Significance
Using budding yeast, we address how Cas9 protein and its guide RNA (gRNA) create double-strand chromosome breaks (DSBs), and explore whether binding of Cas9::gRNA influences subsequent DSB repair by nonhomologous end-joining. We created pairs of gRNAs that are complementary to opposite DNA strands but direct cleavage at the same chromosomal location. The resulting repair profiles (insertion/deletions) are different for the two ostensibly identical DSBs. Most notably, there are frequent +1 insertions that are templated after cleavage creates a 1-nt 5′ overhang that is filled in before ends are ligated. DNA polymerase 4 is required for most +1 insertions and for longer (+2 and +3) insertions. We found similar templating of +1 insertions in published studies of mammalian DSBs created by Cas9.
Keywords: CRISPR/Cas9, templated insertions, DNA polymerase 4, double-strand breaks, nonhomologous end-joining
Abstract
Harnessing CRISPR-Cas9 technology provides an unprecedented ability to modify genomic loci via DNA double-strand break (DSB) induction and repair. We analyzed nonhomologous end-joining (NHEJ) repair induced by Cas9 in budding yeast and found that the orientation of binding of Cas9 and its guide RNA (gRNA) profoundly influences the pattern of insertion/deletions (indels) at the site of cleavage. A common indel created by Cas9 is a 1-bp (+1) insertion that appears to result from Cas9 creating a 1-nt 5′ overhang that is filled in by a DNA polymerase and ligated. The origin of +1 insertions was investigated by using two gRNAs with PAM sequences located on opposite DNA strands but designed to cleave the same sequence. These templated +1 insertions are dependent on the X-family DNA polymerase, Pol4. Deleting Pol4 also eliminated +2 and +3 insertions, which are biased toward homonucleotide insertions. Using inverted PAM sequences, we also found significant differences in overall NHEJ efficiency and repair profiles, suggesting that the binding of the Cas9:gRNA complex influences subsequent NHEJ processing. As with events induced by the site-specific HO endonuclease, CRISPR-Cas9–mediated NHEJ repair depends on the Ku heterodimer and DNA ligase 4. Cas9 events are highly dependent on the Mre11-Rad50-Xrs2 complex, independent of Mre11’s nuclease activity. Inspection of the outcomes of a large number of Cas9 cleavage events in mammalian cells reveals a similar templated origin of +1 insertions in human cells, but also a significant frequency of similarly templated +2 insertions.
The use of CRISPR-Cas9 RNA guided endonuclease has accelerated genome editing in systems ranging from yeast to mammals (1–3). Guided endonucleases target site-specific DNA to create double-strand breaks (DSBs) (4), which can be repaired either by nonhomologous end-joining (NHEJ) or homologous recombination (HR) (reviewed in ref. 5). The mechanisms of these repair processes, including the required core genetic components, have been extensively studied in vivo using the site-specific HO endonuclease of Saccharomyces cerevisiae (6). Thus, we were interested in comparing the DSB repair induced by CRISPR-Cas9 with events promoted by HO endonuclease in budding yeast.
The Streptococcus pyogenes Cas9 endonuclease scans the genome to locate protospacer adjacent motifs (PAMs) of 5′ NGG 3′, which are required for binding and cleavage. Specificity is achieved by a Cas9-bound guide RNA (gRNA) that pairs with sequences adjacent to the PAM, resulting in double-strand DNA cleavage 3-bp 5′ of the PAM sequence (7). In contrast, HO endonuclease recognizes a defined 24-bp cleavage site located in the S. cerevisiae mating type (MAT) locus. When placed under control of a galactose-inducible promoter, HO cleaves euchromatic targets within 30–60 min, creating 4-nt 3′ overhanging ends (8–10).
NHEJ can result in either faithful relegation of the broken ends or the mutagenic insertions or deletions of base pairs at the junctions (indel formation). In budding yeast, perfect relegation of the 3′ overhanging DSB ends requires the “classic” evolutionarily conserved end-joining machinery, including the Ku70-Ku80 proteins as well as DNA ligase 4 and its associated Lif1 (Xrcc4 homolog) and Nej1 cofactors (11, 12). Continuous expression of HO eliminates precise rejoining by recutting, thereby forcing the recovery of mutated HO recognition sequences, including short, templated fill-ins and 3-bp deletions, all of which result from offset base pairings and subsequent processing of overhanging ends (12). The fill-ins depend on the Mre11-Rad50-Xrs2 (MRX) complex as well as on the Polx family DNA polymerase, Pol4, whereas the 3-bp deletions are MRX- and Pol4-independent. Additional microhomology-mediated end-joining events, resulting in larger deletions, can also be recovered with the same genetic requirements, but there is also a Ku-independent microhomology-mediated end-joining (MMEJ) process (13).
The CRISPR-Cas9 system can be used to create targeted indels by designing gRNAs homologous to the genomic site of interest. Biochemical experiments have shown that Cas9 predominantly cleaves 3-bp 5′ from the PAM, resulting in a DSB, with Cas9 remaining bound to its target for hours after cleavage (14). In vitro, the 3′ end of the DSB that is not complementary to the gRNA is more accessible, raising the possibility of strong biases in DSB end-processing dictated by the Cas9 protein (15, 16).
We were especially interested in whether we could infer the consequences of persistent Cas9 binding to the target site by creating the same cleavage in two different ways, by designing gRNAs to PAMs located symmetrically on either strand. Using several pairs of inverted PAM sequences (iPAMs), we found frequent +1 insertions, in which the added base is dependent on the orientation of Cas9::gRNA binding. These insertions appear to have arisen from Cas9 cleavages resulting in 1-nt 5′ overhanging ends that are filled in by DNA polymerase Pol4. Moreover, there are substantial differences in the overall patterns of indels, depending on the strand to which Cas9 is paired and bound.
Materials and Methods
Parental Strains and Selection.
Strains JKM179 (hoΔ MATα hmlΔ::ADE1 hmrΔ::ADE1 ade1-100 leu2-3,112 lys5 trp1::hisG′ ura3-52 ade3::GAL::HO) and JKM139 (hoΔ MATa hmlΔ::ADE1 hmrΔ::ADE1 ade1-100 leu2-3,112 lys5 trp1::hisG′ ura3-52 ade3::GAL::HO) (17) were used in experiments targeting the MAT locus. These strains lack the heterochromatic HML and HMR donor sequences that would allow repair of a DSB at MAT by gene conversion, and thus all repairs are through NHEJ. Parental strain BY4733 (MATa his3Δ200 trp1Δ63 leu2Δ0 met15Δ0 ura3Δ0) was used for CAN1 and LYS2 Cas9-targeting experiments. ORFs were deleted by replacing the target with a prototrophic or antibiotic-resistance marker via single-step transformation of S. cerevisiae colonies with PCR fragments (18).
Plasmid Construction.
