Significance
Embryonic stem cells (ESCs) are derived from the inner cell mass of a preimplantation blastocyst, as has been well established in rodents and primates. To date, however, the derivation and stable propagation of pluripotent ESCs from domestic ungulates remain unsuccessful. The production of ESCs from large livestock species is important for genomic testing and selection, genome engineering, and studying human diseases. Here, we report that stable bovine ESCs can be efficiently derived in a culture condition based on Wnt-pathway inhibition. These well-characterized ESC lines not only will enrich our understanding of pluripotency programs in the ungulate species but also will provide a useful resource for the creation of transgenic ungulate models of human diseases.
Keywords: bovine, embryonic stem cell, pluripotency, inner cell mass
Abstract
Embryonic stem cells (ESCs) are derived from the inner cell mass of preimplantation blastocysts. From agricultural and biomedical perspectives, the derivation of stable ESCs from domestic ungulates is important for genomic testing and selection, genome engineering, and modeling human diseases. Cattle are one of the most important domestic ungulates that are commonly used for food and bioreactors. To date, however, it remains a challenge to produce stable pluripotent bovine ESC lines. Employing a culture system containing fibroblast growth factor 2 and an inhibitor of the canonical Wnt-signaling pathway, we derived pluripotent bovine ESCs (bESCs) with stable morphology, transcriptome, karyotype, population-doubling time, pluripotency marker gene expression, and epigenetic features. Under this condition bESC lines were efficiently derived (100% in optimal conditions), were established quickly (3–4 wk), and were simple to propagate (by trypsin treatment). When used as donors for nuclear transfer, bESCs produced normal blastocyst rates, thereby opening the possibility for genomic selection, genome editing, and production of cattle with high genetic value.
Under specific culture conditions embryonic stem cells (ESCs) can be captured and expanded from the inner cell mass (ICM) of a blastocyst-stage embryo. Once stabilized, ESCs can proliferate unlimitedly in culture while retaining pluripotency, the ability to generate a multitude of cell types and tissues (1, 2).
Stable bovine ESCs (bESCs) will not only enrich our understanding of embryonic pluripotency and early development in livestock species but also will facilitate agricultural- and biotechnological-related applications, e.g., production of genetically superior cattle by genomic selection and/or genome editing. Despite years of research, the derivation of stable pluripotent ESCs in bovine species remains challenging (3–5). Most, if not all, of the reported bESC lines do not pass standard pluripotency tests, i.e., in vitro embryoid body formation, in vivo teratoma assay, and/or chimera formation. Moreover, they show poor derivation efficiencies, limited proliferation capacities, and loss of pluripotency markers after extensive passages (6–16).
In addition to ESCs, epiblast stem cells (EpiSCs) derived from postimplantation epiblasts also exhibit some of the hallmarks of pluripotency (17, 18). ESCs and EpiSCs have distinct molecular features and embody different pluripotent states termed “naive” and “primed,” respectively (2, 19–21). Although both are sourced from preimplantation embryos, mouse ESCs are the gold standard of naive pluripotency, while human ESCs more resemble mouse EpiSCs and exist in the developmentally more advanced primed pluripotent state (17, 18, 22). Both human ESCs and mouse EpiSCs are notorious for their poor single-cell clonality, which is undesirable for gene editing. In addition, their derivation efficiencies vary. Recently, a new culture condition was used to derive a novel type of EpiSCs that share molecular features with gastrula-stage epiblasts and was designated as “region-selective pluripotent stem cells” (rsPSCs) due to their unique property of preferentially engrafting to the posterior part of the gastrula-stage mouse epiblast (23). Remarkably, the rsPSC condition, based on a simple serum-free culture supplemented with fibroblast growth factor 2 (FGF2) and an inhibitor of the canonical Wnt–β-catenin signaling pathway (IWR1), allowed clonal expansion of both human ESCs and mouse EpiSCs and perfect EpiSC derivation efficiency from both pre- and postimplantation murine embryos.
In this study, we applied the human region-selective ESC culture conditions, custom TeSR1 base medium (growth factor-free) supplemented with FGF2 and IWR1 (CTFR) (23), for the derivation of bESCs. CTFR medium enabled highly efficient derivation of ESCs from bovine blastocysts, which are pluripotent, amenable to single-cell dissociation, and maintain long-term stable morphology, karyotype, transcriptome, population-doubling time, pluripotency-marker gene expression, and epigenetic features. Moreover, bESCs displayed transcriptional and epigenetic characteristics of primed pluripotency. In addition, we demonstrated the possibility of using long-term cultured bESCs as nuclear-transfer (NT) nuclei donors. The highly efficient derivation of bovine ESCs holds great potential for producing cattle with desired genetic value through genomic selection and/or genome editing as well as for in vitro breeding schemes through genomic selection, germ cell differentiation, and in vitro fertilization. These applications would lead to a significant reduction of the generational interval and would accelerate genetic progress. Moreover, stable bESCs represent a powerful platform for gaining novel insights into molecular features underpinning the bovine pluripotency program.
Results
CTFR Medium Supports Derivation and Stable Long-Term Culture of Pluripotent bESCs.
bESCs were derived, propagated, cultured, and subjected to several rounds of freezing and thawing in CTFR medium (CTFR-bESCs). ESC lines could be established by the end of week 3 after ICM or whole-embryo plating and remained stable for more than 50 passages (Fig. 1). Unlike human ESCs and mouse EpiSCs, CTFR-bESCs did not show clearly defined colony margins. CTFR-bESCs stained positive for alkaline phosphatase (AP) (Fig. 1A). To assess the genetic stability of the CTFR-bESCs after long-term culture, we performed karyotyping analysis in two different CTFR-bESC lines at passage 34 (P34). Our results showed a normal chromosome content (2N = 60) in more than 70% of the examined metaphase cells (Fig. S1A). Also, CTFR-bESCs maintained a stable population-doubling time across multiple passages (Fig. S1B).
