRET-based experiments testing whether AIM2PYD filaments regulate the dsDNA-mediated assembly of AIM2FL. (F) The time-dependent assembly of FRET-labeled AIM2FL (72 nM) on various dsDNA lengths (10 µg/mL) was monitored in the presence or absence of preformed AIM2PYD filaments (2.5 µM). The lines are fits to a single-exponential equation.
Considering that the oligomerization of PYD is critical for dsDNA binding by AIM2 (16), we next asked whether existing AIM2 filaments could also enhance dsDNA binding of AIM2 monomers by providing an oligomerization template. To test this idea we determined the dsDNA-mediated assembly kinetics of FRET-labeled AIM2FL in the presence of preassembled AIM2PYD filaments (Fig. 4E). We found that AIM2PYD filaments indeed accelerate the dsDNA-mediated assembly of AIM2FL (Fig. 4F and Fig. S2D). The enhancements were more pronounced for the shorter dsDNA (Fig. S2D; e.g., 32-fold for 24 bp vs. fourfold for 300 bp), suggesting that, unlike the forward pathway, this positive feedback loop employs a low-pass filter function in which long dsDNA signals are attenuated (Fig. S1B, b). Together, we concluded that multiple positive feedback loops reinforce the assembly of AIM2 on cytoplasmic dsDNA.
The Length of Bound dsDNA Controls the Ability of AIM2 Filaments to Accelerate the Polymerization of ASC.
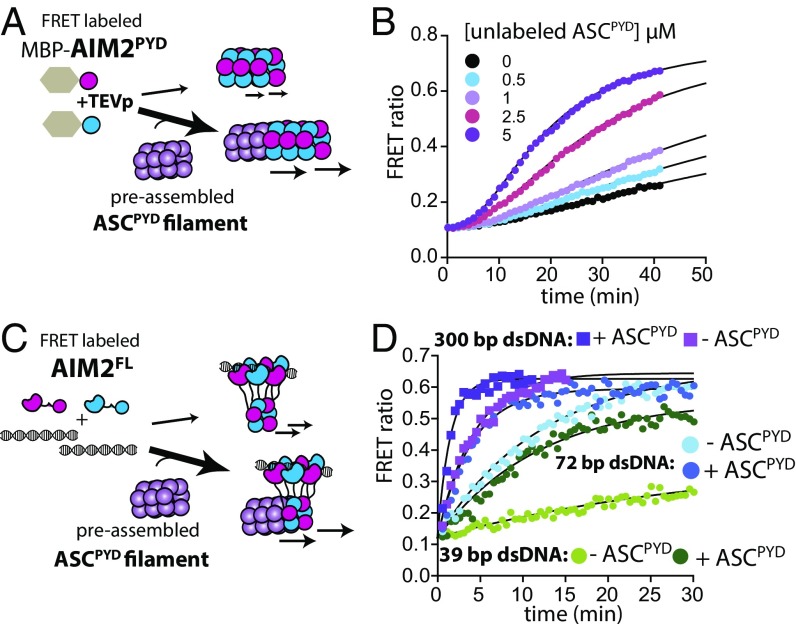
ASC is the central hub in the inflammasome pathways, and its assembly is not only induced by upstream receptors but also by self-propagation (9, 18). It was previously reported that AIM2PYD can accelerate the polymerization of ASCPYD in a concentration-dependent manner (9); however, it is not clear whether AIM2 has to be oligomeric to induce the downstream polymerization. Moreover, the ability of ASCPYD to accelerate its own polymerization has not been directly demonstrated. To address these questions we monitored the polymerization of FRET-labeled ASCPYD in the presence of increasing concentrations of AIM2PYD and ASCPYD (Fig. 5 A and C). Here, we first confirmed that we can recapitulate the previous observation (9) that AIM2PYD directly accelerates the polymerization of ASCPYD (Fig. 5B). Of note, we found that the presence of unlabeled AIM2PYD and ASCPYD above their respective threshold polymerization concentrations reduced the assembly th of labeled ASCPYD up to 15 min, indicating that oligomers are required to accelerate the nascent filament formation (three- and fourfold, respectively; Fig. 5 B and D and Fig. S2 E and F). Moreover, a catalytic amount of 600-bp dsDNA-bound AIM2FL also accelerated ASCPYD assembly to a similar extent, which is also consistent with the previous observation (9) (Fig. 5 E and F; see also Fig. S2G). Of note, the assisted assembly of ASC is still slower than the assembly of AIM2 on dsDNA by 10-fold (Fig. 2B vs. Fig. S2G; see Dataset S1 for values), suggesting that ASC assembly is likely the rate-limiting step. Our observation indicates that for the length of dsDNA to be a factor in the cellular activity it has to play a role in the slower step. We thus monitored ASCPYD polymerization in the presence of preassembled AIM2FL filaments on various dsDNA lengths (Fig. 5 E and F). Indeed, the ability of AIM2FL to accelerate ASC polymerization also changed up to fourfold in a dsDNA length-dependent manner (Fig. 5F and Fig. S2G). Together, we concluded that both AIM2 and ASC filaments accelerate the assembly of ASC monomers and the high-pass filter against short dsDNA is still in place for the second signaling step.