Plasmid pJD1, carrying Cas9 driven by the GAL1 promoter, was provided by the Church laboratory (3). Each gRNA targeting the MAT locus, with an upstream SNR52 promoter and an SUP4 terminator 3′ sequence, was synthesized as a gBlock by Integrating DNA Technologies and was cloned into plasmid pRS426 (URA3) by gap repair. gBlocks were designed with 50-bp flanking homology to pRS426 and then recombined in vivo after cotransformation of 0.02 pmol gRNA gBlock and 5 μg of BamHI digested pRS426 (19).
gRNAs gCAN1-1W, gCAN1-1C, and gLYS (1–4) were cloned into a LEU2-marked plasmid (bRA77), in which Cas9 is expressed under a galactose-inducible promoter. In brief, gRNAs were ligated into a BplI digested site, cloned in Escherichia coli. Plasmids were verified by sequencing (GENEWIZ) and transformed into our yeast strains, as described in more detail previously (20). The inserted DNA sequences are listed in SI Appendix, Table S1.
Galactose Induction and Viability.
Cas9 plasmids were transformed into parental strains using the conventional lithium acetate protocol (21). Experiments targeting the CAN1 and LYS2 locus used the parental strain BY4733. Plasmids containing a galactose-inducible Cas9 and a constitutively expressed gRNA were introduced by transformation using the conventional lithium acetate protocol (21). Cells were plated on YPD plates and replica-plated onto leucine drop-out plates for transformants. Cells containing the plasmid were serially diluted from 100,000 cells to 100 cells. Cells were plated onto leucine drop out plates containing either galactose to induce Cas9 expression or dextrose as a control. Experiments were performed in duplicate and repeated at least three times. Canavanine-resistant colonies were selected by replica plating of survivors onto canavanine-containing plates. The proportions of Lys2+ and Lys2− colonies were identified by replica-plating survivors onto lysine drop-out plates.
For assays targeting the MAT locus, cells were grown at 30 °C overnight in synthetic medium lacking uracil and leucine and 2% dextrose, refreshed in selective medium containing 2% raffinose for 6 h, and induced in selective medium containing 2% galactose. Viability was determined as the average number of colonies growing with galactose relative to those on dextrose plates after 3 d at 30 °C. Viability assays were performed with three biological replicates. Viabilities were corrected by discounting the small proportion of cells that did not contain a matα1 mutation. The viability of strains with unrepaired HO endonuclease-induced DSBs was determined as described previously (12).
DNA Sequence Analysis.
Using flanking primers to the DNA region of interest, PCR was used to amplify DNA from survivor colonies. PCR products were purified and Sanger-sequenced by GENEWIZ and Eton Bioscience. The data represent samples from at least three independent inductions of Cas9 cleavage. Barcoded primers were used to amplify LYS2 from a pool of gLYS2-1C, gLYS2-1W, gLYS2-4C, and gLYS2-4W survivors. Amplified PCR samples were purified and sequenced using Illumina sequencing technology.
Illumina Amplicon Sequencing.
Illumina P7 and P5 adapter sequences and 8-mer index sequences were added by an additional limited-cycle round of PCR. Each 50-μL PCR consisted of 25 μL of 2× KAPA HiFi HotStart ReadyMix (Kapa Biosystems), 5 μL of Index 1 Primer and 5 μL of Index 2 Primer (Nextera XT Index Kit; Illumina), 10 μL of nuclease-free water (Ambion; Thermo Fisher Scientific), and 5 μL of amplicon DNA template. Thermal cycler conditions included an initial denaturation at 95 °C for 3 min, followed by eight cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 30 s, and a final extension at 72 °C for 5 min. PCR products were purified with AMPure XP beads (Beckman Coulter), eluted in 10 mM Tris⋅HCl pH 8, and quantified with a Qubit BR dsDNA Kit (Thermo Fisher Scientific). Final products were pooled, denatured, and sequenced on an Illumina MiSeq in accordance with the manufacturer’s recommendations using v3 chemistry and 2 × 300 paired-end reads.
Data Analysis.
Demultiplexed paired-end reads were preprocessed with the bbduk tool from BBTools v37.33 (22). Reads were filtered to remove phiX contaminants, reads with ambiguous base calls, and reads with an average quality score below Q30 in the first 100 bp (parameters k = 23 hdist = 1 maxns = 0 maq = 30 maqb = 100). Reads were then adapter-trimmed, and reads shorter than 50 bp were removed (parameters ktrim = r k = 23 hdist = 1 ftr2 = 1 mL = 51 tpe = t). Remaining paired reads were merged using PEAR v0.9.10 (23) (parameters -p 0.001 -v 35 -m 0 -n 50). Amplicon primers and barcodes were removed from merged reads with bbduk (parameters k = 23 hdist = 1). Merged reads were collapsed for exact duplicates and aligned to the LYS2 reference sequence (NC_001134.8) using Geneious version 9.1.8 (www.geneious.com) (24).
Results
NHEJ of Cas9 DSBs Reveals Frequent +1-bp Templated Insertions.
