Significance
Our analysis shows that sex can be associated with the degree to which HLA molecules propagate selection and expansion of T cells as characterized by their T cell receptor variable beta chain (TCRBV). Furthermore, CD8 T cells, especially in men with autoimmune diseases such as multiple sclerosis or rheumatoid arthritis, are capable of expanding in unison with other CD8 T cells, even without expressing TCRBVs with biochemical similarity in pivotal HLA-binding regions. Our findings add to the understanding of sex bias in diseases with immune system involvement: autoimmunity, infection, and cancer. These results also reveal pathology-associated TCRBVs of interest for future studies and support the argument for sex-separated analysis of HLA disease associations in general.
Keywords: T cell receptor repertoire, immunological sex bias, autoimmunity, multiple sclerosis, rheumatoid arthritis
Abstract
HLA associations, T cell receptor (TCR) repertoire bias, and sex bias have independently been shown for many diseases. While some immunological differences between the sexes have been described, they do not fully explain bias in men toward many infections/cancers, and toward women in autoimmunity. Next-generation TCR variable beta chain (TCRBV) immunosequencing of 824 individuals was evaluated in a multiparametric analysis including HLA-A -B/MHC class I background, TCRBV usage, sex, age, ethnicity, and TCRBV selection/expansion dynamics. We found that HLA-associated shaping of TCRBV usage differed between the sexes. Furthermore, certain TCRBVs were selected and expanded in unison. Correlations between these TCRBV relationships and biochemical similarities in HLA-binding positions were different in CD8 T cells of patients with autoimmune diseases (multiple sclerosis and rheumatoid arthritis) compared with healthy controls. Within patients, men showed higher TCRBV relationship Spearman’s rhos in relation to HLA-binding position similarities compared with women. In line with this, CD8 T cells of men with autoimmune diseases also showed higher degrees of TCRBV perturbation compared with women. Concerted selection and expansion of CD8 T cells in patients with autoimmune diseases, but especially in men, appears to be less dependent on high HLA-binding similarity than in CD4 T cells. These findings are consistent with studies attributing autoimmunity to processes of epitope spreading and expansion of low-avidity T cell clones and may have further implications for the interpretation of pathogenic mechanisms of infectious and autoimmune diseases with known HLA associations. Reanalysis of some HLA association studies, separating the data by sex, could be informative.
The T cell receptor (TCR) repertoire is highly diverse (1) and can generate up to 1015 theoretical TCR sequences (2, 3) with the effective size of the CD8 TCR repertoire in vivo being made up of 105 to 108 sequences (2, 4–6). The repertoire starts developing prenatally (7) and is subject to continuous adaptation during the course of a lifetime due to antigen exposure and other environmental influences (8, 9). While its complementary determining region 3 (CDR3) is hypervariable and binds to the MHC-presented antigen (10), the binding sites of the TCR to the MHC are germline encoded within the CDR1 and CDR2 of the TCR variable chains (11, 12). Many studies over the last decades have shown that evolutionarily conserved anchor amino acids in these CDRs determine how single TCR constructs bind to their respective MHC (13). Concerning the TCR variable beta chains (TCRBVs), 31 individual genes are known to encode a functional, nonpseudogenic, nonsegregated TCR, each with an individual CDR1 and CDR2 (14, 15). The HLA background is genetically predetermined and invariable (16), leading to the accepted notion that the HLA shapes the TCR repertoire during their continuous interaction (17–20). Very recently, it was shown that mutations in TCR binding amino acids of MHC molecules specifically influence the binding to and usage of TCR variable alpha (TCRAV) and TCRBV chains (19), and that the presence of specific TCR rearrangements is an indication of the HLA background of the host and also of previous exposure to pathogens (20). Dysregulation in the TCR–antigen–MHC complex can lead to autoimmunity (21) in the case of uncontrolled immune cell expansion or cross-reactivity between foreign and autoantigens. Hypoexpansions or low TCR avidity might lead to susceptibility to infections. Additionally, to combat tumor-associated immune system suppression, cancer patients can now be treated with T cell-relevant immune checkpoint inhibitors such as blockade of CTLA-4 (22) or PD-1 (23).
Most immune system dysfunctions show strong sex bias: men are more susceptible to many infectious diseases and cancers of nonreproductive organs, whereas autoimmune diseases are much more common in women (24, 25). Even in psychological disorders such as major depression (26) and in neurodegenerative diseases such as Alzheimer’s, recent studies have shown immune system involvement (27) and sex bias (28). Many studies have been conducted concerning either sex (25), HLA (29, 30), or TCR (31) associations with diseases. However, even though some sex differences in immune regulation have been reported (25, 32), it is still not completely clear why this bias exists. Multiparametric analysis of the influence of sex on HLA-mediated shaping of the TCR repertoire has been hindered by low subject numbers in most studies. The advent of next-generation and high-throughput TCR immunosequencing in large cohorts of HLA-typed subjects now allows for a combined analysis of the TCR repertoire dependence on sex and HLA alleles (20).
Results
Influence of HLA-A and -B on TCRBV Usage Depends on the Biological Sex.