Fig. 5.
The length of AIM2-bound dsDNA regulates the polymerization of ASCPYD. (A) A scheme for FRET-based experiments testing whether AIM2PYD filaments regulate ASCPYD filament assembly. (B) The time-dependent assembly of FRET-labeled ASCPYD (2.5 µM) was monitored with increasing concentrations of unlabeled MBP-AIM2PYD that had been allowed to polymerize by preincubating with TEVp for 1 h. (C) A scheme for FRET-based experiments testing whether the preassembled ASCPYD filament accelerates its own assembly. (D) The time-dependent assembly of FRET-labeled ASCPYD (2.5 µM) was monitored with increasing concentrations of unlabeled MBP-ASCPYD that had been allowed to polymerize by preincubating with TEVp for 1 h. (E) A scheme for FRET-based experiments testing whether the length of AIM2FL-bound dsDNA regulates its ability to induce ASC polymerization. (F) The time-dependent increase in FRET emission ratio of donor- and acceptor-labeled ASCPYD (2.5 µM) was monitored in the presence of preassembled AIM2FL oligomers (36 nM) on indicated dsDNA lengths (10 µg/mL).
ASC Filaments Reinforce AIM2 Assembly via Positive Feedback Loops.
AIM2 is not the only receptor that induces the polymerization of ASC. Indeed, during pathogen invasion the activation of AIM2 often coincides with those of other inflammasome receptors such as NLRP3 and NLRC4 (26, 27). We thus considered whether the induction of ASC polymers by other means would provide positive or negative impact on the polymerization of upstream AIM2. To test this idea we tested whether preassembled ASC filaments can feedback-regulate the assembly of AIM2 filaments. Here, we found that the presence of unlabeled ASCPYD above its threshold concentration for filament assembly accelerated the polymerization of FRET-labeled AIM2PYD up to sevenfold (100 min; Fig. 6 A and B and Fig. S2H). We also found that ASCPYD filaments accelerated the assembly of FRET-labeled AIM2FL on dsDNA (Fig. 6 C and D and Fig. S2I). As with AIM2PYD (Fig. S2D), ASCPYD accelerated the assembly of AIM2FL on ≤150-bp dsDNA more significantly (Fig. S2I), indicating that this feedback loop also employs a low-pass filter. Taken together, we concluded that the assembly of the AIM2-ASC inflammasome is reinforced by an array of positive feedback loops.
Fig. 6.
ASCPYD filaments provide positive feedbacks to the assembly of AIM2. (A) A scheme for FRET-based experiments testing whether ASCPYD filaments accelerate the assembly of AIM2PYD. (B) The time-dependent assembly of FRET-labeled AIM2PYD (2.5 µM) was monitored with increasing concentrations of unlabeled MBP-ASCPYD that had been preincubated with TEVp for 1 h. (C) A scheme for FRET-based experiments testing whether ASCPYD filaments accelerate the dsDNA-mediated assembly of AIM2FL. (D) The time-dependent assembly of FRET-labeled AIM2FL (72 nM) on various dsDNA lengths (10 µg/mL) in the presence or absence of preformed ASCPYD filaments (2.5 µM). The lines are fits to a single-exponential equation.
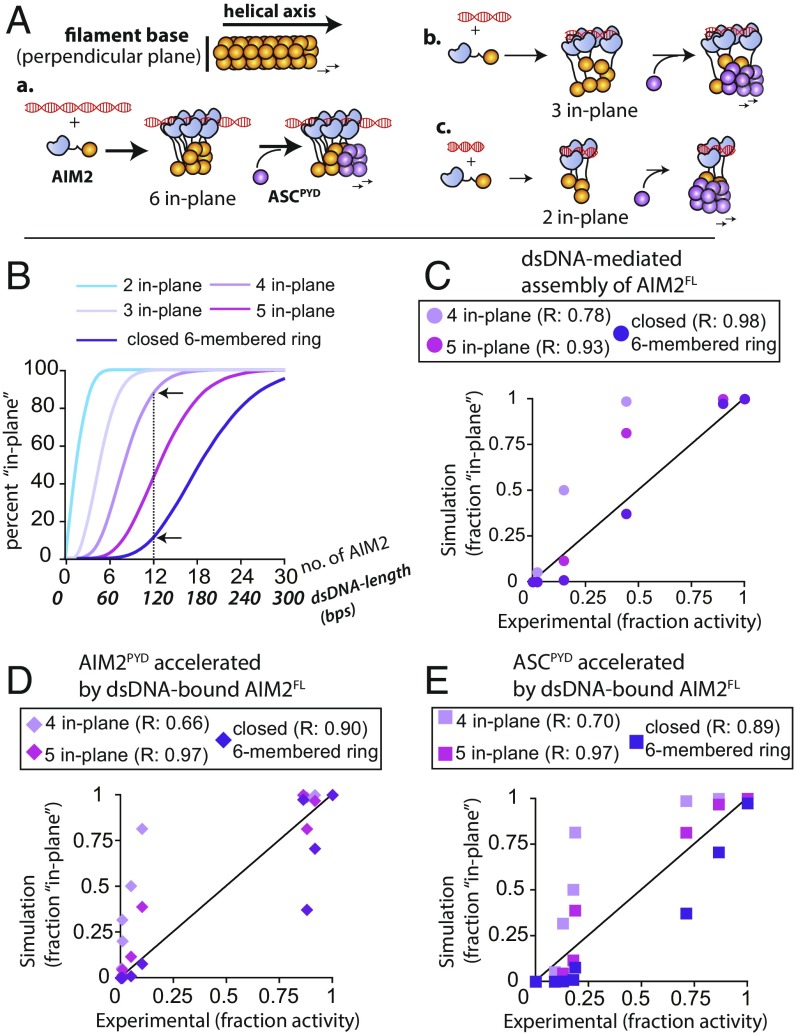
dsDNA Length May Regulate the Probability of Assembling the Helical Base of the AIM2 Filament.