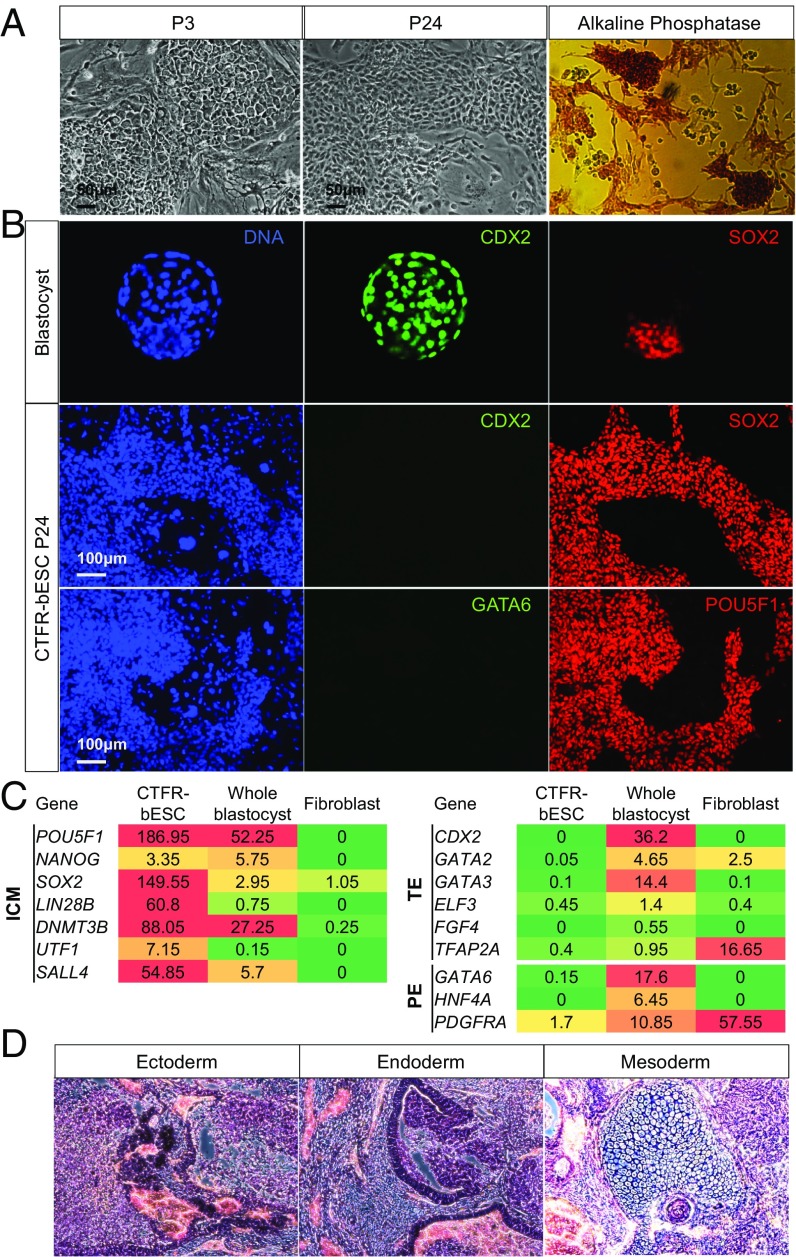
Fig. 1.
Derivation and characterization of CTFR-bESCs. (A) Bright-field images and AP staining showing typical colony morphologies of CTFR-bESCs (note that the feeder layer is negative for AP). P3, passage 3; P24, passage 24. (Scale bars, 50 μm.) (B) IF staining for SOX2, POU5F1, GATA6, and CDX2 in bovine blastocysts [Top Row (magnification: 20× objective)] and CTFR-bESCs (Middle and Bottom Rows). (C) Expression analysis of ICM, TE, and PE lineage-specific markers in CTFR-bESCs, whole blastocysts, and fibroblasts. Transcriptome analysis was done using RNA-seq. Samples include two independent CTFR-bESC lines (P10), two independent pools of whole blastocysts (10 each), and two lines of bovine fibroblasts. The color scale goes from red (high expression) to green (low/no expression). (D) Representative images showing H&E staining of histological sections derived from teratomas generated by CTFR-bESCs. CTFR-bESC–derived teratomas contained tissues of all three germ lineages: ectoderm, endoderm, and mesoderm. (Magnification: 10×.)
Immunofluorescence (IF) analysis revealed that long-term cultured CTFR-bESCs expressed the pluripotency transcription factors SOX2 and POU5F1 (also known as “OCT4”) but not the trophectoderm (TE) and primitive endoderm (PE) markers CDX2 and GATA6, respectively (Fig. 1B and Fig. S1F). IF analysis of bovine blastocysts showed that SOX2+ cells located exclusively to the ICM, while the CDX2 signal was detected only in TE cells (Fig. 1B). A SOX2+/CDX2− staining pattern was consistently observed in CTFR-bESCs from P4 onwards, indicating that CTFR culture favored the proliferation of the ICM over TE cells.
We performed transcriptome analysis of two independently established CTFR-bESC lines, bovine blastocysts, and bovine fibroblasts via RNA-sequencing (RNA-seq). Our results showed that ICM markers were expressed [reads per kilobase of transcript per million reads mapped (RPKM) ≥0.4] in CTFR-bESCs and bovine blastocysts but not (RPKM <0.4) in bovine fibroblasts (Fig. 1C and Fig. S1E). Both TE and PE markers were expressed in the blastocysts but were absent in CTFR-bESCs. These results indicate that the global gene-expression profile of CTFR-bESCs was more similar to the ICM than to TE/PE or fibroblast cells. Of note is that CTFR-bESCs’ transcriptome profile remained stable even after long-term culture (Fig. S1 C–E).
Next we performed a teratoma assay to test CTFR-bESCs pluripotency. To this end, we independently injected two CTFR-bESC lines intramuscularly into immunodeficient NOD SCID mice. Both cell lines were able to form teratomas (Fig. S1G) containing tissues from all three primary germ layers: ectoderm, mesoderm, and endoderm, as evidenced by H&E staining (Fig. 1D) and IF analysis: ectoderm (TUJ1), endoderm (FOXA2), and mesoderm (ASM) (Fig. S1H).
These results indicate that bESCs derived in CTFR medium are pluripotent and can maintain a stable karyotype and transcriptome after extended in vitro culture.
Histone Methylation Landscape of CTFR-bESCs.
The lack of culture conditions that support long-term propagation of pluripotent ESCs has impeded our molecular understanding of pluripotency in large livestock species. Taking advantage of the stable pluripotent CTFR-bESCs, we examined the global distribution of H3K4me3 and H3K27me3 marks to gain insights into the epigenetic regulation of the bovine pluripotency program. To this end, we implemented a low-input ChIP-sequencing (ChIP-seq) protocol developed and validated in the P.J.R. laboratory. An average of 31 million uniquely mapped reads were used for peak calling (Fig. S2A), which resulted in 8,816, 2,553, and 3,886 genes associated with H3K4me3, H3K27me3, or both (bivalent domains), respectively. RNA-seq analysis showed that most of the H3K4me3-only genes (94%; RPKM ≥0.4) were expressed, while 47% of the H3K27me3-only and 64% of the bivalent genes were expressed (Fig. 2A). Notably, gene-expression levels were higher for H3K4me3-only genes than for H3K27me3-only and bivalent genes (average RPKM = 34, 5, and 8, respectively).
