Significance
In mammalian cells, a protein called STING (stimulator of IFN genes) is typically viewed as a factor dedicated to defense strategies induced by DNA viruses. The antiviral activities of STING are linked to its ability to induce the expression of genes that combat viral replication cycles. In our study, we have discovered a transcription-independent function for STING in the restriction of viruses that contain RNA genomes. We found that cells lacking STING are sensitive to RNA virus infections and that during these infections, STING inhibits the translation machinery to prevent viral protein synthesis. This study therefore establishes that STING has dual functions in host defense, regulating antiviral gene expression or interfering with translation, to restrict replication of distinct classes of viruses.
Keywords: STING, RIG-I, cGAS, interferon, translation
Abstract
In mammalian cells, IFN responses that occur during RNA and DNA virus infections are activated by distinct signaling pathways. The RIG-I–like-receptors (RLRs) bind viral RNA and engage the adaptor MAVS (mitochondrial antiviral signaling) to promote IFN expression, whereas cGAS (cGMP–AMP synthase) binds viral DNA and activates an analogous pathway via the protein STING (stimulator of IFN genes). In this study, we confirm that STING is not necessary to induce IFN expression during RNA virus infection but also find that STING is required to restrict the replication of diverse RNA viruses. The antiviral activities of STING were not linked to its ability to regulate basal expression of IFN-stimulated genes, activate transcription, or autophagy. Using vesicular stomatitis virus as a model, we identified a requirement of STING to inhibit translation during infection and upon transfection of synthetic RLR ligands. This inhibition occurs at the level of translation initiation and restricts the production of viral and host proteins. The inability to restrict translation rendered STING-deficient cells 100 times more likely to support productive viral infections than wild-type counterparts. Genetic analysis linked RNA sensing by RLRs to STING-dependent translation inhibition, independent of MAVS. Thus, STING has dual functions in host defense, regulating protein synthesis to prevent RNA virus infection and regulating IFN expression to restrict DNA viruses.
In all well-studied organisms, viral infections are detected through the actions of host-encoded receptors that recognize cytosolic nucleic acids (1, 2). Mammalian cells utilize distinct nucleic acid binding proteins to detect viruses containing RNA or DNA genomes. These proteins are members of the larger family of pattern recognition receptors (PRRs), which are responsible for unleashing the antiviral activities of the innate immune system (3). Central to our understanding of antiviral immunity are members of the IFN family of secreted proteins. During infections, cytosolic PRRs detect viral DNA or RNA and induce signaling pathways that promote the expression and secretion of IFNs. Upon release into the extracellular space, IFNs signal via dedicated receptors present on infected and uninfected cells to induce the expression of hundreds of IFN-stimulated genes (ISGs). Many ISGs antagonize viral replication cycles and thereby induce an antiviral cellular state (4).
The major IFN-inducing PRRs that detect viral RNAs include members of the retinoic acid-inducible gene I (RIG-I)–like receptor (RLR) and Toll-like Receptor (TLR) families of proteins. Of these families, the TLRs are restricted in expression and are best known to function on professional phagocytes (3). In contrast, the RLRs are expressed ubiquitously and are therefore considered to be universal sentinels of viral infection (3). When one of the RLR family members detects viral RNA, these receptors engage the protein mitochondrial antiviral signaling (MAVS), which is located on mitochondria, peroxisomes and mitochondria-associated endoplasmic reticulum (ER) membranes (5). RLR-bound MAVS coordinates the activation of inflammatory transcription factors that drive IFN expression and the establishment of an antiviral cellular state (6). Because RLRs bind viral RNA directly, it is generally believed that these receptors are dedicated to the detection of viruses with RNA genomes. Experimental evidence supporting this conclusion is extensive, as the only RNA viruses that do not induce an RLR–MAVS-dependent IFN response are those that encode virulence determinants that antagonize RLR signaling (7). However, evidence exists that self- or viral RNAs present in cells infected with DNA viruses can also be detected by these receptors (8).