The signaling activity of AIM2 depends on the length of foreign dsDNA in cells (12, 17). Previously, we proposed that dsDNA length-dependent binding would act as the dominant underpinning mechanism (16). However, we found here that dsDNA length regulates not only the initial assembly but also all subsequent assembly activities (Figs. 2–6). Moreover, it appears that the ability of AIM2 to form filaments depends on the length of dsDNA (Fig. S1H). While contemplating a unifying mechanism by which dsDNA can differentially regulate the filament-mediated signaling activity we recalled that both AIM2PYD and ASCPYD assemble into six-start helical filaments and interact coaxially (i.e., six protomers constitute the filament base; Fig. 7A and refs. 9 and 16). We also noted that assembling an intact helical base is important for the signaling activities of the supramolecular platforms assembled by PYD-like domains [e.g., the Rig-I⋅mitochondrial antiviral signaling (MAVS) complex and the PIDDosome], as such a well-defined base would provide the most number of coaxial protein–protein interaction surfaces (19, 28, 29). We thus hypothesized that the role of dsDNA length is to regulate the probability of assembling the intact base of the AIM2 filament (Fig. 7A), as the length dictates the number of bound molecules per duplex (16). To test this idea, we conducted a Monte Carlo simulation that determines the probability of contiguously placing a given number of PYDs on the perpendicular plane of a six-start filament (Fig. S3A; see Dataset S2 for the code). We then converted the number of AIM2PYD used in each simulation to dsDNA length in base pairs per the binding footprint of AIM2FL [10 bp/AIM2FL (16); Fig. 7B]. For example, our results suggest that there is a 10% chance of completing the filament base by 12 AIM2 molecules bound to 120-bp dsDNA, while there is an 88% chance of placing at least four molecules in the base (arrows in Fig. 7B). We then compared the fraction of AIM2PYDs in-plane to the normalized fractional assembly activities.
Fig. 7.
Monte Carlo simulation of AIM2 nucleoprotein filament assembly. (A) A cartoon describing the potential role of dsDNA length in assembling the intact base of the AIM2 filament. (B) The probability of contiguously placing the minimum number of AIM2PYDs in the filament base was plotted against a various fixed number of AIM2 molecules/dsDNA length (one AIM2 per 10 bp). (C–E) Plots of fraction activities vs. fraction of AIM2PYDs in the contiguous perpendicular plane. The fraction activity for each experiment was calculated by dividing a given dataset with its highest determined value. The correlation coefficients were determined from the equation y = x.
Judging by the correlation coefficients (R values), the dsDNA length-dependent assembly of AIM2FL fitted best with the probability of completing the six-membered base by the given number of PYDs (thus the corresponding dsDNA length; Fig. 7C). However, the acceleration of AIM2PYD and ASCPYD polymerizations by dsDNA-bound AIM2FL correlated best with the probability of placing at least five AIM2PYDs in-plane (Fig. 7 D and E). The reduced requirements for the latter could arise from feedback interactions and/or additional PYD molecules filling in the sixth protomer. Overall, our results suggest that dsDNA length dependence of the AIM2-ASC inflammasome arises because the duplex length dictates the probability of assembling the intact (or near complete) helical base of the AIM2 filament.
Quantitatively Modeling the Assembly of the AIM2-ASC Inflammasome.