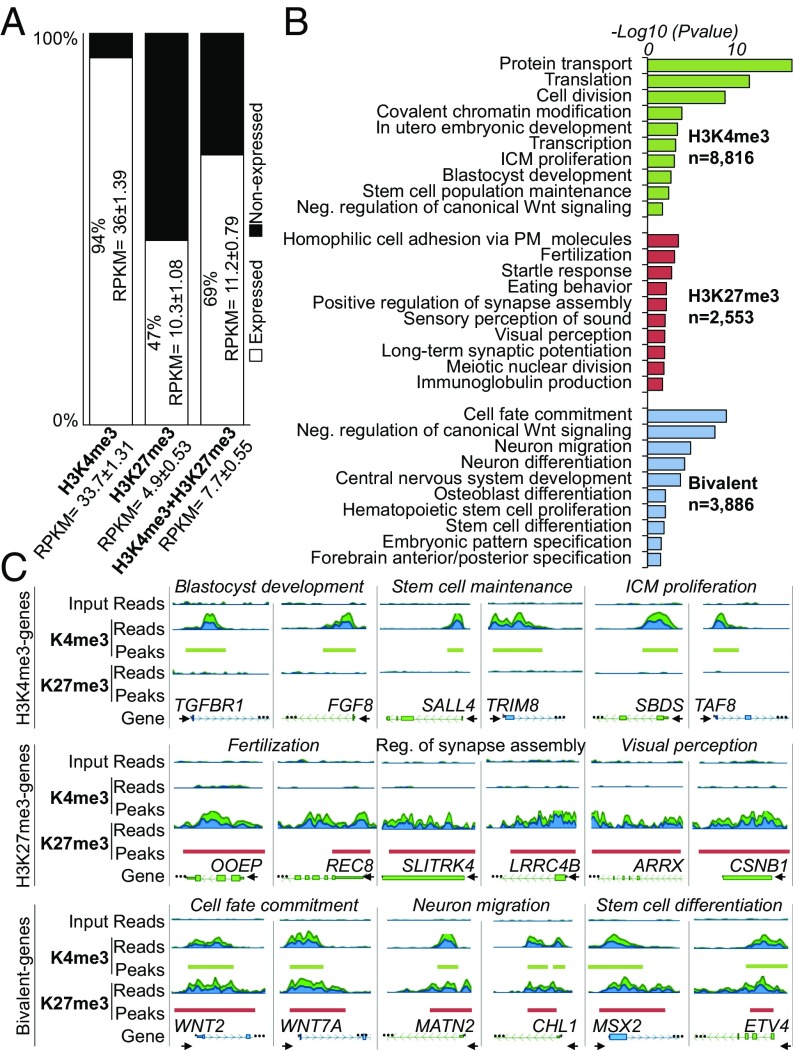
Fig. 2.
Histone methylation landscape of CTFR-bESCs. (A) Transcriptional status of genes containing H3K4me3, H3K27me3, or bivalent domains. Genes with RPKM ≥0.4 were considered expressed, and genes with RPKM <0.4 were considered nonexpressed. Mean RPKM ± SEM values of expressed genes are shown inside the bar plot, and mean RPKM ± SEM values for all genes (expressed and nonexpressed) are shown on the x axis. (B) Functional characterization of genes containing H3K4me3 (n = 8,816), H3K27me3 (n = 2,553), or bivalent domains (n = 3,886). The top-10 GO terms are shown. The bar plot shows the −log10 of the P value for selected GO term biological processes from DAVID. (C) Genome browser snapshot of genes containing H3K4me3 (TGFBR1, FGF8, SALL4, TRIM8, SBDS, and TAF8), H3K27me3 (OOEP, REC8, SLITRK4, LRRC4B, ARRX, and CSNB1), or bivalent domains (WNT2, WNT7A, MATN2, CHL1, MSX2, and ETV4). H3K4me3-, H3K27me3-, and bivalent-selected genes were associated with three different GO terms. The start of the gene is denoted by a black arrow.
Gene ontology (GO) terms associated with H3K4me3-only genes included not only housekeeping cellular functions such as protein transport, cell division, transcription, and translation but also specific pluripotency-related functions including ICM proliferation, stem cell population maintenance, and blastocyst development (Fig. 2B). Also, consistent with the use of a small-molecule Wnt inhibitor (IWR1) in CTFR-bESC culture, a noteworthy enriched term was the negative regulation of the canonical Wnt-signaling pathway. Enriched GO terms of H3K27me3-only genes related to specific functions that are generally silenced in pluripotent stem cells (PSCs), such as fertilization, startle response, Ig production, and visual perception. Last, enriched GO terms for bivalent genes were mainly related to cell-fate decisions, e.g., cell-fate commitment, neuron migration and differentiation, central nervous system development, and stem cell differentiation (Fig. 2B).
Visual assessment of genes associated with three selected GO terms for H3K4me3, H3K27me3, or bivalent genes are shown in Fig. 2C. We found that all genes containing the H3K4me3 mark presented well-defined peaks in their promoter regions, while H3K27me3 genes displayed broader peaks (some in the promoter region, but most of them covered much of the gene bodies). Bivalent genes showed colocalization of H3K4me3 and H3K27me3 marks near their promoter regions. Notably, the H3K27me3 mark in bivalent genes was more confined to the transcription start site, showing sharp peak shapes similar to the H3K4me3 mark (Fig. 2C).