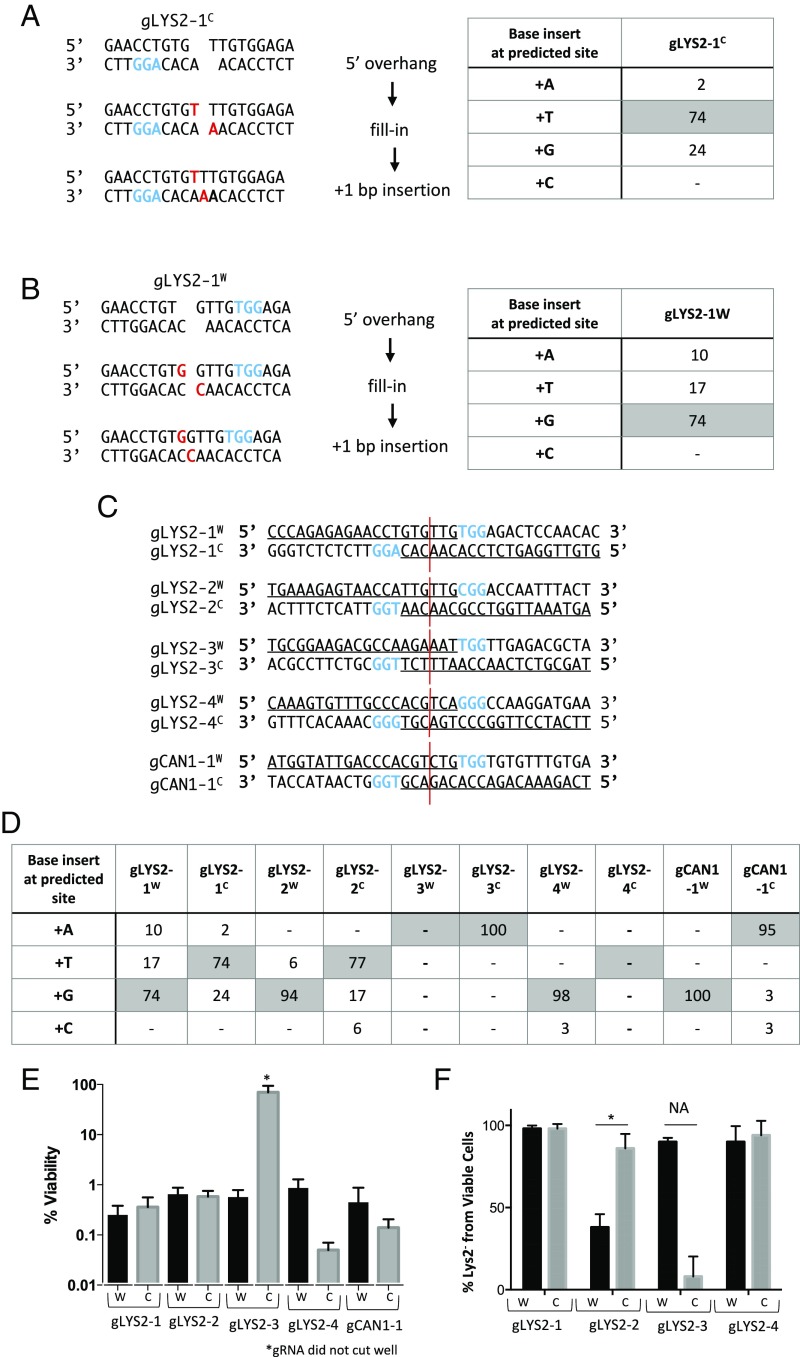
In several reports of Cas9-generated indels in mammalian cells and in vitro studies, there are frequent instances of +1 insertions (25–29). Close inspection of these sequences led us to hypothesize that the insertions were nonrandom and in fact could be explained if Cas9 sometimes leaves a 1-nt 5′ overhang by cleaving 3 nt from the PAM on the gRNA complementary strand and 4 nt away from the PAM on the noncomplementary strand that would then be filled in and ligated (Fig. 1 A and B). We found that this was also the case in budding yeast. One example is shown in Fig. 1A, using a gRNA (gLYS2-1C) that cleaves within the LYS2 locus. In these experiments, a LEU2-marked plasmid carrying both a gRNA expressed constitutively and a galactose-inducible Cas9 was transformed into strain BY4733 and then cells plated on synthetic medium lacking leucine and with galactose as the carbon source (30). Viability was ∼0.4%, with >95% of the viable colonies Lys2− (Fig. 1E). Survivors were analyzed by sequencing individual survivors and by Illumina sequencing of pools of >1,200 survivors from each of three separate experiments (SI Appendix, Tables S2 and S3). Approximately 21% of the survivors proved to be +1 insertions of T, as would be predicted had Cas9 cleavage left a 1-nt 5′ overhanging base that was subsequently filled in and ligated. There were other +1 insertions of G or A at the same site, but the apparently templated insertion represented >73% of all such events (Fig. 1A).
Fig. 1.
Templated insertions and nonrandom deletions at CRISPR-Cas9 DSBs. (A) Templated 1-nt insertions following Cas9-induced DSB, resulting in a 5′ 1-nt overhang-mediated gLYS2-1C. The table shows the percentage of +1 insertions, with shaded values representing expected templated insertions. (B) Templated 1-nt insertions following Cas9 induced DSB resulting in a 5′ 1-nt overhang mediated gLYS2-1W. The table shows the percentage of +1 insertions, with shaded values representing expected templated insertion. (C) Five pairs of gRNAs targeting either the LYS2 or CAN1 locus were designed to cleave five different genomic sites. Paired gRNAs were designed to create a DSB in the same DNA location. (D) Percentage of +1 insertions following cleavage from 10 different gRNAs targeting different iPAMs. Shaded values represent the predicted templated insertions for each guide. (E) Percent viability from gRNAs targeting the LYS2 or CAN1 locus. W and C reflect PAM location on Watson and Crick strands as shown. (F) Percentage of Lys2− for each gRNA targeting LYS2 from viable cells. A statistically significant difference in the percentage of Lys2− cells was found for gRNA pair 2 (P = 0.002). NA denotes statistics not applicable because gRNA did not cut well.
The observation that +1 insertions were apparently frequently templated with gRNA LYS2-1C led us to create an ostensibly identical DSB using a PAM on the opposite (Watson) strand, directed by gLYS2-1W. We term this pair of PAMs iPAMs (Fig. 1B). For gLYS2-1W, the expected insertion would be a G adjacent to the fourth base from the PAM. Indeed, nearly 74% of the +1 insertions were G, with a minority of T or A inserts (Fig. 1B and SI Appendix, Tables S2, S3, and S4).
This same disproportionate +1 insertion of a predicted, templated base was also seen for four other iPAM gRNA pairs, three in LYS2 and one in CAN1 (Fig. 1D). Among the eight cases with +1 insertions at the predicted site, the predicted base accounted for ≥73% of all such insertions, far more frequent than the 25% expected from random insertions (SI Appendix, Table S4). There were no +1 insertions in one case (gLYS2-3W) and none at the predicted site for gLYS2-4W (Fig. 1D). For gLYS2-2W and gCAN1-1W, nearly all of the +1 insertions at the predicted site were the expected base, but there were other insertions after the third base from the PAM (SI Appendix, Tables S2 and S3). The rate of survival for LYS2-4C was much lower than for other gRNAs, and nearly 48% of survivors were not indels, but base pair substitutions (Fig. 1E and SI Appendix, Tables S2 and S3).
We also noted that in each of the iPAMs, there were also a small proportion of +2 and +3 insertions (SI Appendix, Table S4). These were not random insertions but again were mostly adjacent to the fourth base from the PAM. In a few cases, these insertions can be explained by assuming that they also were templated after a cleavage, leaving 2-nt 5′ overhangs. One example of this is the insertion of CG adjacent to the sequence CGTACGGG in 5.8% of the events for LYS2-4W (SI Appendix, Fig. S1). However, the great majority of these insertions appear to be homonucleotide insertions favoring the base that was templated in a +1 insertion (SI Appendix, Table S4B). As discussed below, there were also asymmetries in the deletion outcomes between the two iPAM gRNAs suggesting that the orientation of Cas9 on its target exerts a strong effect on the outcomes.
+1 and Other Insertions Depend Mostly on Pol4.