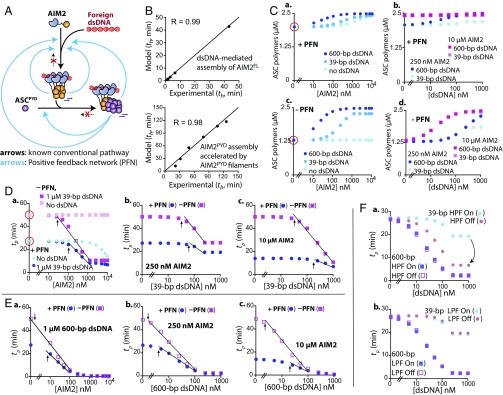
We found here that the AIM2-ASC inflammasome filters short dsDNA as noise at each signaling step, generates sustained amplification, and employs multiple positive feedback loops. These results warrant a revision of the conventional linear signaling model (Fig. 1A) to a complex network-like assembly (Fig. 8A). To understand and predict how such a signaling circuit would operate we modeled the assembly using differential equations (Fig. S3B). Our approach here was to establish a parsimonious model that could globally fit our data without excessive assumptions. This approach resulted in a model that predicts the assembly ths consistent with our experiments (average R = 0.95 ± 0.05, n = 11; Fig. 8B; see also Fig. S4A). Thus, although more complicated formulations are possible (e.g., ref. 30), we reasoned that our model provides a robust framework for decoding the design principles of the assembly network.
Fig. 8.
Modeling the assembly of the AIM2-ASC inflammasome. (A) The assembly network of the AIM2-ASC inflammasome. (B) Sample plots of model vs. observed (experimental) th. (C) Plots of ASC polymer amounts upon varying AIM2 and dsDNA at t = ∞. ASC concentration in C–F is fixed at 2.5 µM. (D) Plots of ASC tps vs. varying AIM2 and 39-bp dsDNA in the presence and absence of PFN. The lines in D and E indicate [AIM2] or [dsDNA] ranges that induce the most dynamic changes in ASC tp, while the arrows indicate the concentrations at which such changes begin. (E) Plots of ASC tps vs. varying AIM2 and 600-bp dsDNA in the presence and absence of PFN. (F) Plots of ASC tps vs. varying 39- and 600-bp dsDNA in the presence and absence of high-pass filters (a, HPF) and low-pass filters (b, LPF).
Positive Feedback Network Ensures Consistent Total Output.
One of the major findings here is that the assembly employs multiple positive feedback loops (i.e., positive feedback network, PFN). To test the role of PFN, we generated a hypothetical minimal system in which the entire PFN is removed and dsDNA is required for signaling (e.g., all of the blue arrows in Fig. 8A are removed) and monitored how it would respond differently to noise (short dsDNA) vs. signal (long dsDNA). Below, we show exemplary cases, and the results for different ASC concentrations and additional dsDNA lengths can be found in Dataset S3, which also agree/scale with the presented results.
The in vivo expression level of AIM2 is typically low; however, it is overexpressed by interferons during pathogen invasion (31). However, the amount of cytoplasmic dsDNA would vary depending on the source (e.g., bacterial genome, nearby necrosis, or dysfunctional mitochondria). It is noteworthy that AIM2 displays a digital response in cells (6), implying that the pathway may have an intrinsic mechanism to generate a predictable yet stable output regardless of input variance. Thus, we first asked whether PFN has a role in shaping the final outcome of the AIM2-ASC pathway. Here, without dsDNA or AIM2, we found that PFN increases the total amount of ASC polymers by 35% at time = ∞ vs. in the absence (e.g., total output, red circles in Fig. 8C, a vs. Fig. 8C, c). Additional input (AIM2 or dsDNA) marginally raised the final ASC polymer population when PFN was present (≤20%; Fig. 8C, a and b). These observations are consistent with our biochemical experiments in which the polymerization of ASC is not thermodynamically driven but is kinetically controlled due to the lack of intrinsic disassembly (e.g., nonequilibrium; Figs. 2 and 3). By contrast, increasing AIM2 or dsDNA allowed the minimal system to eventually catch up to similar ASC polymer amounts as the intact system, with the longer dsDNA being more effective (Fig. 8C, c and d). As expected from the severed dsDNA-free assembly, increasing AIM2 without dsDNA for the minimal system had no effect (Fig. 8C, c). These results indicate that PFN ensures the total output is consistent regardless of input variance, which is a hallmark of a digital response (3, 5, 32). By contrast, the variable output in the absence of PFN indicates that the pathway now operates in more analog (gradual) fashion (3, 5, 32).
PFN Preconditions the Assembly to Trigger Ultrasensitive Digital Responses.
Next, we examined the role of PFN in regulating ASC polymerization kinetics with short and long dsDNA (noise vs. signal). Of note, because the final ASC polymer amounts are variable (Fig. 8C), we compared the time required to polymerize (tp) 1 µM of 2.5 µM ASC with or without PFN (i.e., ([2.5 µM] − [ASCthreshold])/2 = 1 µM; [ASCthreshold] = 0.5 µM, as determined in Fig. 1F).