Although mouse and human ESCs represent distinct pluripotent states, they do share some common transcriptional and epigenetic features characteristic of pluripotent cells. We obtained lists of genes that are known to harbor H3K4me3, H3K27me3, or bivalent domains in human and mouse ESCs and compared them with those in CTFR-bESCs. We found that 62% (n = 4,898) of H3K4me3-only bovine genes also contained the H3K4me3 mark in human and/or mouse ESCs (Fig. S2 B and C). Most of these genes were shared by both species (33%, n = 2,563), but a greater number of genes were shared with human ESCs (23%, n = 1,822) than with mouse ESCs (6%, n = 513). Forty-four percent of the bovine bivalent genes were also shared by mouse and/or human ESCs, and half of these were shared by all three species (n = 757). Similarly, bovine bivalent genes showed more similarity to those of human ESCs than mouse ESCs (13%, n = 447 and 9%, n = 317, respectively). Interestingly, only 4% of the H3K27me3-only bovine genes (n = 1,697) were shared with those in the other two species. This low level of overlap was likely due to the low number of H3K27me3 genes in mouse (n = 137) and human (n = 424) ESCs. Overall, these results suggest that bESCs have an epigenetic landscape similar to ESCs from other mammalian species, further confirming their pluripotent status.
CTFR-bESCs Display Transcriptional and Epigenetic Features Characteristic of the Primed Pluripotent State.
To investigate the pluripotency state of CTFR-bESCs, we analyzed the expression of typical naive and primed pluripotency markers identified in mouse and human ESC studies (24, 25). We found that most of the examined primed pluripotency markers were expressed (RPKM ≥0.4) in CTFR-bESCs (19/22, 86%), but fewer naive markers were expressed (14/22, 64%). Also, the average RPKM values of all the analyzed markers were higher for the primed than for the naive pluripotency marker genes (RPKM = 25 ± 8.4 and RPKM = 17 ± 13, respectively) (Fig. 3A).
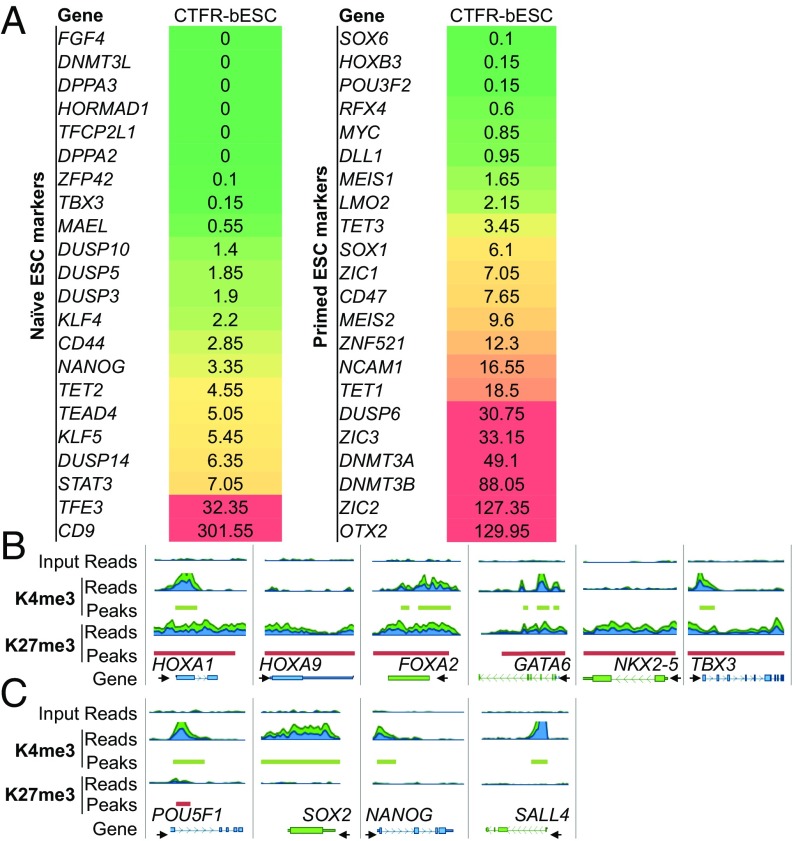
Fig. 3.
CTFR-bESCs show molecular signatures characteristic of primed pluripotency. (A) Transcriptome analysis of selected naive and primed pluripotency markers in CTFR-bESCs. RNA-seq was performed, and RPKM values were used to define expressed (RPKM ≥0.4; red) and nonexpressed (RPKM <0.4; green) genes. The means of two biological replicates are shown. (B) Genome browser snapshots of histone methylation profiles of primed and naive pluripotency markers in CTFR-bESCs. (C) Genome browser snapshots of H3K4me3 and H3K27me3 marks in core pluripotency genes (POU5F1, SOX2, NANOG, SALL4) in CTFR-bESCs.
The presence of an H3K4me3 peak in the promoter region of core pluripotency transcription factors, including POU5F1, SOX2, SALL4, and NANOG genes, is common to both naive and primed PSCs. Consistently, CTFR-bESCs also showed sharp H3K4me3 peaks at the promoter regions of these genes (Fig. 3C). Next, we focused on genes that display distinct histone methylation patterns in naive- and primed-state PSCs in the mouse and human species (26, 27). Consistent with the gene-expression analysis, the epigenetic signatures for all the examined genes in CTFR-bESCs reflect a primed pluripotency state, e.g., bivalent domains are present in HOXA1, FOXA2, GATA6, and TBX3 genes, and there is an accumulation of the H3K27me3 mark in HOXA9 and NKX-2 genes (Fig. 3B).
Taken together, these results show that prominent pluripotent epigenetic features are shared among ungulates, rodents, and primates and that CTFR-bESCs harbor a primed pluripotency program.
CTFR-bESCs’ Derivation Is Highly Efficient.
CTFR-bESCs’ derivation efficiency was measured as the percentage of embryos successfully establishing a CTFR-bESC line at P3 over the total number of embryos plated. To optimize the CTFR-bESCs’ derivation efficiency, we evaluated three different methods: whole blastocyst, mechanical isolation of the ICM by microdissection, and isolation of the ICM by immunosurgery. We did not find that derivation efficiency differed significantly among the three methods, and the efficiency was comparable to or even higher than that reported for mouse ESCs (Fig. 4A) (23, 28, 29). The initial outgrowths from the three methods were slightly different. The immunosurgery-derived ICM outgrowths were smaller and showed more homogeneous cell morphology than those from the other two methods. By the end of the second week, however, cells showed a similar homogenous morphology regardless of the derivation method used (Fig. S3A). These results indicate that even with simple whole-blastocyst plating, CTFR-bESCs with homogeneous cell morphology could be derived at high efficiency within a short period of time.