While some DNA viruses may be detected by RLRs, the dominant means of DNA virus detection is not by these receptors. Rather, most DNA viruses are detected by the PRR cGAS (cGMP–AMP synthase). cGAS is a DNA binding protein that, like RLRs, is present in the cytosol of most mammalian cells (9). Upon DNA binding, cGAS stimulates the generation of a cyclic dinucleotide cyclic-GMP–AMP (cGAMP), which engages an endoplasmic reticulum (ER)-localized protein stimulator of IFN genes (STING) (10). In a manner analogous to MAVS, cGAMP-bound STING coordinates the activation of inflammatory transcription factors to induce IFN expression and the establishment of an antiviral cellular state (11–13). The ability of STING to induce IFN is coincident with this protein becoming phosphorylated and exiting the ER (11, 14, 15). Thus, IFN responses induced during RNA and DNA virus infections are generally considered to be managed by the RLR–MAVS or cGAS–STING pathways, respectively.
While the distinction between the RNA and DNA sensing pathways appears true when considering IFN expression, these distinctions are less clear when actual antiviral activities are examined. For example, compared with wild-type (WT) cells, fibroblasts lacking STING support increased replication of vesicular stomatitis virus (VSV) and dengue virus (11, 12, 16, 17). In addition, multiple flaviviruses, coronaviruses, and influenza virus employ strategies to antagonize STING signaling (11, 16–21). In several of the studies cited above, STING was reported to influence IFN expression during RNA virus infection. In contrast to these observations, several reports indicate that STING does not regulate RNA-induced IFN expression (11, 13). Thus, how STING influences host defense during RNA virus infection is unclear.
The activities of IFN are not the only means by which cells restrict viral replication. Indeed, during the first 24 h of infection, cells that are genetically unresponsive to IFN are no more sensitive to VSV infection than WT counterparts (22, 23). The actions of IFNs may therefore be most important to protect uninfected cells from subsequent infections, as pretreatment of cells with IFN is a well-established mechanism of restricting virus replication (24). Cell-intrinsic mechanisms of host defense, which may be most important at early stages of infection, are less understood but may involve autophagy and the regulation of protein synthesis. Notably, STING has the additional ability to promote antibacterial autophagy to restrict the replication of intracellular Mycobacterium tuberculosis, and the cytosolic PRR Protein Kinase R (PKR) suppresses the protein synthesis machinery after it binds to viral double-stranded RNA (dsRNA) (25, 26). The role of STING in other potential defense responses during viral infection is unclear.
In this study, we have explored the antiviral activities of STING during RNA virus infection. We found that STING is required to restrict the replication of diverse RNA viruses in fibroblasts but that STING is not required to induce IFN expression during these infections. Using VSV as a model, we found that the mechanism of viral entry was not regulated by STING, but STING-deficient cells were 100 times more likely to support productive viral infections than WT counterparts. The antiviral activities of STING were not linked to its ability to influence the basal expression of IFNs in uninfected cells or autophagy. Rather, we identified a requirement of STING for translation inhibition during VSV infection or upon transfection of synthetic RLR ligands. Protein synthesis inhibition by STING occurs at the level of translation initiation and restricts the production of viral and host proteins. Further analysis revealed a genetic pathway that links RLRs to STING, independent of MAVS. Thus, STING has dual functions in host defense, regulating protein synthesis to prevent RNA virus infection and regulating IFN expression to restrict DNA viruses.
Results
STING Is Required to Control the Replication of Diverse RNA Viruses.
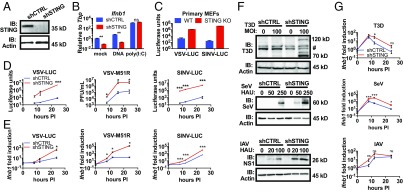
To analyze the role of STING in RNA virus replication, immortal mouse embryonic fibroblasts (MEFs) were transduced with short hairpin RNAs (shRNA) targeting STING (shSTING). MEFs expressing a nontargeting hairpin were used as control cells and were hereafter referred to as shCTRL. This approach resulted in the complete depletion of the STING protein from cells transduced with shSTING, as assessed by western analysis (Fig. 1A). To verify the function of MEFs depleted of STING, we transfected these cells with a cGAS ligand (double stranded DNA) or an RLR ligand [poly(I:C)]. As expected, shSTING cells did not induce Ifnβ1 transcripts in response to transfected DNA, whereas poly(I:C)-induced expression of Ifnβ1 was unaffected by the absence of STING (Fig. 1B). These results are consistent with the specific and essential function of STING in DNA-induced IFN responses and validate the use of these cells as a model to study STING activities.