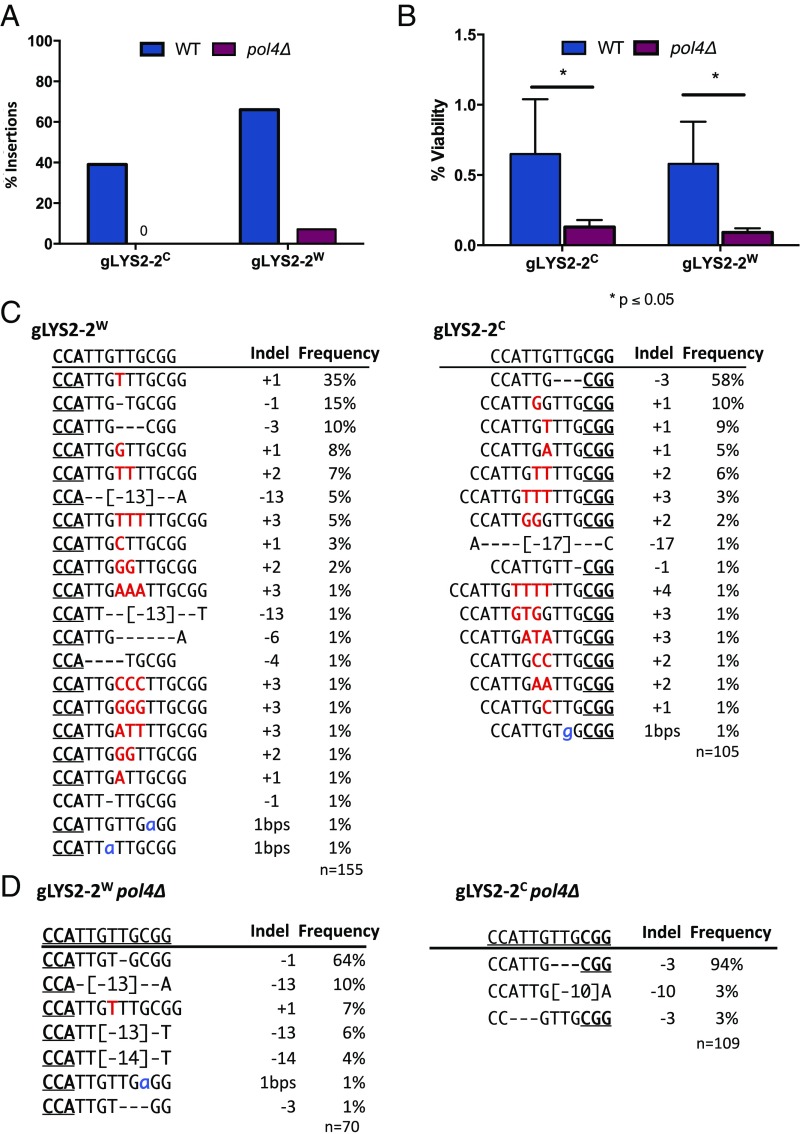
If there are 1-nt 5′ overhanging ends, then there should be a DNA polymerase to fill in (5′ to 3′ on the recessed strand) before ligation can occur. We had previously implicated the PolX family DNA polymerase Pol4 in filling in misaligned DSB 3′ overhanging ends created by HO endonuclease (12). Here we show that most of the +1 templated events do indeed require Pol4 (Fig. 2). For both gLYS2-2W and gLYS2-2C, the deletion of POL4 eliminated nearly all of the insertions, including the insertions of multiple bases (Fig. 2). For gLYS2-2W, in the absence of Pol4, the frequency of 3-bp microhomology-bounded TTG deletions increased from 58% to 94% (Fig. 2D). For gLYS2-2C, the number of −1 deletions increased from 15% to 64%. These differences again demonstrate the biased outcomes produced by two methods of cleaving the same site. With few to no insertions in the absence of Pol4, viability for both gRNAs decreased significantly (Fig. 2B). Pol4 likely is also involved in filling-in microhomology-based deletions formed by annealing resected 3′-ended single strands. Additional evidence of the role of Pol4 in generating +1 and +2 insertions was found for insertions created at several sites within the MATa or MATα loci (SI Appendix, Fig. S2), where 78% of indels were +1, +2, or +3 insertions in wild-type strains and only 2.5% contained insertions in the absence of Pol4.
Fig. 2.
Pol4 is required for templated and nontemplated insertions. (A) Insertions during repair of gLYS2-2W and gLYS2-2C DSBs are mostly lost in the absence of POL4. (B) Effect on NHEJ survival in the absence of POL4. (C) Indel profile from colonies recovered following DSB from gLYS2-2W and gLYS2-2C. (D) Indel profile in the absence of pol4Δ for gLYS2-2W and gLYS2-2C.
iPAMs Reveal Other Nonrandom Consequences of the Orientation of Cas9 Relative to the Cleavage Site.
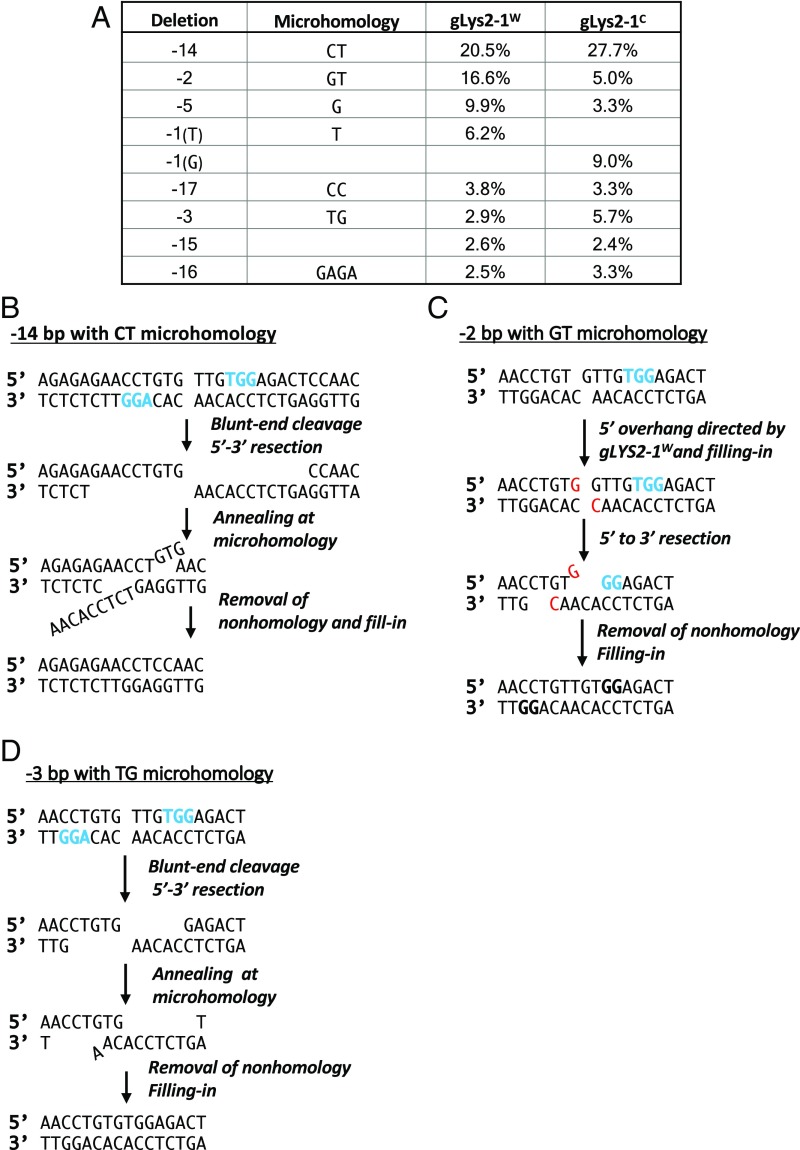
In addition to the +1 insertions described above, most of the indels that were recovered appear to result from the pairing of microhomologies exposed by 5′ to 3′ resection of the DSB. In the cases of gLYS2-1C and -1W, the most abundant event for both gRNAs is a 14-bp deletion, which can be explained by the pairing of CT sequences following 5′ to 3′ resection of the DSB ends (Fig. 3B). Similar processing of blunt-end cleavages can account for a 3-bp deletion bounded by a TG microhomology (Fig. 3D); however, their relative abundance is quite different for the two gRNAs. A 2-bp deletion bounded by GT (Fig. 3C) accounts for 16.2% of the events with gLYS2-1W, but only 5% of the events in gLYS2-1C. The higher prevalence of this event for gLYS2-1W might be explained by a 1-nt 5′ overhang following cleavage (Fig. 3C), whereas a 1-nt overhang directed by the opposite iPAM (gLYS2-1C) does not give this result. Similarly, a 5-bp deletion, bounded by a single G, represents 9.9% in gLYS2-1W but only 3.3% in gLYS2-1C. In this case, the processing of a blunt end or either 1-nt 5′ overhang can be envisioned to yield the 5-bp deletion; however, the presumed intermediate created by gLYS2-1C would require excision of a nonhomologous nucleotide on both sides of the annealed structure (SI Appendix, Fig. S3A). Some of the other biases also may be attributable to the different outcomes created by 1-nt overhangs directed by one PAM or the other, or to the fact that subsequent steps are influenced by the orientation of Cas9. For example, whereas a blunt-end cleavage of the gLYS2-2C target will yield a 2-bp GT deletion, the creation of −1 T deletions likely derives from a 1-nt 5′ overhanging cut (SI Appendix, Fig. S3B).