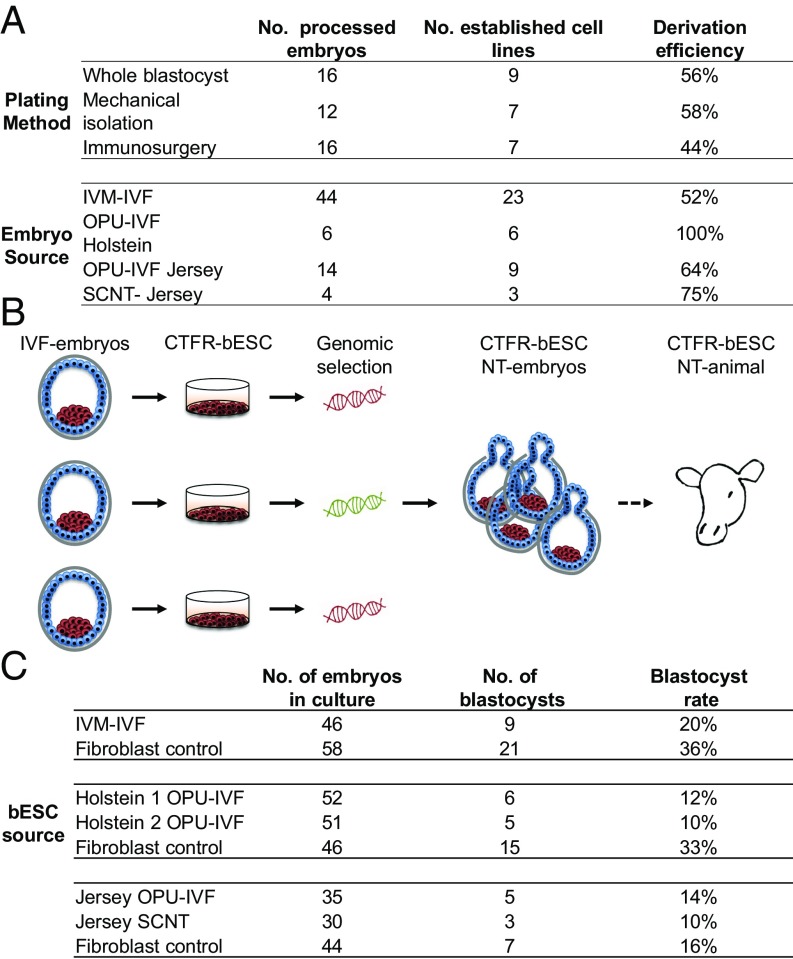
Fig. 4.
Potential applications of CTFR-bESCs for genomic selection. (A) CTFR-bESC derivation efficiency using three different plating methods (whole blastocyst, mechanical isolation of ICM, and immunosurgery-derived ICM) and different embryo sources (IVM-IVF, OPU-IVF, SCNT, and Holstein and Jersey breeds). Derivation efficiency was measured as the percentage of blastocysts that gave rise to a stable CTFR-bESC line at P3 over the total number of embryos seeded per method. (B) A schematic diagram showing the strategy of using CTFR-bESCs for genomic selection to produce animals of superior genetic value through highly efficient CTFR-bESC derivation and NT. (C) CTFR-bESCs generated from different sources can be used as nuclear donors for cloning.
We next examined whether different embryo sources could affect the derivation efficiency. We found that when using ovum pick-up in vitro-fertilized (OPU-IVF)–derived embryos of the Holstein and Jersey breeds, derivation efficiency was 100% and 64%, respectively, while when using embryos produced by in vitro fertilization (IVF) of oocytes aspirated from slaughterhouse-derived ovaries [in vitro-matured (IVM)-IVF oocytes] the efficiency was 52%. Somatic cell nuclear transfer (SCNT)-derived embryos produced CTFR-bESCs with 75% efficiency (Fig. 4A). Overall, these results show that CTFR-bESCs can be produced efficiently from different embryo sources.
CTFR-bESCs Can Be Used as Nuclear Donors for NT Cloning.
We envision that a potential application of CTFR-bESCs is for genomic selection followed by the production of NT-derived cattle (Fig. 4B). To this end, we tested the possibility of generating NT embryos using CTFR-bESCs (obtained from different embryo sources) and found that all of them were able to produce NT blastocysts at efficiencies ranging from 10 to 20%, a lower blastocyst-formation rate than with control fibroblasts (29%) (Fig. 4C). A possible explanation for the lower blastocyst rate is that CTFR-bESCs have a shorter G1 phase of the cell cycle than fibroblasts. Indeed, FACS analysis revealed that CTFR-bESCs had only half as many cells in G1 as fibroblasts (Fig. S3B). Also, we observed a higher percentage of cells in S phase [a cell-cycle stage that is incompatible with NT embryo development (30)] in CTFR-bESCs than in fibroblasts. Finally, the CTFR-bESC NT embryos could again be used to derive secondary CTFR-bESCs that displayed stable SOX2 and POU5F1 IF-staining patterns identical to those in the original cells (Fig. S3C).
Discussion
Despite years of research, no stable bESC line that can withstand the rigor of extended passaging in culture while maintaining in vivo pluripotency has been reported. In the current study, we tested a CTFR condition (23) for the derivation of bESCs. Our results show that bESCs can be established efficiently under the CTFR condition by simple whole-blastocyst plating of embryos derived from various sources and genetic backgrounds. CTFR-bESCs are amenable to long-term cultivation and display stable genetic, transcriptional, epigenetic, and functional pluripotency features, including teratoma formation. CTFR-bESCs exhibit molecular features characteristic of primed pluripotency. In addition, we demonstrate the efficacy of using CTFR-bESCs as nuclei donors for the production of NT-cloned blastocysts, a necessary first step toward the generation of live CTFR-bESC NT cattle.
The CTFR culture condition allows robust derivation and long-term propagation of pluripotent bESCs. We consider that the combination of factors, and especially the addition of IWR1, was crucial for the derivation of bESCs. IWR1 blocks the translocation of β-catenin to the nucleus by stabilizing AXIN 1/2 (31). The addition of the canonical WNT inhibitor has proven effective in deriving and maintaining mouse EpiSCs and primed human PSCs (23, 32). In bovine species, maintenance of an inactive canonical WNT-signaling pathway is important for normal preimplantation and early postimplantation embryo development (33, 34). Activation of the canonical WNT-β–catenin signaling pathway by the addition of 2-amino-4-(3,4-(methylenedioxy)benzylamino)-6-(3-methoxyphenyl)pyrimidine (AMBMP) during in vitro preimplantation development shows detrimental effects for bovine blastocyst development and reduces the total number of cells in the ICM (34). Additionally, in vitro treatment of bovine embryos (from day 5 to day 7 after fertilization) with a protein inhibitor of the canonical WNT-signaling pathway (DKK1), which is naturally secreted by the female reproductive tract and is involved in maternal-to-embryo communication, has been shown to improve embryo survival significantly after transfer to recipients (33). Therefore, it is plausible that the addition of the canonical WNT inhibitor was critical for the successful derivation and propagation of bESCs. Indeed, withdrawal of IWR1 from the culture medium resulted in the loss of pluripotency markers’ expression (Fig. S4). In addition, no bESC line could be established if IWR1 was not present in the medium.