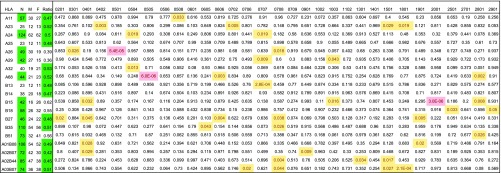
To assess whether the HLA background influences the frequency of T cells expressing certain TCRBVs in a sex-associated manner, TCRBV immunosequencing data from 673 individuals were analyzed (for details see Datasets S1 and S2). The resulting dataset, therefore, contained the following parameters: sex (2; male, female), HLA-A and HLA-B genes (19; 8 HLA-A, 7 HLA-B, 4 most common haplotypes in the Caucasian population), and 31 TCRBV genes represented as percentage of the productive TCR repertoire in a sample. Only HLA with at least 40 individuals in the dataset or a balanced female-to-male ratio (percentage of female donors between 45% and 55%) and only functional TCRBV genes according to the International ImMunoGeneTics (IMGT) nomenclature (14, 15) were included. Nonfunctional TCRBV genes, pseudogenes, segregating, or open-reading frame genes (with potential mutations in the leader sequence) were not analyzed. A multiparametric analysis using multivariate linear models assessing the interaction of HLA and sex showed that the manner in which HLA genes influence TCRBV usage can differ between men and women (Fig. 1). Controlled for the known immunosequencing covariates, we found sex-associated differences in the propagation of TCRBV selection and expansion by the HLA [raw data in Dataset S2, P values in Dataset S3, and effect sizes (partial eta squared) in Dataset S4]. With the number of individuals in our study, 42 HLA/TCRBV combinations (P < 0.05) were found in this dataset, of which three remained significant after correcting for multiple comparisons among those 589 assessed combinations, where TCRBV usage by a specific HLA in one sex was elevated compared with the other sex, independent of a potential influence of HLA or sex alone (Fig. S1 A–C). The Bonferroni-corrected sex-specific TCRBV expressions were challenged in a sensitivity analysis, where only healthy donors or only donors with determined (not inferred) HLA were assessed. The sex-specific differences could also be observed, when age or ethnicity (which was not available for all donors) or other relevant hidden covariates [as determined by probabilistic estimation of expression residuals (PEER) analysis] (33, 34) were included in the model (Fig. S1 D and E).
Fig. 1.
Sex bias in HLA-associated shaping of the TCRBV usage. Shown are uncorrected P values from a multivariate linear regression model (covariates in Dataset S2), assessing the interaction of HLA and sex on TCRBV usage. HLA/TCRBV combinations highlighted in red indicate P < 0.05 after Bonferroni correction for multiple comparisons; yellow color suggests additional, potentially sex-specific HLA/TCRBV combinations, which did not pass Bonferroni correction. Left columns indicate the total number (N) as well as the male (M) and female (F) subject numbers for each HLA (raw data in Dataset S3), and green color indicates that this inclusion criterion was passed (one criterion was enough for inclusion).
CD8 T Cells in Men with Autoimmune Diseases Are Less Dependent on Biochemical CDR1/2 Similarity for Concerted Selection/Expansion.
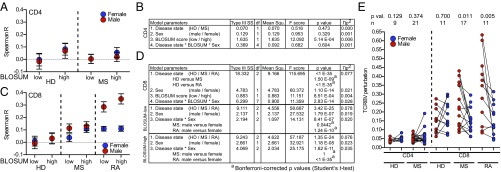
Having shown that the HLA background can alter TCRBV usage in a sex-dependent manner and given that MHC molecules interact with the TCR mainly via the CDR1 and -2, we hypothesized that there could be a functional relationship between how similar TCRBV chains are to one another (in these regions) and how they are selected in the thymus or expanded in the periphery via their HLA interaction. To assess this, the germline-encoded CDR1 and -2 sequences of the 31 functional TCRBV transcripts were compared via a Blocks Substitution Matrix (BLOSUM62) to estimate biochemical amino acid similarities (35). Resulting alignment matrices are used to score single amino acids at a specific position in two different sequences with respect to their biophysical and biochemical similarities. The scores of each pairing are then computed and indicate how similar two sequences are to each other. The current nomenclature groups TCRBV chains according to the nucleotide similarity of their complete DNA sequence. As our question did not relate to evolutionary homology per se, but rather to biological function with regard to TCR–MHC binding, we only assessed the known binding amino acids (position 28, and 29 in CDR1 as well as 46, 48, and 54 in CDR2) with the BLOSUM62 matrix (13) (Fig. S2A). The resulting 465 individual similarity scores between −9 and 29 