Fig. 3.
Formation of indels by annealing of adjacent microhomologies. (A) Frequency of prominent deletions from cleavages directed by gLYS2-1W and gLYS2-1C PAM sequences (blue). (B) Origin of the common 14-bp deletion flanked by a CT microhomology. (C) Formation of a 2-bp deletion with GT microhomology. (D) Formation of a 3-bp deletion bounded by TG microhomology.
Similar differences are seen with three other iPAMs in LYS2 and one iPAM in CAN1. We note that gLYS2-3C was inefficient in cleavage (Fig. 1E), but we could recover sufficient Lys2− examples with an altered sequence to compare with gLYS2-3W (SI Appendix, Table S3). One other especially notable difference was found with gLYS2-2W and 2C, where—although they both generate efficient cleavage and survivors at a rate of 1%—>60% of the survivors for gLYS2-2W are Lys+, while only 20% are Lys+ for LYS2-2C (Fig. 1F). The Lys2+ survivors all proved to have in-frame deletions (3 or 6 bp) or insertions (3 bp), or else to have base pair substitutions (SI Appendix, Tables S2 and S3), but a great majority of the Lys+ survivors at this site are 3-bp deletions that would arise after 5′ to 3′ resection of the ends and annealing of the single-stranded, 3′-ended AAC and TTG sequences at the two DSB ends (SI Appendix, Fig. S4). This event is much more strongly favored in one orientation than the other (Discussion). In this case, the difference cannot be explained by differences that might have been envisioned from different 1-nt 5′ overhangs directed by the two PAMs.
A significant proportion of the survivors had no indels, but instead had base pair substitutions that apparently rendered the gRNA ineffective. This is especially evident for gLYS2-4C, for which the survival rate was <10% of the other gRNAs used and >47% of the survivors had base pair substitutions. The low rate of viability cannot be attributed to an off-target cleavage by this gRNA, because viability proved to be nearly 100% when a 12-bp deletion of the protospacer—obtained from a survivor using gLYS2-4W—was transformed with Cas9 and gLYS2-4C. The high rate of base pair substitution is reminiscent of survivors of HO endonuclease cleavage when normal NHEJ is impaired, and the rate of end repair was only ∼5% of the wild-type rate (12). In addition to Sanger sequencing we also used Illumina to sequence a larger pool of NHEJ survivors for gLYS2-4C and gLYS2-4W. None of the +1 insertions in gLYS2-4C survivors were at the predicted site (Fig. 1D and SI Appendix, Tables S2 and S3). This discrepancy and the variety of base pair substitutions most likely can be attributed to the very low viability seen for gLYS2-4C and the likely NHEJ-independent origin of many of the mutations recovered. Why Cas9 cleavage with gLYS2-4C appears to block NHEJ whereas gLYS2-4W does not, will require further investigation.
There was one instance (gLYS2-1W) of a GTGGGGTGTGGG sequence that is reminiscent of telomere sequences (SI Appendix, Table S3). Such captures of telomere sequences at DSB sites have been previously noted (31).
In SI Appendix, Table S3 we also include results from a single gRNA in CAN1, showing again a strong bias toward templated +1 insertions (29%) as well as homonucleotide insertions (+AA, +AAA and even +AAAAA, as well as +TT and +TTTTT).
Cas9-Mediated Indels Are Strongly Dependent on the MRX Complex.