Without dsDNA and AIM2 we found that PFN allows ASC to accelerate its own assembly by 23 min (red circles in Fig. 8D, a). Moreover, with PFN, increasing AIM2 1,000-fold minus dsDNA further enhanced ASC tp twofold (13 min; Fig. 8D, a), while ASC tp was inert without PFN because it requires dsDNA for signaling (Fig. 8D, a). Of note, PFN reduced the variance in ASC tp upon increasing AIM2 in the presence of short dsDNA, and vice versa (i.e., heights and slopes of the lines are smaller when PFN is present in Fig. 8D). For example, increasing AIM2 in the presence of 1 µM 39-bp dsDNA (a large amount of noise) accelerated ASC tp fourfold with PFN (20 min; Fig. 8D, a, dark blue). However, without PFN, increasing AIM2 accelerated tp fivefold (40 min; Fig. 8D, a, magenta). Moreover, increasing 39-bp dsDNA in the presence of low AIM2 accelerated ASC tp 1.4-fold with PFN (8 min), while the enhancement was 1.8-fold (22 min) without PFN (Fig. 8D, b). In the presence of high AIM2 (Fig. 8D, c), increasing 39-bp dsDNA accelerated ASC tp twofold with PFN (7 min), while the rate enhancement was almost fivefold without PFN (40 min). Moreover, AIM2 or dsDNA amounts required to influence ASC tp were consistently higher when PFN was present (the x-axis positions of the arrows in Fig. 8D).
However, as expected from the correct signal, 600-bp dsDNA accelerated ASC tp more readily (Fig. 8E). Notably, as observed from 39-bp, PFN also reduced the overall changes in ASC tp upon increasing the concentrations of AIM2 and 600-bp dsDNA (line slopes and heights are smaller with PFN in Fig. 8E). For example, increasing AIM2 plus 1 µM 600-bp accelerated ASC tp 50-fold with PFN, while the acceleration was 80-fold without PFN (Fig. 8E, a). With fixed low AIM2 (Fig. 8E, b), increasing 600-bp dsDNA then improved ASC tp 13-fold when PFN was intact (25 min), while the acceleration was 18-fold without PFN (47 min). In the presence of high AIM2 (Fig. 8E, c), the acceleration was 27-fold with PFN (14 min), while it was 80-fold without PFN (48 min). As observed from 39-bp dsDNA, AIM2 and dsDNA concentrations required to influence ASC tp were also consistently higher with PFN for 600-bp dsDNA (the x-axis positions of the arrows in Fig. 8E).
In sum, we found that PFN steered ASC tp (output) in three major ways. First, it primed the assembly to proceed faster without additional input (dsDNA and AIM2; red circles in Fig. 8D, a). Second, due to this priming effect, PFN then reduced output dynamics and variance upon changing input concentrations (heights and slopes of the lines in Fig. 8 D and E were smaller with PFN). Third, with PFN not only was a larger amount of input required to influence ASC tp (the x-axis positions of the arrows are shifted to right in Fig. 8 D and E), but most dynamic changes also occurred over smaller input ranges (the lines in Fig. 8 D and E are shorter). These traits are remarkably consistent with the ideas of added noise filtering, analog-to-digital (A/D) conversion, and induction of ultrasensitivity (see also Fig. S5), all of which are the hallmarks of positive feedback (1–5, 24, 32, 33). For instance, consistent with the idea of added noise filtering, PFN dampened the changes in ASC tp when a large amount of short dsDNA entered the system (e.g., Fig. 8D, b and c). Consistent with A/D conversion, the changes in ASC tps were less dynamic with PFN (e.g., Fig. 8E, b and c). Finally, consistent with ultrasensitivity, PFN required higher amounts of dsDNA to influence ASC tp, and such changes also required a smaller change in input (e.g., Fig. 8E, c). Overall, we concluded that PFN positions the assembly in “standby,” ready to trigger a highly sensitive digital response against foreign dsDNA.
Multiple High-Pass Filters Synergistically Attenuate the Short dsDNA Signal.
We proposed previously that dsDNA length-dependent assembly of AIM2 would be important in regulating its cellular activity (16). However, we found here that dsDNA binding is unlikely rate-limiting, and dsDNA length regulates all subsequent assembly activities (Figs. 2–5). Moreover, AIM2 and ASC filaments accelerated the assembly of AIM2 monomers on dsDNA, with the positive impact more pronounced with shorter dsDNA (low-pass filters; Fig. S2 D and I). These observations then raised a question as to how the different filters would work together. We thus simulated a scenario that, after the initial assembly, all dsDNA lengths are as effective as 600-bp not only in AIM2 self-propagation but also downstream signaling. The lack of dsDNA length dependence beyond the initial assembly resulted in faster ASC tps with increasing 39-bp dsDNA, significantly reducing the gap between signal and noise (Fig. 8F, a). However, removing the low-pass filters in the feedback loops slowed the assembly by 30 s (Fig. 8F, b; i.e., the relative benefit of preformed filaments are as low as seen from 600-bp dsDNA in Fig. 6F). These results suggest that the low-pass filters in the feedback loops cannot override the high-pass filters in the forward pathway. We thus concluded that multiple high-pass filters synergistically prevent noise from inducing the assembly of AIM2-ASC inflammasome.