were color coded (values listed in Dataset S5) and clustered using the Ward (36) method. This resulted in four optimal clusters [calculated via gap statistic (37)] (Fig. S2B), grouping TCRBV gene segments into the known homology-based subgroups or “families” (14) (i.e., subgroups 4, 5, 7, and 10, but not TCRBV06). However, the analysis also suggested relationships, which were not expected from the nomenclature (i.e., TCRBV05–08 and the TCRBV07 subgroup). To evaluate the in vivo relevance of these relationships in health and disease, the TCRBV usage by CD4 and CD8 T cells from age-matched male and female healthy donors (HD) or patients suffering from autoimmune diseases [multiple sclerosis (MS) and rheumatoid arthritis (RA)] was analyzed for potential correlations in all possible TCRBV chain combinations (raw data in Dataset S6) resulting in 465 TCRBV relationship Spearman’s rank correlation coefficients (TCRBV rhos, Dataset S5). To allow for a categorical assessment, BLOSUM values were divided at the median into 232 low- and 233 high-similarity scores. Subsequently the TCRBV rhos were used as the dependent variable in linear models for CD4 (Fig. 2 A and B) and CD8 T cells (Fig. 2 C and D) with the main effects “disease state (HD/MS/RA),” “sex (male/female),” and “BLOSUM score (high/low).” As hypothesized, TCRBV rhos of both T cell subsets were influenced by BLOSUM scores. However, only CD8 T cells were additionally influenced by disease state, as well as sex: (i) CD8 T cells from patients with both autoimmune diseases (MS or RA) differed from CD8 T cells from healthy donors. (ii) With regard to sex, men with autoimmune diseases showed significantly higher rhos in TCRBV combinations with low BLOSUM scores than women, which in the case of RA could also be observed in TCRBV combinations with high BLOSUM scores. To further explore the hypothesis that an MHC–TCR stimulus of the same affinity could lead to higher CD8 expansions of similar TCRBV chains in men with autoimmune diseases than in women, TCRBV chain perturbation was assessed (4, 38). In an age-matched, paired analysis, male subjects with autoimmune diseases (MS or RA) presented with higher “productive clonality” (a measurement of TCRBV perturbation), than female subjects in the CD8 T cell compartment (Fig. 2E).
Fig. 2.
CD8 T cells in men with autoimmune diseases are less dependent on biochemical CDR1/2 similarity for concerted selection/expansion. (A) TCRBV relationships Spearman’s correlation coefficients (TCRBV rhos) of CD4 T cells of healthy donors (HD) and MS patients grouped according to BLOSUM-low and BLOSUM-high scores and divided by sex [male (red) versus female (blue)]. Shown are mean and 95% confidence interval. (B) Parameters of the linear model for CD4 T cells. Shown are the type III sum of squares (type III SS), degrees of freedom (df), mean square (mean squ.), F score, significance (P value), and partial eta squared (ɳp2) as effect size. (C) TCRBV rhos of CD8 T cells of healthy donors (HD), and MS and RA patients grouped according to A. (D) Parameters of the linear model for CD8 T cells according to B. Two nested models are shown (BLOSUM-low and BLOSUM-high) within CD8 T cells. Pairwise comparison (superscript a) of the mean is shown for groups of interest (Bonferroni-corrected Student’s t test). (E) TCRBV perturbation, measured as productive clonality. Shown are paired comparisons (Wilcoxon matched pairs signed rank test) between age-matched male and female individuals (CD4 and CD8 T cells; HD, MS, or RA).
In Vivo Relationships Between TCRBV Transcripts Are Cause and Not Result of TCRBV Perturbations.
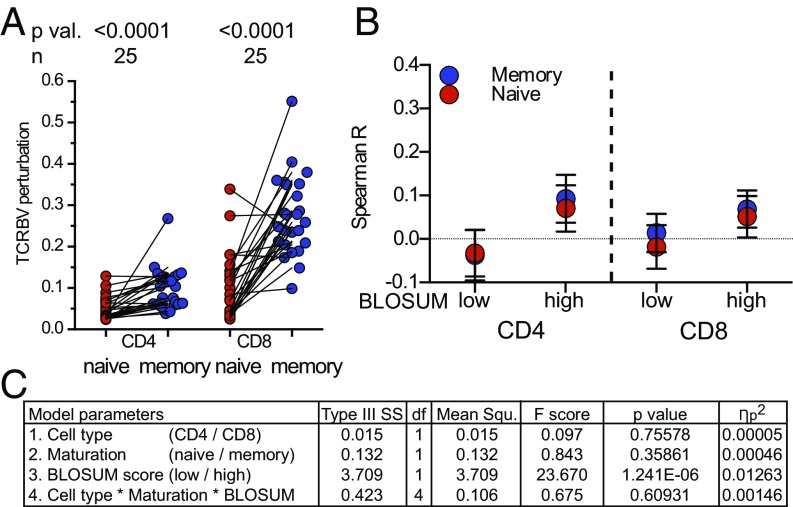
Twenty-five pairs of naïve and memory CD4 and CD8 T cells from healthy donors (100 samples in total, raw data in Dataset S7) were analyzed to address the following questions: (i) Is the higher TCRBV perturbation in CD8 T cells in men cause or effect of the relationships seen in Fig. 2? (ii) Can a high degree of TCRBV perturbation, which would be expected in memory T cells, influence the relationships between TCRBV gene transcripts? As expected, memory T cells presented with higher TCRBV perturbation compared with naïve T cells (Fig. 3A). However, this neither influenced the relationship between BLOSUM scores and TCRBV rhos in CD4 T cells nor CD8 T cells (Fig. 3 B and C). Additionally, the relationships between TCRBV transcripts found in naïve cells could also be found in memory cells (Fig. S3 A and B, TCRBV rhos listed in Dataset S5).