Mouse ESCs are considered the gold standard for naive PSCs, whose molecular features resemble those of nascent epiblast cells within the murine ICM (35). The primed state of pluripotency is associated with postimplantation epiblast cells and could be stabilized in cultured mouse EpiSCs. In addition to rodents, naive-like and primed PSCs, which are mostly defined by respective molecular signatures, have been described in primate species, including humans (2, 20). All reported naive-like primate PSCs have not passed the stringent germline chimera assay, and genuine primate naive PSCs remain elusive. It is possible that species differences may account for the difficulty in obtaining cells equivalent to mouse ESCs in nonrodent species. The CTFR-bESCs derived in this study share many defining transcriptional features with mouse EpiSCs and primed human PSCs, for example, the high expression of genes implicated in lineage commitment, e.g., OTX2 and ZIC2 (36), the moderate expression levels of the pluripotency-related transcription factors Homeobox protein NANOG and KLF4, and the negligible expression of many naive pluripotency marker genes such as FGF4, DNMT3L, DPPA2, DPPA3, HORMAD1, TFCP2L1, ZFP42 (REX1), and TBX3 (23, 26, 27). Epigenetically, CTFR-bESCs share many features with their mouse and human primed counterparts, such as the presence of bivalent domains in HOXA1, FOXA2, GATA6, and TBX3 genes and the enrichment of the H3K27me3 mark in the HOXA9 and NKX-2 genes (26, 27).
The co-occupancy of H3K4me3 and H3K27me3 marks on gene promoters is one of the most notable epigenetic features of pluripotent cells (37–40). These bivalent domains typically localize to key developmental genes that are transcriptionally silenced in ESCs but are poised for activation (38, 41). Interestingly, 44% of the CTFR-bESC bivalent genes were also present in human and mouse ESCs (24, 25), suggesting that many molecular features that delineate pluripotency and early lineage commitment are conserved across distant mammalian species. The conserved epigenetic features underlying mammalian pluripotency were further noted in the GO terms for the CTFR-bESC bivalent genes. Four of the top-10 enriched GO terms (negative regulation of the canonical Wnt-signaling pathway, neuron migration, central nervous system development, and neuron differentiation) were also found among the top-five enriched GO terms for bivalent genes in human ESCs according to the database of bivalent genes (BGDB) (42).
The CTFR-bESCs described in this study were easy to derive from whole blastocysts, fast to obtain, highly efficient to establish, and easy to passage (single-cell dissociation using trypsin). These are all desirable features that will facilitate the creation of genetically superior cattle and the industrial production of valuable pharmaceuticals, as they allow efficient genomic selection through bESC derivation and facile genome editing and are amenable for NT cloning to generate live animals. Another potential use of these cells is the in vitro differentiation to gametes, facilitating in vitro breeding schemes that could result in multiple rounds of genomic selection, gamete production and fertilization, and bESC derivation to achieve genetically superior cattle within a significantly shorter generational interval.
In summary, by using a serum-free culture condition, we have derived stable pluripotent bESC lines that can be propagated over the long term in culture (for more than 70 passages and more than 1 y at the time of writing) while maintaining stable morphology, normal karyotype, pluripotent transcriptome and epigenome signatures, and the ability to generate teratomas containing cells and tissues from all three primary germ lineages. Moreover, CTFR-bESCs were used successfully as nuclear donors to produce cloned blastocysts. The derivation of stable bESCs opens avenues for various agricultural and biotechnological applications. The culture condition and protocol developed in this study can potentially be applied to many other farm animal species for the generation of stable ESCs.
Materials and Methods
Embryo Production and Processing.
The IVM-IVF embryos used in this study were produced as previously described (43). OPU-IVF– and SCNT-derived embryos were produced by Trans Ova Genetics using their standard procedures and were shipped overnight to the University of California, Davis for bESC derivation.
Microsurgery and Immunosurgery of Bovine Blastocysts.
Isolated ICMs were obtained by microsurgery. Briefly, zona pellucida (ZP)-depleted blastocysts were placed in a Petri dish containing PBS, and microsurgery was performed using a microblade connected to micromanipulation equipment (NT88-V3; Nikon/Narishige) attached to an inverted microscope (TE2000-U; Nikon).
Immunosurgery was carried out by incubating the embryos in KSOM medium with 20% anti-bovine serum (Jackson ImmunoResearch) for1 h at 37 °C followed by repeated washes with synthetic oviductal fluid (SOF)⋅Hepes and incubation in KSOM medium supplemented with 20% guinea pig complement (Innovative Research) for 1 h at 37 °C.
Derivation and Culture of CTFR-bESCs.
Individual whole blastocysts or isolated ICMs were placed in separate wells of a 12-well dish seeded with a monolayer of gamma-irradiated mouse embryonic fibroblasts (MEFs), were cultured in CTFR medium [a custom basal medium similar to mTeSR medium (STEMCELL Technologies)] that is completely devoid of growth factors FGF2 and TGFb and contains low fatty acid BSA (MP Biomedicals NZ), similar to basal medium in the published recipe (44), and supplemented with 20 ng/mL human FGF2 and 2.5 μM IWR1, and were incubated at 37 °C and 5% CO2 (23).
After 48 h, blastocysts/ICMs that failed to adhere to the feeder layer were physically pressed against the bottom of the culture dish with a 22-gauge needle under a stereoscope to facilitate attachment. Thereafter, the medium was changed daily. Outgrowths (after 6–7 d in culture) were dissociated and passaged using TrypLE (12563011; Gibco) and were reseeded in the presence of the Rho kinase (ROCK) inhibitor Y-27632 (10 μM) into newly prepared wells containing MEFs and fresh medium (Fig. 1).