Discussion
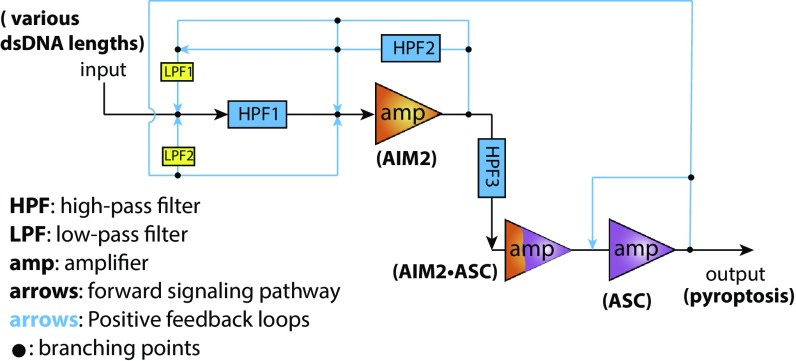
Although the assembly of suprastructures has emerged as a new paradigm in intracellular signaling pathways (19), the design principles of their underpinning signaling circuits remain poorly understood. We show here that the filamentous assembly of the AIM2-ASC inflammasome effectively employs three key principles essential for a high-fidelity digital response: synergistic noise filtering, multistage amplification, and PFN. Of note, we can draft an electronic circuit employing these features (Fig. 9), highlighting the shared design principles between biological and artificial networks.
Fig. 9.
A draft of an electronic circuit conceptualized by the design principles of the AIM2-ASC pathway. HPFs represent the preference for larger dsDNA in each signaling step (i.e., dsDNA length dependence in 1: dsDNA mediated assembly, 2: trans-activation of AIM2, and 3: ASC polymerization). AIM2, AIM2⋅ASC, and ASC filaments are described as amplifiers given their persistence. LPFs represent the enhanced assembly on shorter dsDNA in the presence preassembled 1: AIM2 and 2: ASC filaments. Pathways without any filter indicate dsDNA-free assembly. All of the feedback loops are connected to the same forward direction (polarity) to generate positive feedback.
Considering that the major outcome of the AIM2-ASC pathway is cell death (8), noise filtering is deemed to be critical. Of note, dsDNA length plays a key role in regulating the activity of the AIM2-ASC inflammasome in cells (12, 17). Here, we find that the inflammasome filters short dsDNA as noise in three key steps: receptor assembly, receptor self-propagation, and downstream signaling (Figs. 2B, 4D, and 5F). We found that the additivity of these filters is critical for generating the overall dsDNA length-dependent response (Fig. 8F, a). These additional filters are necessary because formation of AIM2⋅dsDNA is unlikely rate-limiting (e.g., Fig. 2B vs. Fig. S2G). We reason that the assembly of filaments is ideal for implementing synergistic noise filters. It is because not only must all primary signaling components be physically connected during an active cascade, but also the assembly of their congruent architecture (16) would play a key role in both affinity and signaling activity.
Once correct signals initiate a pathway, the next important step in achieving high fidelity is amplification (4, 5). We find here that once dsDNA triggers assembly AIM2, ASC, and AIM2⋅ASC filaments persist, generating a three-stage, essentially infinite amplification (Fig. 2). Moreover, although dsDNA can be accessed and degraded, AIM2 filaments still persist, leaving a lasting memory of infection at the molecular level (hysteresis; Fig. 3 and Fig. S2A). We reason that filamentous assemblies are ideal for generating high stability, as passive disassembly would only initiate from either end [e.g., actin filaments (21)]. The high stability of the AIM2-ASC inflammasome underscores that the assembly commits to the death process once initiated. Moreover, it may also have a role in various autoinflammatory disorders whose hallmarks include persistence (8, 13, 14).
We found here that PFN reinforces the assembly of the AIM2-ASC inflammasome (Figs. 4–6), allowing the host signaling pathway to actively tailor its response instead of passively relaying on the cytoplasmic dsDNA signal. Moreover, PFN positions the assembly on “standby,” poised to trigger ultrasensitive digital responses against invading pathogens. It is also noteworthy that positive feedback loops are known to promote binary responses, hysteresis, and noise attenuation in both biological and electrical pathways (5, 24, 33). Considering that the major outcome of the AIM2-ASC inflammasome is death (a binary response), the implementation of PFN is bound to be essential for the design of the AIM2-ASC inflammasome pathway. We also reasoned that the filamentous assemblies are ideal for implementing PFN due to the congruent architecture (9, 16).