Fig. 3.
In vivo relationships between TCRBV chains are similar in naïve and memory T cell subsets. (A) TCRBV perturbation quantified by the sequencing parameter productive clonality of naïve and memory CD4 and CD8 T cell subsets from 25 healthy subjects. (B) TCRBV rhos of naïve (red) and memory (blue) CD4/CD8 T cells grouped according to low and high BLOSUM scores. Shown is mean and 95% confidence interval (TCRBV rhos in Dataset S5 and raw data in Dataset S7). (C) Parameters of the linear model with TCRBV rhos as dependent and cell type, maturation, BLOSUM scores as main effects with the added interaction of those three main effects.
Discussion
The most surprising finding of our study was that the biological sex of an individual can influence the HLA association with T cell selection and expansion. Most autoimmune diseases more frequently affect women (25), whereas men are more susceptible to many infectious diseases, e.g., leishmaniasis or hepatitis A (39). HLA associations with cancer and the fact that men develop cancer in nonreproductive organs much more frequently than women has also been known for a long time (24). As cancer can also be viewed as a failure of the immune system to control neoplastic tissue, low expression in certain TCRBV families could be an indication of this defect as well. Apart from susceptibilities and disease incidences, general sex differences in parameters of the adaptive immune system have been found before. Women have higher amounts of circulating IgMs (40) and more CD4 T cells (41). These differences are thought to be mainly mediated by steroid hormones, with estrogens often promoting and androgens suppressing immune responses during infections, after vaccination, or in case of autoimmunity (reviewed in refs. 42–44). Recent studies also suggest an influence of glucocorticoid receptors in the suppression of autoimmunity in pregnant MS patients (32). Therefore, the observed sex differences in HLA-mediated shaping of the TCR repertoire could in part be caused or facilitated by the steroid hormone milieu, influencing lymphocyte activation during immune responses (32, 45), but also their maturation during thymic selection (46). Our data support these studies by providing evidence that the observed relationships between TCRBV transcripts play a similar role in thymic selection (reflected in naïve cells), as well as during expansion in the periphery due to antigenic stimuli (reflected in memory cells).
It has previously been shown that the HLA background shapes the TCR repertoire by favoring or neglecting certain T cell subpopulations represented by TCRBV usage (19). Certain HLAs are better at presenting auto- or foreign antigens to TCRBV-specific subgroups of T cells. Over time this can lead to an individually diverse “immunological fingerprint” shaped by genetics and exposure to antigens. This better or worse HLA–TCR interaction will have consequences for infectious diseases (susceptibility, progression, adverse effects), and also for antitumor responses, autoimmunity, or adverse medical reactions. Unexpectedly, we found differences between men and women with respect to the influence of HLA genes on TCRBV transcripts. In light of these findings, it makes sense that the TCR repertoire at birth is similar between cohorts with vastly different HLA backgrounds (47), but is then shaped in the course of life due to exposure to and HLA presentation of different antigens as shown in twin studies (17, 48). Consistent with this idea, it has recently been shown that T cell expansions show high degrees of homology on an amino acid level, but not on a DNA sequence level in individuals with similar HLA backgrounds (49). One possible explanation might have been a different expression level of the HLA molecules on the cells of men and women, but a recently published large study showed that at least the soluble form of the β-2 microglobulin in serum of men and women is comparable (50), which would fit the above-mentioned hypothesis that the observed differences could be a hormone-mediated downstream effect taking place after the MHC–TCR binding.
The second finding that CD8 T cell subpopulations with low biochemical similarities in their TCRBV chains correlate more strongly in patients with autoimmune diseases (MS and RA) in general, but particularly in men, is consonant with the overall higher CD8 T cell TCRBV perturbation in men compared with women. We hypothesize that a defined antigen stimulus would thereby lead to more clonal expansions in CD8 T cells in men, as the presentation by a given HLA molecule could also trigger T cell expansions with low binding affinity between TCR and MHC. Additionally, it is known that CD4 and CD8 in their function as coreceptors bind to the MHC molecules with different binding affinities (reviewed in ref. 51): CD4 has a weak binding affinity to MHC class II (52), whereas CD8 has a high binding affinity to MHC class I (53). These studies have also suggested that MHC class II ligands modulate TCR signaling directly via the TCR, whereas MHC class I ligands do not influence TCR signaling via the TCR, but via CD8 itself (54), and CD8 T cells are less dependent on continuous antigen presentation (55). Therefore, our data showing that low CDR similarity can still lead to parallel expansions in unrelated TCRBVs fit the hypothesis that CD8 T cells are less dependent on a strong MHC–TCR binding. Furthermore, antigen-triggered expansions could also occur in TCRs despite suboptimal binding to MHC as long as the CD8 binding affinity is strong enough (56). Many hypotheses concerning autoimmunity involve an immune reaction against self-antigens via antigen spreading, molecular mimicry, or because low-avidity clones become activated, even though the threshold for activation should not have been reached, especially in cases of dual TCR expression by CD8 T cells (ref. 57 and reviewed in ref. 58). Our finding that CD8 T cells of MS patients in general, but particularly in men with MS expand in concert in regions of low CDR1/2 similarity is consistent with the finding that men present with higher disease burden, if they suffer from MS (59). We found similar correlations in CD8 T cells of patients with RA, but in contrast to MS, the clinical picture in RA is not as clear concerning sex differences and disease severity, potentially due to the fact that the measurement of disease activity in RA involves secondary outcome parameters with inherent sex bias (60). Our study provides evidence that biological sex modifies associations between HLA molecules and T cell TCRBV subpopulations and might, therefore, have implications regarding prior studies of HLA associations. These studies could possibly be revisited and male and female participants analyzed separately to ensure that some significant results are not lost due to statistical adjustments.