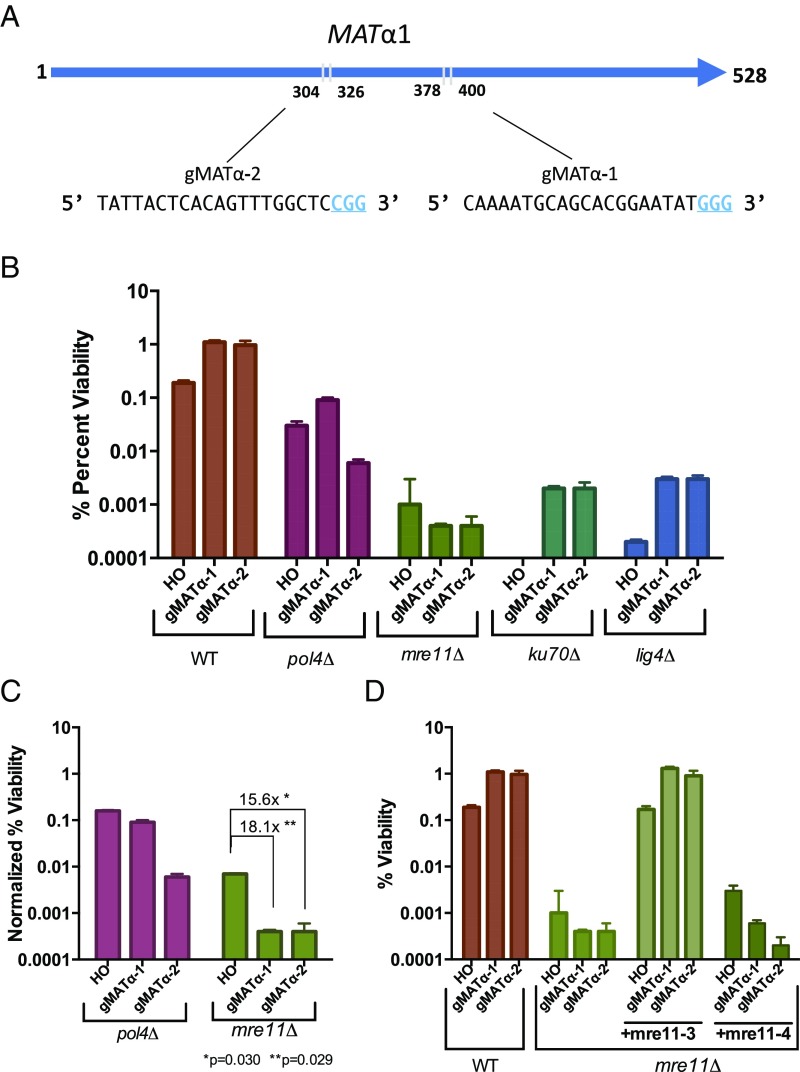
As previously reported (12), indels within the HO cleavage site in MATα, even an in-frame 3-bp deletion or insertion, results in a matα1 mutation, causing sterility. We designed two gRNAs (α#1 and α#2) to cut in the same MATα1 gene, 23 and 96-bp away from the HO cleavage site (Fig. 4A). Cells were transformed with two plasmids, one containing a galactose-inducible Cas9 gene and the other expressing the appropriate gRNA. As expected, with continued Cas9 cleavage in the MATα1 gene, nearly all of the survivors were sterile (α#1: 39/40; α#2: 38/40); the few nonsterile colonies proved to have not been mutated. The viability in GAL::CAS9 strains was ∼5 times higher than with GAL::HO (Fig. 4B); this difference is likely a reflection of the slower kinetics of cleavage by Cas9 and the possibility that cells replicate before cleavage has occurred. As with HO DSBs, Cas9-mediated NHEJ strongly requires DNA ligase 4 and the Ku proteins, as dnl4Δ and yku70Δ derivatives displayed an ∼100-fold decrease in viability (Fig. 4B).
Fig. 4.
MRE11 is critical for Cas9-induced NHEJ repair. (A) Two gRNAs were used to target the MATα locus in the Yα sequence. (B) Viability for HO and CRISPR-Cas9 with gMATα-1 or gMATα-2 gRNAs in donorless strains that cannot repair the DSB by homologous recombination. (C) Viability relative to wild-type for HO and CRISPR-Cas9 MAT gMATα-1 or gMATα-2 gRNA. Error bars represent SEM. (D) NHEJ repair requires the MRX complex, independent of Mre11’s nuclease activity. Error bars represent SEM.
The Mre11-Rad50-Xrs2 (MRX) complex is an evolutionarily conserved complex that has been implicated in many steps of DSB repair, including control of end-resection and bridging of broken DNA ends (32–34). In budding yeast, MRE11 is important for most, but not all NHEJ events; in its absence, templated insertions were lost but some deletions, bounded by microhomology, were still recovered (12). With HO-induced DSBs, MRX proved to be important in the templated +2 and +3 insertions but was not required for a 3-bp deletion that also involved misaligned base-pairing of the overhanging ends (12). Here we examined the role of Mre11 in Cas9-induced NHEJ. Deletion of MRE11 resulted in a >1,000-fold reduction in survivors; moreover, among the mre11 Δ survivors, only 8 of 367 had actually altered the cleavage site sequence, and the rest were presumed to be rare cells that failed to induce cleavage of the target site, as observed previously in rare survivors of HO cleavage (12). In those eight cases, the altered target sequence harbored either a spontaneous mutation in the PAM or PAM-adjacent sequences (two instances) or an indel, all of which were 1-bp deletions (SI Appendix, Fig. S5). Thus, NHEJ of Cas9-cleaved DSBs was significantly more dependent on MRX than on the HO breaks (P < 0.05) (Fig. 4C), which suggests that Mre11 is required not only for fill-in events, but for nearly all deletions. This difference most likely reflects the fact that HO ends provide a 4-nt 3′ overhang to facilitate microhomology-mediated annealing, whereas Cas9 cleavages are mostly blunt-ended and thus may require the MRX complex either to initiate resection of the ends or to tether the ends together.
To test whether Mre11 nuclease activity or DNA end-bridging by the MRX complex is critical for Cas9 NHEJ repair, we investigated the repair efficiency in mre11Δ strains cotransformed with centromere-containing plasmids expressing either the nuclease-dead mre11-3 allele or the mre11-4 allele that fails to form a MRX complex (32), transcribed from their native promoter. Mre11-3 rescued the strain viability, whereas the mre11-4 allele did not (Fig. 4D). Importantly, the mre11-3 allele rescued mre11Δ NHEJ viability for HO induction as well (Fig. 4D). These results suggest that Mre11’s nuclease activity is not required for its role in NHEJ of Cas9-generated ends, but the integrity of the MRX complex is required to allow efficient end-joining.
Discussion
Although CRISPR-Cas9 has made it possible to create mutations and gene editing in many organisms, budding yeast still provides an important platform to analyze the mechanisms of Cas9-mediated events in great detail. Here we have studied NHEJ repair profiles in response to Cas9-mediated DBSs and have reached several important conclusions.
First, we show that Cas9 induces a significant fraction of +1 indels that are apparently templated from a 1-bp 5′ overhang at the Cas9 cleavage site. Overall, among the five iPAMs and the additional sequences we have analyzed, >75% of the +1 insertions were the specific base and the specific location predicted from the model shown in Fig. 1. We note again that the finding of +1 insertions is not restricted to budding yeast, but is readily seen in the spectra of indels in mammalian cells (35–37). To extend our analysis, we specifically analyzed the nonrandom end-joining events described in an extensive study of different gRNAs by van Overbeek et al. (25) (Dataset S1). We found that, as in yeast, mammalian cells exhibit a very strong bias for the +1 insertion to be templated by the fourth base from the PAM. Indeed, among 151 examples of Cas9-induced indels described by van Overbeek et al. (25), in which there were at least 1,000 +1 events, on average 91% of the +1 insertions at the expected insertion site were the base predicted from the sequence (range, 36%–100%). In only 2 of 151 cases was the most frequent insertion not the predicted templated event. These data suggest that the great majority of the +1 inserts are templated, but the DNA polymerase(s) involved in creating this +1 insertion may be significantly error-prone (see below).