Once established, CTFR-bESC lines were grown at 37 °C and 5% CO2 in wells containing MEFs and were passaged every 4–5 d at a 1:10 split ratio. To increase cell survival, optionally, the ROCK inhibitor Y-27632 (10 μM) was added to the wells 1 h before passage and was also added to the newly prepared wells containing MEFs and fresh culture medium during the 24 h until the next media change. Medium was changed daily between passages. The ROCK inhibitor is dispensable for routine maintenance and passaging of CTFR-bESCs.
IF.
Cells and embryos were immunostained and imaged as previously described (45) using the following primary antibodies: anti-GATA6 (sc-7244, 1:300; Santa Cruz Biotechnology), anti-SOX2 (ANS79-5M, 1:300; BioGenex), anti-CDX2 (MU392A-UC, 1:300; BioGenex), and anti-POU5F1 (sc-8628, 1:300; Santa Cruz Biotechnology).
RNA-Seq.
A confluent well of a six-well plate of two CTFR-bESC lines (A: cells P13, P23, P35, P45; B: cells at P12, P24, P35, and P46) were used for RNA-seq. Whole blastocysts and fibroblasts were used as controls. Total RNA was isolated using the Qiagen RNeasy Mini Kit and then was reverse-transcribed using iScript RT Supermix (Bio-Rad). Libraries were constructed using the TruSeq RNA Sample Prep Kit (Illumina) and were sequenced on an Illumina HiSeq 2500 system according to the manufacturer’s instructions. Sequenced reads were mapped to the bovine UMD3.1 genome assembly and Ensembl 78 genebuild annotation using CLC Genomics Workbench 7.0 (CLC bio). RPKM values were calculated for each gene.
ChIP-Seq.
Two lines of CTFR-bESCs at P12 were separated from the MEFs using Feeder Removal MicroBeads (130-095-531; Miltenyi Biotech) following the manufacturer’s protocol (20,000 cells). CTFR-bESCs were cross-linked using 0.25% formaldehyde (28906; Pierce), and the cross-linking was stopped by adding 125 mM of glycine (provided in the True MicroChIP Kit, C01010130; Diagenode). ChIP was performed using the True MicroChIP Kit using the antibodies anti-H3K27me3 (ABE 44; Millipore) and anti-H3K4me3 (provided with the True MicroChIP Kit). Sequencing libraries were prepared following the manufacturer’s instructions using the ThruPLEX DNA-seq Kit (R400406; Rubicon) with 16 cycles in the library amplification step. Libraries were sequenced at the Vincent J. Coates Genomics Sequencing Laboratory at the University of California, Berkeley in an Illumina HiSeq 4000 platform where sequencing was performed as 100 bp paired-end. Raw reads were checked for sequencing quality using FastQC and then were aligned to the annotated bovine genome (UMD 3.1 assembly) using bwa aln (46, 47). Peak calling was done using MACS2 (48) with narrow settings for H3K4me3 (-g 2.67e9 -q 0.01 -m 2 100 -B–SPMR) and broad settings for H3K27me3 (-g 2.67e9 -q 0.05 -m 2 100-broad -B–SPMR–fix-bimodal–extsize 200). Peaks were visualized using the Golden Helix GenomeBrowse tool (Golden Helix, Inc., available at www.goldenhelix.com). Called peaks were further analyzed using Hypergeometric Optimization of Motif EnRichment (HOMER) (49) to find peak associations with gene features, and GO analysis was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (50, 51).
Somatic Cell NT.
For testing the capacity of established CTFR-bESC lines to make NT blastocysts, several CTFR-bESC lines were used as nuclear donors for reconstructing enucleated oocytes using standard SCNT methodology (45, 52). Primary fibroblasts were used as controls.
Karyotyping, the teratoma formation assay, AP staining, population-doubling time, and cell-cycle analysis by flow cytometry are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank M. Schwarz and P. Schwarz for administrative help; the Salk Stem Cell Core for providing cell culture reagents; Trans Ova Genetics for providing OPU-derived embryos and performing SCNT from bESCs; and M. C. Ku, N. Hah, and T. Nguyen for RNA-seq experiments. Work in the P.J.R. laboratory was supported by NIH/Eunice Kennedy Shriver National Institute of Child Health and Human Development Grant R01-HD070044, United States Department of Agriculture (USDA)–National Institute of Food and Agriculture (NIFA)–Agriculture and Food Research Initiative (AFRI) Multistate Project W3171, and by a University of California, Davis Academic Senate New Research Grant. Y.S.B. was supported by National Science Foundation-Graduate Research Fellowship Program Award 1148897. R.V.S. was supported by São Paulo Research Foundation (FAPESP) Grants 2013/08135-2 and 2015/25111-5. Work in the J.C.I.B. laboratory was supported by the Universidad Católica San Antonio de Murcia, the G. Harold and Leila Y. Mathers Charitable Foundation, The Moxie Foundation, Fundación Dr. Pedro Guillén, and NIH Grant DP1-DK113616.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE110040).
See Commentary on page 1962.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716161115/-/DCSupplemental.