Materials and Methods
Study Cohorts.
This study included samples from five cohorts: (cohort 1) healthy subjects (20); (cohort 2) subjects suffering from epilepsy, multiple sclerosis, or a common cold, as well as corresponding healthy subjects (ref. 61 and raw data in Datasets S2 and S6); (cohort 3) subjects suffering from rheumatoid arthritis and corresponding healthy subjects (62); (cohort 4) healthy subjects (63); and (cohort 5) healthy subjects (64). Details of these cohorts and their contribution to the analyses are listed in Dataset S1.
Study Approval.
Studies on human samples were approved by the local ethics committee (University of Muenster: Ethik-Kommission der Ärztekammer Westfalen-Lippe und der Medizinischen Fakultät der Westfälischen Wilhelms-Universität, registration number: 2010–245-f-S). Written informed consent was given by all participants in the study and all experiments were performed according to the Declaration of Helsinki.
Sequencing.
TCR immunosequencing was done according to the immunoSEQ assay (Adaptive Biotechnologies) as previously described (38, 61, 65); for details please see SI Materials and Methods.
Statistics.
In Fig. 1, the interaction of sex and HLA on TCRBV usage was analyzed with multivariate linear regression models with adjustment for covariates influencing TCRBV usage. Eight covariates (see below and Dataset S2) were used for all comparisons. The linear regression model included 31 TCRBV transcripts as dependents, the interaction of sex and one HLA as fixed factors, together with covariates (TCRBV = HLA + sex + HLA × sex + covariates; one model per HLA, resulting in 31 models with 589 HLA × sex interactions). Significance was corrected for multiple comparisons according to the Bonferroni method (66) using R (function p.adjust). Significances not passing Bonferroni correction, but exhibiting an uncorrected P value ≤0.05, are considered exploratory. Covariates are as follows: fraction productive, the fraction of in-frame templates; productive rearrangements, count of unique and functional rearrangements; productive clonality, normalized TCRBV repertoire diversity measure using log (base 2) of the total number of unique productive rearrangements to normalize the entropy and subtracting the result from the value 1; max productive frequency, the highest frequency of a rearrangement; productive entropy, diversity measure for the TCRBV repertoire according to Shannon’s entropy (67), calculated over all productive rearrangements; cell type, isolated cell subset (PBMC, CD4, or CD8 T cells); sequencing source material (cgDNA), genomic DNA (gDNA), or cDNA (cDNA); and batch, sequencing batch as assessed by time of sequencing (24 batches). Covariates “age” (of donor at time point of sampling) and “ethnicity” (self-reported ethnicity of a donor, Caucasian, or non-Caucasian) were included in a sensitivity analysis (Fig. S1E), as these variables were not available for all donors. The sensitivity assessment also included one analysis performed only with “healthy” individuals and one only with individuals, where the HLA background was determined, not “inferred.” HLA-A/B background was fully available for cohorts 1 and 3 and was inferred for part of samples of cohort 2, based on HLA-A/B-specific CDR3 sequences according to Emerson (20). To determine possible hidden factors in addition to known covariates, we used the PEER package (33, 34). From 25 hidden factors provided by the algorithm, 9 were included in the sensitivity analysis, as these were in the αC range of the known covariates (αC < 100) (Fig. S1D). The SPSS syntax used for Fig. 1 is given in Dataset S2.
In Fig. 2, all combinations of 31 TCRBVs resulted in 465 scores ranging from −9 to 26, each of them the sum of the CDR1 and -2 binding sites’ BLOSUM62 scores (Fig. S2). BLOSUM62 scores were calculated using the function “pairwiseAlignment” from the Biostrings package within Bioconductor (68, 69) and are presented as a heatmap clustered according to Ward (36). Optimal number of clusters was determined with gap statistic (37) by using the functions “clusGap” and “fviz_gap_stat” from the factoextra package. The 465 scores were divided at the median value of 3 into two groups of low (BLOSUM-low, score −9–2, n = 232) and high (BLOSUM-high, score 3–26, n = 233) biochemical similarity. Multivariate linear models were applied to CD4 and CD8 datasets with TCRBV rhos as dependent and the main effects “sex,” “disease state,” and “BLOSUM score,” as well as their interaction. Subgroup analyses (nested models) were applied to BLOSUM-low and BLOSUM-high with the main effects of sex and disease state, as well as their interaction.