The proportion of Cas9 cleavages that are not blunt-ended is not known. In vitro, the great majority of ends are certainly blunt (38); however, in vivo, a 1-nt 5′ extension may facilitate recovery after NHEJ events, possibly through more efficient recruitment of an end-joining factor (39). In addition, an all-atom dynamics simulation predicted that Cas9 DNA cleavage could have 1-nt 5′ overhangs (40). In budding yeast, blunt end ligations, as measured by plasmid ligation after transformation, are markedly less efficient than those with complementary overhanging ends (41).
Second, the filling-in of 5′ overhangs to produce +1 insertions generally requires the X family DNA polymerase, Pol4. In budding yeast, Pol4 has been shown to be required for gap-filling between misaligned 5′ overhanging ends, although—in contrast to gap-filling of misaligned 3′ overhangs, which totally depend on Pol4—other polymerases appear to be able to substitute for Pol4 at reduced efficiency (39). Thus, the residual insertions that we find without Pol4 may depend on other terminal transferase polymerases with the ability to add one or more bases. Pol4 lacks proofreading activity and thus is error-prone (42). This may explain the appearance of other bases instead of the templated base in a significant minority of junctions. The frequency of recovering alternative bases is quite different in different gRNA contexts.
In addition to +1 insertions, Pol4 appears to be required when there are multiple base insertions at the break. The origin of these two-, three-, and four-base insertions in budding yeast might be explained by the fact that Pol4 belongs to the X family of DNA polymerases, some of which have terminal transferase activity (43–45). Surprisingly, these multiple-base insertions are overwhelmingly nonrandom, with >85% of them being homonucleotide runs, the majority of which match the base most frequently inserted as the templated +1 insertions (SI Appendix, Table S4B). For example, for gLYS2-2C, the templated base is T, and the most frequent 2- and 3-bp insertions are also TT and TTT. Less often, there were +1 G insertions, and there were also GG and GGG inserts. These may reflect some sort of slippage at the 1-nt overhang or some sort of terminal transferase activity. Among the multiple-base homonucleotide insertions, 26% were runs of A, 54% incorporated T, 12% were Gs, and 8% were Cs. Some bias likely reflects the iPAM sequences chosen. Among the five iPAMs that we analyzed, there were no predicted +1C insertions. It is also possible that Pol4 could copy the gRNA.
Our analysis of indel data from an extensive study in human cells (25–27) also revealed the presence of +2 insertions at the same site at which +1 insertions are created. Overall, +2 insertions appeared at a frequency of ∼8% of the number of +1 insertions at the same site. We examined 82 instances in which there were 200 or more +2 insertions (Dataset S2). Among this set, when there were +1 insertions, the most frequent insertion was the expected templated event in all but six cases. More than 90% of all +1 insertions appear to be templated. Overall, 85% of the time, the +2 inserts began with the same base as seen in +1 cases (ranging from 30% to 100%). We do not see evidence of homonucleotide +2 insertions, as seems to be the case in budding yeast; however, in 57 of these 82 cases, the most frequent +2 insertion can be accounted for if it too was templated, ostensibly by a +2 5′ overhang. Indeed, 76% of all insertions were the predicted +2 insertion in these 57 cases. Among the 22 cases in which the most frequent dinucleotide insertion was not that expected from a 2-nt 5′ overhang, the first base inserted was the expected base in all but eight cases. Determining whether the additions of one or two bases will require PolX will be of great interest.
Third, the use of palindromic PAM sequences allowed us to clearly demonstrate that the orientation of Cas9 on DNA strongly influences the spectrum of indels. Most striking is the case of gLYS2-2W and gLYS2-2C directed against LYS2. With a PAM on the nontranscribed strand, >50% of the outcomes had a 3-bp deletion that would result from annealing TTG sequences on either side of the (blunt and resected) DSB, and another 10% were +1 templated insertions of a G (Fig. 2C). In contrast with a PAM on the transcribed strand, only 10% had the 3-bp deletion and 30% were the expected templated +1 insertion (+T) (Fig. 2C). These differences in outcome could reflect in vitro studies showing that Cas9 remains tightly bound to the site of cleavage, but the two sides of the DSB may be differently accessible (15). A large difference in the microhomology-mediated annealing of the flanking TTG sequences would also be expected if one of the two orientations led to a much larger proportion of 5′ overhanging cuts. Whereas blunt end cleavage would lead to both ends having TTG on the top strand, allowing perfect annealing of the termini after resection, cleavages producing 1-nt 5′ overhanging (resulting in TT and GTTG on the top strand) would not have precisely complementary microhomologies at the two ends. Whether there is an influence of the orientation of Cas9 relative to transcription also merits systematic investigation; however, from the five iPAMs that we investigated, there does not seem to be a clear pattern indicating that one orientation or the other is more efficient in cleavage per se or in generating indels after cleavage (Fig. 1C). It is possible that differences in the spectrum of indels could reflect differences in the affinity of the two iPAM gRNAs, based on, for example, GC content or some other sequence-specific influence on Cas9’s residency at the DSB ends.
In the case of gLYS2-2C, the prominent −T deletion (15% of survivors) can be readily explained if the cleavage had a 5′ overhang; however, an analogous process beginning with a blunt end would instead produce a −G, which was not seen (SI Appendix, Fig. S3B).
Finally, NHEJ events after Cas9-mediated cleavage are nearly completely dependent on the MRX complex. Two separation-of-function mutants have led us to the conclusion that NHEJ does not depend on the nuclease activity of MRX, but does depend on the integrity of the complex. We suggest that the end-tethering promoted by the MRX complex becomes essential when the DSB ends are blunt. We note that NHEJ of Cas9 cleavages are not more sensitive to the absence of Ku or DNA ligase 4.
Consistent with other studies of Cas9-mediated indels, most of the deletions that we recovered are mediated by microhomology present near the break site (46–49). Keeping this in mind may be helpful when designing gRNAs for NHEJ-induced loss of function. For example, almost 60% of cells expressing gLYS2-2C targeting LYS2 retained protein function and still grew on Lys2 dropout plates, due to a 3-bp deletion resulting from 3-bp microhomology, maintaining the protein in-frame (Fig. 2C).
In conclusion, we find that the types of indels recovered by a particular gRNA-directed cleavage is strongly dependent on DNA sequence context around the cleavage site. The basis of these biases remains an important subject to investigate.