Both AIM2 and ASC were once thought to be autoinhibited (17). However, we and others have shown differently (16, 34). Here, we found several kinetic mechanisms that would sufficiently suppress the spurious activity of the AIM2-ASC inflammasome under normal conditions. First, polymerization is a prerequisite not only for forward signaling but also for the feedback loops (Figs. 2–6). Second, without dsDNA both AIM2 and ASC display threshold behaviors, where there is no basal assembly below their threshold concentrations (Fig. 1). Third, the dsDNA-free assembly kinetics of AIM2 is significantly slower than that of ASC (see Dataset S1 for values; Figs. 1–6). This rate-limiting first step would then effectively bar the receptor from influencing ASC polymerization without foreign dsDNA (e.g., increasing AIM2 by 1,000-fold leads to only twofold faster ASC assembly; Fig. 8D, a). We propose that assembling suprastructures that share congruent architectures also plays a role in this regard, because the number of interactions and iterations required to place each monomer on a correct position in the congruent filaments would involve a large kinetic barrier (i.e., filament base; Fig. 7). Additionally, we speculate that the balance between the intrinsic polymerization kinetics of ASC and its degradation rates would play a key role in keeping the ASC polymer population in check under normal conditions.
Signaling by the sequential assembly of filamentous oligomers has emerged as a shared paradigm in cytoplasmic nucleic acid sensing pathways (28). Of note, Rig-I-like receptors (RLRs) and MAVS protein assemble into congruent filaments to initiate IFN responses (28). It is possible that RLR–MAVS complexes are more dynamic and less reinforced than the AIM2-ASC inflammasome, given that interferons do not directly induce cell death (28). However, RLR–MAVS complexes might also assemble in a digital fashion and generate a sustained response until cell death pathways are activated [RLRs and MAVS are also implicated in inducing cell death (35, 36)]. Future mechanistic studies are warranted to decipher whether there are shared design principles between AIM2-ASC and RLR–MAVS signaling platforms.
The activation of the AIM2-ASC pathway often coincides with those of other inflammasomes (26, 27). Additionally, pyrin-only proteins eventually enter the cascade to provide much needed negative feedback (37). Future investigation will reveal not only how other host receptors and regulators modulate the digital assembly network of the AIM2-ASC inflammasome but also how the design of the AIM2-ASC circuit compares to those of other inflammasomes.
Materials and Methods
Biochemical Experiments.
FRET and FA measurements were performed in 384-well microplates (Corning) using the Tecan M1000 plate-reader at room temperature (23 ± 3 °C). All assays were performed in 40 mM Hepes-KOH, pH 7.4, 160 mM KCl, 5% glycerol, 0.1% triton-X-100, 1 mM EDTA, and 5 mM DTT; 1 mg/mL TEVp was added to remove MBP from AIM2 and ASC constructs. For AIM2FL, MBP was removed before measurements by preincubating with TEVp for 30 min. All reported values are the average of at least three independent experiments, and the errors are determined using SDs. To obtain the apparent polymerization rates (kpoly), we first determined the half-time (th) of each polymerization reaction using a variant of the Hill equation: observed FRET ratio at time (t) = (max observed FRET ratio)/(1 + (th/t)n), where n is an exponent that allows fitting to the lag-phase. kpoly was then calculated by ln(2)/th. Of note, while analyzing our results we found that we could use double-exponential equations to fit our data (Figs. 3–5, in particular); however, we decided to use the overall half-times (t.h and kpolys) and single-exponential fits for quantitation to facilitate more intuitive understanding and assessing of the assembly mechanism. All measured values are listed in Dataset S1. nsEM experiments were conducted using a Philips BioTwin CM120 (FEI) as described previously (16). Reagents were prepared according to ref. 16; see also SI Materials and Methods.
Computational Experiments.
The Monte Carlo simulation for calculating the probability of assembling the intact filament base was written in Python (see also Fig. S3A and Dataset S2). The quantitative modeling of the assembly pathway was initially performed using Excel. Integration and global fitting were performed using Python scripts (see Fig. S3B). See also SI Materials and Methods for details.
Supplementary Material
Acknowledgments
This work was supported by Jerome L. Greene Foundation and American Cancer Society Research Scholars Grant DMC-RG-15-224 (to J.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712860115/-/DCSupplemental.