In Fig. 3, a multivariate model was applied to CD4 and CD8 naïve and memory T cell datasets with the main effects “cell type,” “maturation,” and BLOSUM score as well as their interaction.
The following statistical software was used: IBM SPSS Statistics V24, GraphPad Prism V6 and R V3.4.1 using the packages PEER, Bioconductor, Biostrings, made4, dendextend, factoextra, cluster, pheatmap, corrplot, stats, and their respective dependencies.
Supplementary Material
Acknowledgments
This study was funded by DFG Grant SFB/CRC128 Project B1 (to N.S.), A5 (to K.D.), and Z2 (to H.W.); single Grant GR3946_3/1 (to C.C.G.); the Kompetenznetz Multiple Sklerose (Competence Network for Multiple Sclerosis) funded by the Federal Ministry of Education and Research (Bundesministerium für Bildung und Forschung) Grants FKZ 01GI1308B 01GI0907 and GER-TYS-12-10-401 (Immuntherapieregister zur Verbesserung der Arzneimittelsicherheit in der Multiple Sklerose Therapie Innerhalb des Krankheitsbezogenen Kompetenznetzes MS) (to H.W.); the Interdisziplinäre Zentrum für Klinische Forschung Münster (Wie3/009/16) (to N.S. and H.W.); the European Research Council (Novel etiology of autoimmune disorders: The role of acquired somatic mutations in lymphoid cells, M-IMM project); Academy of Finland; Finnish special governmental subsidy for health sciences, research and training; the Sigrid Juselius Foundation; and the Finnish Cancer Institute (S.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716146115/-/DCSupplemental.
References
- 1.Kappler J, et al. The major histocompatibility complex-restricted antigen receptor on T cells in mouse and man: Identification of constant and variable peptides. Cell. 1983;35:295–302. doi: 10.1016/0092-8674(83)90232-5. [DOI] [PubMed] [Google Scholar]
- 2.Arstila TP, et al. A direct estimate of the human alphabeta T cell receptor diversity. Science. 1999;286:958–961. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- 3.Lythe G, Callard RE, Hoare RL, Molina-París C. How many TCR clonotypes does a body maintain? J Theor Biol. 2016;389:214–224. doi: 10.1016/j.jtbi.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robins HS, et al. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci Transl Med. 2010;2:47ra64. doi: 10.1126/scitranslmed.3001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qi Q, et al. Diversity and clonal selection in the human T-cell repertoire. Proc Natl Acad Sci USA. 2014;111:13139–13144. doi: 10.1073/pnas.1409155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li HM, et al. TCRβ repertoire of CD4+ and CD8+ T cells is distinct in richness, distribution, and CDR3 amino acid composition. J Leukoc Biol. 2016;99:505–513. doi: 10.1189/jlb.6A0215-071RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haynes BF, Heinly CS. Early human T cell development: Analysis of the human thymus at the time of initial entry of hematopoietic stem cells into the fetal thymic microenvironment. J Exp Med. 1995;181:1445–1458. doi: 10.1084/jem.181.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Batliwalla F, Monteiro J, Serrano D, Gregersen PK. Oligoclonality of CD8+ T cells in health and disease: Aging, infection, or immune regulation? Hum Immunol. 1996;48:68–76. doi: 10.1016/0198-8859(96)00077-8. [DOI] [PubMed] [Google Scholar]
- 9.Britanova OV, et al. Age-related decrease in TCR repertoire diversity measured with deep and normalized sequence profiling. J Immunol. 2014;192:2689–2698. doi: 10.4049/jimmunol.1302064. [DOI] [PubMed] [Google Scholar]
- 10.Dai S, et al. Crossreactive T cells spotlight the germline rules for alphabeta T cell-receptor interactions with MHC molecules. Immunity. 2008;28:324–334. doi: 10.1016/j.immuni.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moss PAH, Bell JI. Sequence analysis of the human alpha beta T-cell receptor CDR3 region. Immunogenetics. 1995;42:10–18. doi: 10.1007/BF00164982. [DOI] [PubMed] [Google Scholar]
- 12.Yin L, et al. A single T cell receptor bound to major histocompatibility complex class I and class II glycoproteins reveals switchable TCR conformers. Immunity. 2011;35:23–33. doi: 10.1016/j.immuni.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu Rev Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lefranc MP, et al. IMGT, the International ImMunoGeneTics database. Nucleic Acids Res. 1998;26:297–303. doi: 10.1093/nar/26.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dean J, et al. Annotation of pseudogenic gene segments by massively parallel sequencing of rearranged lymphocyte receptor loci. Genome Med. 2015;7:123. doi: 10.1186/s13073-015-0238-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Leeuwen A, Termijtelen A, Shaw S, van Rood JJ. Recognition of a polymorphic monocyte antigen in HLA. Nature. 1982;298:565–567. doi: 10.1038/298565a0. [DOI] [PubMed] [Google Scholar]
- 17.Hoover ML, et al. Insulin-dependent diabetes mellitus is associated with polymorphic forms of the T-cell receptor beta chain gene. Hum Immunol. 1986;17:176 (abstr). [Google Scholar]