References
- 1.Ying QL, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu J, Izpisua Belmonte JC. Dynamic pluripotent stem cell states and their applications. Cell Stem Cell. 2015;17:509–525. doi: 10.1016/j.stem.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Blomberg LA, Telugu BP. Twenty years of embryonic stem cell research in farm animals. Reprod Domest Anim. 2012;47:80–85. doi: 10.1111/j.1439-0531.2012.02059.x. [DOI] [PubMed] [Google Scholar]
- 4.Ezashi T, Yuan Y, Roberts RM. Pluripotent stem cells from domesticated mammals. Annu Rev Anim Biosci. 2016;4:223–253. doi: 10.1146/annurev-animal-021815-111202. [DOI] [PubMed] [Google Scholar]
- 5.Soto DA, Ross PJ. Pluripotent stem cells and livestock genetic engineering. Transgenic Res. 2016;25:289–306. doi: 10.1007/s11248-016-9929-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao S, et al. Isolation and culture of primary bovine embryonic stem cell colonies by a novel method. J Exp Zool A Ecol Genet Physiol. 2009;311:368–376. doi: 10.1002/jez.535. [DOI] [PubMed] [Google Scholar]
- 7.Cibelli JB, et al. Transgenic bovine chimeric offspring produced from somatic cell-derived stem-like cells. Nat Biotechnol. 1998;16:642–646. doi: 10.1038/nbt0798-642. [DOI] [PubMed] [Google Scholar]
- 8.Kim D, Jung YG, Roh S. Microarray analysis of embryo-derived bovine pluripotent cells: The vulnerable state of bovine embryonic stem cells. PLoS One. 2017;12:e0173278. doi: 10.1371/journal.pone.0173278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitalipova M, Beyhan Z, First NL. Pluripotency of bovine embryonic cell line derived from precompacting embryos. Cloning. 2001;3:59–67. doi: 10.1089/15204550152475563. [DOI] [PubMed] [Google Scholar]
- 10.Muñoz M, et al. Conventional pluripotency markers are unspecific for bovine embryonic-derived cell-lines. Theriogenology. 2008;69:1159–1164. doi: 10.1016/j.theriogenology.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 11.Saito S, et al. Generation of cloned calves and transgenic chimeric embryos from bovine embryonic stem-like cells. Biochem Biophys Res Commun. 2003;309:104–113. doi: 10.1016/s0006-291x(03)01536-5. [DOI] [PubMed] [Google Scholar]
- 12.Saito S, Strelchenko N, Niemann H. Bovine embryonic stem cell-like cell lines cultured over several passages. Rouxs Arch Dev Biol. 1992;201:134–141. doi: 10.1007/BF00188711. [DOI] [PubMed] [Google Scholar]
- 13.Stice SL, Strelchenko NS, Keefer CL, Matthews L. Pluripotent bovine embryonic cell lines direct embryonic development following nuclear transfer. Biol Reprod. 1996;54:100–110. doi: 10.1095/biolreprod54.1.100. [DOI] [PubMed] [Google Scholar]
- 14.Talbot NC, Powell AM, Rexroad CE., Jr In vitro pluripotency of epiblasts derived from bovine blastocysts. Mol Reprod Dev. 1995;42:35–52. doi: 10.1002/mrd.1080420106. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, et al. Generation and characterization of pluripotent stem cells from cloned bovine embryos. Biol Reprod. 2005;73:149–155. doi: 10.1095/biolreprod.104.037150. [DOI] [PubMed] [Google Scholar]
- 16.Wu X, et al. Establishment of bovine embryonic stem cells after knockdown of CDX2. Sci Rep. 2016;6:28343. doi: 10.1038/srep28343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tesar PJ, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 18.Brons IG, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 19.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 20.Weinberger L, Ayyash M, Novershtern N, Hanna JH. Dynamic stem cell states: Naive to primed pluripotency in rodents and humans. Nat Rev Mol Cell Biol. 2016;17:155–169. doi: 10.1038/nrm.2015.28. [DOI] [PubMed] [Google Scholar]
- 21.De Los Angeles A, et al. Hallmarks of pluripotency. Nature. 2015;525:469–478. doi: 10.1038/nature15515. [DOI] [PubMed] [Google Scholar]
- 22.Hanna J, et al. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci USA. 2010;107:9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J, et al. An alternative pluripotent state confers interspecies chimaeric competency. Nature. 2015;521:316–321. doi: 10.1038/nature14413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan G, et al. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Gafni O, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 27.Theunissen TW, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brook FA, Gardner RL. The origin and efficient derivation of embryonic stem cells in the mouse. Proc Natl Acad Sci USA. 1997;94:5709–5712. doi: 10.1073/pnas.94.11.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bryja V, et al. An efficient method for the derivation of mouse embryonic stem cells. Stem Cells. 2006;24:844–849. doi: 10.1634/stemcells.2005-0444. [DOI] [PubMed] [Google Scholar]
- 30.Tian XC, Kubota C, Enright B, Yang X. Cloning animals by somatic cell nuclear transfer–Biological factors. Reprod Biol Endocrinol. 2003;1:98. doi: 10.1186/1477-7827-1-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim H, et al. Modulation of β-catenin function maintains mouse epiblast stem cell and human embryonic stem cell self-renewal. Nat Commun. 2013;4:2403. doi: 10.1038/ncomms3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugimoto M, et al. A simple and robust method for establishing homogeneous mouse epiblast stem cell lines by wnt inhibition. Stem Cell Rep. 2015;4:744–757. doi: 10.1016/j.stemcr.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Denicol AC, et al. The WNT signaling antagonist Dickkopf-1 directs lineage commitment and promotes survival of the preimplantation embryo. FASEB J. 2014;28:3975–3986. doi: 10.1096/fj.14-253112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denicol AC, et al. Canonical WNT signaling regulates development of bovine embryos to the blastocyst stage. Sci Rep. 2013;3:1266. doi: 10.1038/srep01266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boroviak T, Loos R, Bertone P, Smith A, Nichols J. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat Cell Biol. 2014;16:516–528. doi: 10.1038/ncb2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buecker C, et al. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14:838–853. doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 38.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 39.Sachs M, et al. Bivalent chromatin marks developmental regulatory genes in the mouse embryonic germline in vivo. Cell Rep. 2013;3:1777–1784. doi: 10.1016/j.celrep.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharov AA, Ko MS. Human ES cell profiling broadens the reach of bivalent domains. Cell Stem Cell. 2007;1:237–238. doi: 10.1016/j.stem.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 41.Tee WW, Reinberg D. Chromatin features and the epigenetic regulation of pluripotency states in ESCs. Development. 2014;141:2376–2390. doi: 10.1242/dev.096982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Q, Lian S, Dai Z, Xiang Q, Dai X. BGDB: A database of bivalent genes. Database (Oxford) 2013;2013:bat057. doi: 10.1093/database/bat057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ross PJ, et al. Activation of bovine somatic cell nuclear transfer embryos by PLCZ cRNA injection. Reproduction. 2009;137:427–437. doi: 10.1530/REP-08-0419. [DOI] [PubMed] [Google Scholar]
- 44.Ludwig TE, et al. Feeder-independent culture of human embryonic stem cells. Nat Methods. 2006;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- 45.Canovas S, Cibelli JB, Ross PJ. Jumonji domain-containing protein 3 regulates histone 3 lysine 27 methylation during bovine preimplantation development. Proc Natl Acad Sci USA. 2012;109:2400–2405. doi: 10.1073/pnas.1119112109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leggett RM, Ramirez-Gonzalez RH, Clavijo BJ, Waite D, Davey RP. Sequencing quality assessment tools to enable data-driven informatics for high throughput genomics. Front Genet. 2013;4:288. doi: 10.3389/fgene.2013.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, et al. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 51.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ross PJ, Cibelli JB. Bovine somatic cell nuclear transfer. Methods Mol Biol. 2010;636:155–177. doi: 10.1007/978-1-60761-691-7_10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.