References
- 1.Alon U. Network motifs: Theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 2.Angeli D, Ferrell JE, Jr, Sontag ED. Detection of multistability, bifurcations, and hysteresis in a large class of biological positive-feedback systems. Proc Natl Acad Sci USA. 2004;101:1822–1827. doi: 10.1073/pnas.0308265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Das J, et al. Digital signaling and hysteresis characterize ras activation in lymphoid cells. Cell. 2009;136:337–351. doi: 10.1016/j.cell.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grubelnik V, Dugonik B, Osebik D, Marhl M. Signal amplification in biological and electrical engineering systems: Universal role of cascades. Biophys Chem. 2009;143:132–138. doi: 10.1016/j.bpc.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Lim W, Mayer B, Pawson T. Cell Signaling: Principles and Mechanisms. Garland; New York: 2015. [Google Scholar]
- 6.Liu T, et al. Single-cell imaging of caspase-1 dynamics reveals an all-or-none inflammasome signaling response. Cell Rep. 2014;8:974–982. doi: 10.1016/j.celrep.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 7.Tay S, et al. Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature. 2010;466:267–271. doi: 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Lu A, et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts TL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 13.Man SM, Karki R, Kanneganti TD. AIM2 inflammasome in infection, cancer, and autoimmunity: Role in DNA sensing, inflammation, and innate immunity. Eur J Immunol. 2016;46:269–280. doi: 10.1002/eji.201545839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu B, et al. The DNA-sensing AIM2 inflammasome controls radiation-induced cell death and tissue injury. Science. 2016;354:765–768. doi: 10.1126/science.aaf7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrone SR, et al. Assembly-driven activation of the AIM2 foreign-dsDNA sensor provides a polymerization template for downstream ASC. Nat Commun. 2015;6:7827. doi: 10.1038/ncomms8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin T, et al. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai X, et al. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156:1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kagan JC, Magupalli VG, Wu H. SMOCs: Supramolecular organizing centres that control innate immunity. Nat Rev Immunol. 2014;14:821–826. doi: 10.1038/nri3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu A, et al. Plasticity in PYD assembly revealed by cryo-EM structure of the PYD filament of AIM2. Cell Discov. 2015;1:15013. doi: 10.1038/celldisc.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stratmann SA, Morrone SR, van Oijen AM, Sohn J. The innate immune sensor IFI16 recognizes foreign DNA in the nucleus by scanning along the duplex. eLife. 2015;4:e11721. doi: 10.7554/eLife.11721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turkin A, et al. Speeding up biomolecular interactions by molecular sledding. Chem Sci (Camb) 2016;7:916–920. doi: 10.1039/c5sc03063c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cinquin O, Demongeot J. Roles of positive and negative feedback in biological systems. C R Biol. 2002;325:1085–1095. doi: 10.1016/s1631-0691(02)01533-0. [DOI] [PubMed] [Google Scholar]
- 25.Zhang T, Brazhnik P, Tyson JJ. Computational analysis of dynamical responses to the intrinsic pathway of programmed cell death. Biophys J. 2009;97:415–434. doi: 10.1016/j.bpj.2009.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denes A, et al. AIM2 and NLRC4 inflammasomes contribute with ASC to acute brain injury independently of NLRP3. Proc Natl Acad Sci USA. 2015;112:4050–4055. doi: 10.1073/pnas.1419090112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karki R, et al. Concerted activation of the AIM2 and NLRP3 inflammasomes orchestrates host protection against Aspergillus infection. Cell Host Microbe. 2015;17:357–368. doi: 10.1016/j.chom.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sohn J, Hur S. Filament assemblies in foreign nucleic acid sensors. Curr Opin Struct Biol. 2016;37:134–144. doi: 10.1016/j.sbi.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su TW, et al. Structural insights into DD-fold assembly and caspase-9 activation by the Apaf-1 apoptosome. Structure. 2017;25:407–420. doi: 10.1016/j.str.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 30.Frieden C, Goddette DW. Polymerization of actin and actin-like systems: Evaluation of the time course of polymerization in relation to the mechanism. Biochemistry. 1983;22:5836–5843. doi: 10.1021/bi00294a023. [DOI] [PubMed] [Google Scholar]
- 31.Veeranki S, Duan X, Panchanathan R, Liu H, Choubey D. IFI16 protein mediates the anti-inflammatory actions of the type-I interferons through suppression of activation of caspase-1 by inflammasomes. PLoS One. 2011;6:e27040. doi: 10.1371/journal.pone.0027040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qian H. Cooperativity in cellular biochemical processes: Noise-enhanced sensitivity, fluctuating enzyme, bistability with nonlinear feedback, and other mechanisms for sigmoidal responses. Annu Rev Biophys. 2012;41:179–204. doi: 10.1146/annurev-biophys-050511-102240. [DOI] [PubMed] [Google Scholar]
- 33.Hornung G, Barkai N. Noise propagation and signaling sensitivity in biological networks: A role for positive feedback. PLOS Comput Biol. 2008;4:e8. doi: 10.1371/journal.pcbi.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Alba E. Structure and interdomain dynamics of apoptosis-associated speck-like protein containing a CARD (ASC) J Biol Chem. 2009;284:32932–32941. doi: 10.1074/jbc.M109.024273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lei Y, et al. MAVS-mediated apoptosis and its inhibition by viral proteins. PLoS One. 2009;4:e5466. doi: 10.1371/journal.pone.0005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duewell P, et al. RIG-I-like helicases induce immunogenic cell death of pancreatic cancer cells and sensitize tumors toward killing by CD8(+) T cells. Cell Death Differ. 2014;21:1825–1837. doi: 10.1038/cdd.2014.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matusiak M, Van Opdenbosch N, Lamkanfi M. CARD- and pyrin-only proteins regulating inflammasome activation and immunity. Immunol Rev. 2015;265:217–230. doi: 10.1111/imr.12282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.