- 18.Akolkar PN, Gulwani-Akolkar B, McKinley M, Fisher SE, Silver J. Comparisons of T cell receptor (TCR) V β repertoires of lamina propria and peripheral blood lymphocytes with respect to frequency and oligoclonality. Clin Immunol Immunopathol. 1995;76:155–163. doi: 10.1006/clin.1995.1110. [DOI] [PubMed] [Google Scholar]
- 19.Sharon E, et al. Genetic variation in MHC proteins is associated with T cell receptor expression biases. Nat Genet. 2016;48:995–1002. doi: 10.1038/ng.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emerson RO, et al. Immunosequencing identifies signatures of cytomegalovirus exposure history and HLA-mediated effects on the T cell repertoire. Nat Genet. 2017;49:659–665. doi: 10.1038/ng.3822. [DOI] [PubMed] [Google Scholar]
- 21.Garcia KC, et al. Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen. Science. 1998;279:1166–1172. doi: 10.1126/science.279.5354.1166. [DOI] [PubMed] [Google Scholar]
- 22.Ramagopal UA, et al. Structural basis for cancer immunotherapy by the first-in-class checkpoint inhibitor ipilimumab. Proc Natl Acad Sci USA. 2017;114:E4223–E4232. doi: 10.1073/pnas.1617941114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le DT, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clocchiatti A, Cora E, Zhang Y, Dotto GP. Sexual dimorphism in cancer. Nat Rev Cancer. 2016;16:330–339. doi: 10.1038/nrc.2016.30. [DOI] [PubMed] [Google Scholar]
- 25.Whitacre CC, Reingold SC, O’Looney PA. A gender gap in autoimmunity. Science. 1999;283:1277–1278. doi: 10.1126/science.283.5406.1277. [DOI] [PubMed] [Google Scholar]
- 26.Labonté B, et al. Sex-specific transcriptional signatures in human depression. Nat Med. 2017;23:1102–1111. doi: 10.1038/nm.4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong S, et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 2016;352:712–716. doi: 10.1126/science.aad8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mosconi L, et al. Sex differences in Alzheimer risk: Brain imaging of endocrine vs chronologic aging. Neurology. 2017;89:1382–1390. doi: 10.1212/WNL.0000000000004425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gough SC, Simmonds MJ. The HLA region and autoimmune disease: Associations and mechanisms of action. Curr Genomics. 2007;8:453–465. doi: 10.2174/138920207783591690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blackwell JM, Jamieson SE, Burgner D. HLA and infectious diseases. Clin Microbiol Rev. 2009;22:370–385. doi: 10.1128/CMR.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirsch I, Vignali M, Robins H. T-cell receptor profiling in cancer. Mol Oncol. 2015;9:2063–2070. doi: 10.1016/j.molonc.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engler JB, et al. Glucocorticoid receptor in T cells mediates protection from autoimmunity in pregnancy. Proc Natl Acad Sci USA. 2017;114:E181–E190. doi: 10.1073/pnas.1617115114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stegle O, Parts L, Piipari M, Winn J, Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc. 2012;7:500–507. doi: 10.1038/nprot.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stegle O, Parts L, Durbin R, Winn J. A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput Biol. 2010;6:e1000770. doi: 10.1371/journal.pcbi.1000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward JH. Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 1963;58:236–244. [Google Scholar]
- 37.Tibshirani R, Walther G, Hastie T. Estimating the number of clusters in a data set via the gap statistic. J R Stat Soc Series B Stat Methodol. 2001;63:411–423. [Google Scholar]
- 38.Robins HS, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guerra-Silveira F, Abad-Franch F. Sex bias in infectious disease epidemiology: Patterns and processes. PLoS One. 2013;8:e62390. doi: 10.1371/journal.pone.0062390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grundbacher FJ. Human X chromosome carries quantitative genes for immunoglobulin M. Science. 1972;176:311–312. doi: 10.1126/science.176.4032.311. [DOI] [PubMed] [Google Scholar]
- 41.Amadori A, et al. Genetic control of the CD4/CD8 T-cell ratio in humans. Nat Med. 1995;1:1279–1283. doi: 10.1038/nm1295-1279. [DOI] [PubMed] [Google Scholar]
- 42.Giefing-Kröll C, Berger P, Lepperdinger G, Grubeck-Loebenstein B. How sex and age affect immune responses, susceptibility to infections, and response to vaccination. Aging Cell. 2015;14:309–321. doi: 10.1111/acel.12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fish EN. The X-files in immunity: Sex-based differences predispose immune responses. Nat Rev Immunol. 2008;8:737–744. doi: 10.1038/nri2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klein SL. Sex influences immune responses to viruses, and efficacy of prophylaxis and treatments for viral diseases. BioEssays. 2012;34:1050–1059. doi: 10.1002/bies.201200099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grimaldi CM, Cleary J, Dagtas AS, Moussai D, Diamond B. Estrogen alters thresholds for B cell apoptosis and activation. J Clin Invest. 2002;109:1625–1633. doi: 10.1172/JCI14873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu M-L, et al. Sex bias in CNS autoimmune disease mediated by androgen control of autoimmune regulator. Nat Commun. 2016;7:11350. doi: 10.1038/ncomms11350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramakrishnan NS, Grunewald J, Janson CH, Wigzell H. Nearly identical T-cell receptor V-gene usage at birth in two cohorts of distinctly different ethnic origin: Influence of environment in the final maturation in the adult. Scand J Immunol. 1992;36:71–78. doi: 10.1111/j.1365-3083.1992.tb02942.x. [DOI] [PubMed] [Google Scholar]