Supplementary Material
Acknowledgments
We thank Maria Jasin for suggesting the term “iPAM.” We thank James DiCarlo and George Church for providing the yeast Cas9 expression plasmid. This research was supported by National Institutes of Health (NIH) Grants GM20056, GM76020, and GM105473 (to J.E.H.). B.R.L., A.C.K., and D.P.W. were supported by NIH Genetics Training Grant T32 GM007122. J.E.B. was supported by the Brandeis University Provost’s Undergraduate Research Fund.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716855115/-/DCSupplemental.
References
- 1.Shen B, et al. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013;23:720–723. doi: 10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiCarlo JE, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehta A, Haber JE. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect Biol. 2014;6:a016428. doi: 10.1101/cshperspect.a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haber JE. A life investigating pathways that repair broken chromosomes. Annu Rev Genet. 2016;50:1–28. doi: 10.1146/annurev-genet-120215-035043. [DOI] [PubMed] [Google Scholar]
- 7.Doudna JA, Charpentier E. Genome editing: The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 8.Haber JE. Uses and abuses of HO endonuclease. Methods Enzymol. 2002;350:141–164. doi: 10.1016/s0076-6879(02)50961-7. [DOI] [PubMed] [Google Scholar]
- 9.Nickoloff JA, Chen EY, Heffron F. A 24-base-pair DNA sequence from the MAT locus stimulates intergenic recombination in yeast. Proc Natl Acad Sci USA. 1986;83:7831–7835. doi: 10.1073/pnas.83.20.7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hicks WM, Yamaguchi M, Haber JE. Real-time analysis of double-strand DNA break repair by homologous recombination. Proc Natl Acad Sci USA. 2011;108:3108–3115. doi: 10.1073/pnas.1019660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valencia M, et al. NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature. 2001;414:666–669. doi: 10.1038/414666a. [DOI] [PubMed] [Google Scholar]
- 12.Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma JL, Kim EM, Haber JE, Lee SE. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol Cell Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol. 2016;34:339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 16.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SE, et al. Saccharomyces Ku70, mre11/rad50, and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 18.Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 19.Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997;25:451–452. doi: 10.1093/nar/25.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anand R, Memisoglu G, Haber J. Cas9-mediated gene editing in Saccharomyces cerevisiae. Protoc Exch. April 13, 2017 doi: 10.1038/protex.2017.021a. [DOI] [Google Scholar]
- 21.Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- 22.Bushnell B. 2014 BBMap short read aligner, and other bioinformatic tools, version 37.33. Available at https://sourceforge.net/projects/bbmap/. Accessed June 25, 2017.
- 23.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: A fast and accurate Illumina paired-end reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kearse M, et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Overbeek M, et al. DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Mol Cell. 2016;63:633–646. doi: 10.1016/j.molcel.2016.06.037. [DOI] [PubMed] [Google Scholar]
- 26.Seeger C, Sohn JA. Complete spectrum of CRISPR/Cas9-induced mutations on HBV cccDNA. Mol Ther. 2016;24:1258–1266. doi: 10.1038/mt.2016.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang Q, et al. Small indels induced by CRISPR/Cas9 in the 5′ region of microRNA lead to its depletion and Drosha processing retardance. RNA Biol. 2014;11:1243–1249. doi: 10.1080/15476286.2014.996067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geisinger JM, Turan S, Hernandez S, Spector LP, Calos MP. In vivo blunt-end cloning through CRISPR/Cas9-facilitated non-homologous end-joining. Nucleic Acids Res. 2016;44:e76. doi: 10.1093/nar/gkv1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D, et al. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol. 2016;34:863–868. doi: 10.1038/nbt.3609. [DOI] [PubMed] [Google Scholar]
- 30.Anand R, Beach A, Li K, Haber J. Rad51-mediated double-strand break repair and mismatch correction of divergent substrates. Nature. 2017;544:377–380. doi: 10.1038/nature22046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marzec P, et al. Nuclear receptor-mediated telomere insertion leads to genome instability in ALT cancers. Cell. 2015;160:913–927. doi: 10.1016/j.cell.2015.01.044. [DOI] [PubMed] [Google Scholar]
- 32.Bressan DA, Baxter BK, Petrini JH. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7681–7687. doi: 10.1128/mcb.19.11.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paull TT, Gellert M. A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc Natl Acad Sci USA. 2000;97:6409–6414. doi: 10.1073/pnas.110144297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van den Bosch M, Bree RT, Lowndes NF. The MRN complex: Coordinating and mediating the response to broken chromosomes. EMBO Rep. 2003;4:844–849. doi: 10.1038/sj.embor.embor925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Canver MC, et al. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J Biol Chem. 2017;292:2556. doi: 10.1074/jbc.A114.564625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Veres A, et al. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell. 2014;15:27–30, and correction (2014) 15:254. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang Z, Sunder S, Nallasivam S, Wilson TE. Overhang polarity of chromosomal double-strand breaks impacts kinetics and fidelity of yeast non-homologous end joining. Nucleic Acids Res. 2016;44:2769–2781. doi: 10.1093/nar/gkw013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuo Z, Liu J. Cas9-catalyzed DNA cleavage generates staggered ends: Evidence from molecular dynamics simulations. Sci Rep. 2016;5:37584. doi: 10.1038/srep37584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end-joining in yeast. Annu Rev Genet. 2005;39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- 42.Bebenek K, Garcia-Diaz M, Patishall SR, Kunkel TA. Biochemical properties of Saccharomyces cerevisiae DNA polymerase IV. J Biol Chem. 2005;280:20051–20058. doi: 10.1074/jbc.M501981200. [DOI] [PubMed] [Google Scholar]
- 43.Nick McElhinny SA, et al. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end-joining. Mol Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 44.Li P, et al. Multiple end-joining mechanisms repair a chromosomal DNA break in fission yeast. DNA Repair (Amst) 2012;11:120–130. doi: 10.1016/j.dnarep.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 45.Loc’h J, Rosario S, Delarue M. Structural basis for a new templated activity by terminal deoxynucleotidyl transferase: Implications for V(D)J recombination. Structure. 2016;24:1452–1463. doi: 10.1016/j.str.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 46.Bae S, Kweon J, Kim HS, Kim JS. Microhomology-based choice of Cas9 nuclease target sites. Nat Methods. 2014;11:705–706. doi: 10.1038/nmeth.3015. [DOI] [PubMed] [Google Scholar]
- 47.Li J, et al. Efficient inversions and duplications of mammalian regulatory DNA elements and gene clusters by CRISPR/Cas9. J Mol Cell Biol. 2015;7:284–298. doi: 10.1093/jmcb/mjv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakade S, et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat Commun. 2014;5:5560. doi: 10.1038/ncomms6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang WW, Matlashewski G. CRISPR-Cas9-mediated genome editing in Leishmania donovani. MBio. 2015;6:e00861. doi: 10.1128/mBio.00861-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.