- 48.Zvyagin IV, et al. Distinctive properties of identical twins’ TCR repertoires revealed by high-throughput sequencing. Proc Natl Acad Sci USA. 2014;111:5980–5985. doi: 10.1073/pnas.1319389111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruggiero E, et al. High-resolution analysis of the human T-cell receptor repertoire. Nat Commun. 2015;6:8081. doi: 10.1038/ncomms9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Juraschek SP, et al. Comparison of serum concentrations of β-trace protein, β2-microglobulin, cystatin C, and creatinine in the US population. Clin J Am Soc Nephrol. 2013;8:584–592. doi: 10.2215/CJN.08700812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stone JD, Chervin AS, Kranz DM. T-cell receptor binding affinities and kinetics: Impact on T-cell activity and specificity. Immunology. 2009;126:165–176. doi: 10.1111/j.1365-2567.2008.03015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krummel MF, Sjaastad MD, Wülfing C, Davis MM. Differential clustering of CD4 and CD3zeta during T cell recognition. Science. 2000;289:1349–1352. doi: 10.1126/science.289.5483.1349. [DOI] [PubMed] [Google Scholar]
- 53.Garcia KC, et al. CD8 enhances formation of stable T-cell receptor/MHC class I molecule complexes. Nature. 1996;384:577–581. doi: 10.1038/384577a0. [DOI] [PubMed] [Google Scholar]
- 54.Yachi PP, Ampudia J, Gascoigne NRJ, Zal T. Nonstimulatory peptides contribute to antigen-induced CD8-T cell receptor interaction at the immunological synapse. Nat Immunol. 2005;6:785–792. doi: 10.1038/ni1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rabenstein H, et al. Differential kinetics of antigen dependency of CD4+ and CD8+ T cells. J Immunol. 2014;192:3507–3517. doi: 10.4049/jimmunol.1302725. [DOI] [PubMed] [Google Scholar]
- 56.Tscharke DC, Croft NP, Doherty PC, La Gruta NL. Sizing up the key determinants of the CD8(+) T cell response. Nat Rev Immunol. 2015;15:705–716. doi: 10.1038/nri3905. [DOI] [PubMed] [Google Scholar]
- 57.Ji Q, Perchellet A, Goverman JM. Viral infection triggers central nervous system autoimmunity via activation of CD8+ T cells expressing dual TCRs. Nat Immunol. 2010;11:628–634. doi: 10.1038/ni.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cusick MF, Libbey JE, Fujinami RS. Molecular mimicry as a mechanism of autoimmune disease. Clin Rev Allergy Immunol. 2012;42:102–111. doi: 10.1007/s12016-011-8294-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leray E, et al. Evidence for a two-stage disability progression in multiple sclerosis. Brain. 2010;133:1900–1913. doi: 10.1093/brain/awq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sokka T, et al. QUEST-RA Group Women, men, and rheumatoid arthritis: Analyses of disease activity, disease characteristics, and treatments in the QUEST-RA study. Arthritis Res Ther. 2009;11:R7. doi: 10.1186/ar2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schneider-Hohendorf T, et al. CD8(+) T-cell pathogenicity in Rasmussen encephalitis elucidated by large-scale T-cell receptor sequencing. Nat Commun. 2016;7:11153. doi: 10.1038/ncomms11153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Savola P, et al. Somatic mutations in clonally expanded cytotoxic T lymphocytes in patients with newly diagnosed rheumatoid arthritis. Nat Commun. 2017;8:15869. doi: 10.1038/ncomms15869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emerson R, et al. Estimating the ratio of CD4+ to CD8+ T cells using high-throughput sequence data. J Immunol Methods. 2013;391:14–21. doi: 10.1016/j.jim.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 64.Kanakry CG, et al. Origin and evolution of the T cell repertoire after posttransplantation cyclophosphamide. JCI Insight. 2016;1:e86252. doi: 10.1172/jci.insight.86252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dandekar S, et al. Shared HLA class I and II alleles and clonally restricted public and private brain-infiltrating αβ T cells in a cohort of Rasmussen encephalitis surgery patients. Front Immunol. 2016;7:608. doi: 10.3389/fimmu.2016.00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Salkind N. Teoria Statistica Delle Classi e Calcolo Delle Probabilità. SAGE Publications; Thousand Oaks, CA: 2010. [Google Scholar]
- 67.Shannon CE. A mathematical theory of communication. Bell Syst Tech J. 2013;27:379–423. [Google Scholar]
- 68.Gentleman RC, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huber W, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.