Fig. 1.
STING loss confers a viral replication advantage, independently of IFN signaling. (A) Lysates from shCTRL and shSTING MEFs were separated by SDS/PAGE, and endogenous STING and actin were detected by western analysis. (B) MEFs were transfected with herring testes DNA (2 μg/mL) or poly(I:C) (0.5 μg/mL). Ifnb1 transcripts were analyzed by qRT-PCR. Data are represented as mean ± SEM. (C) Primary, matched MEFs were infected with VSV-LUC and SINV-LUC at MOI 0.1. Infections were monitored by luciferase activity assay at 20 h postinfection. Data are represented as mean ± SEM. (D) The MEFs indicated were infected with VSV-LUC at MOI 0.05, VSV-M51R at MOI 0.1, or SINV-LUC at MOI 0.05. VSV-LUC and SINV-LUC infections were monitored by luciferase assay at the indicated time postinfection (hours PI). VSV-M51R titer was determined by pfu assay (pfu/mL) at the indicated time postinfection. Data are represented as mean ± SEM. (E) The MEFs indicated were infected with the indicated virus at MOI 1.0. Ifnb1 transcript was quantified by qRT-PCR at the indicated times. Data are displayed as fold induction of the indicated gene compared with uninfected. Data are represented as mean ± SEM. (F) The MEFs indicated were infected with the indicated viruses at the indicated viral quantities. After 24 h of infection, lysates were separated by SDS/PAGE. Viral proteins and actin were detected by specific antibodies. # denotes nonspecific bands. (G) The MEFs indicated were infected with T3D (MOI 100), IAV (100 HAU/mL), or SeV (250 HAU/mL). Ifnb1 transcript was quantified by qRT-PCR at the indicated times. Data are displayed as fold induction of the indicated gene compared with uninfected. Data are represented as mean ± SEM. ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.0001 (Student’s t test).
To determine the function of STING in RNA virus infection, we infected shCTRL and shSTING cells with a panel of viruses representing diverse viral families (Table S1). The replication of VSV and Sindbis virus (SINV) was quantified by measuring the expression of luciferase reporter genes embedded in the viral genomes. Sendai virus (SeV) and influenza A virus PR8 (IAV) were also used as well as the type 3 Dearing strain of reovirus (T3D). Replication of these viruses was monitored by western analysis for virus-specific proteins. We also used a mutant VSV, VSV-M51R, which harbors a methionine-to-arginine point mutation in the VSV M protein. This mutant M diminishes the ability of VSV to suppress host gene expression and can induce strong IFN responses (27). VSV-M51R replication was determined by monitoring the production of infectious virus by plaque forming unit (pfu) assays.
All viral infections were more productive in the absence of STING (Fig. 1 D and F). Specifically, VSV, VSV-M51R, and SINV displayed multilog increases in replication in shSTING cells, compared with shCTRL cells (Fig. 1D). Western analysis of SeV, IAV, and T3D replication indicated a greater abundance of viral proteins in shSTING cells, compared with shCTRL cells (Fig. 1F). Similar findings were made in viral infections of primary MEFs derived from WT and STING knockout (KO) mice (Fig. 1C). These findings indicate that STING is required to control the replication of diverse RNA viruses.
During RNA Virus Infection, STING Displays No Activities That Promote IFN Expression.
While STING is well-recognized to promote IFN expression during DNA virus infection, some studies have implicated this protein in IFN expression during RNA virus infection (12, 16, 17). To examine the role of STING in IFN expression during RNA virus infection, we infected our MEF lines with the viruses indicated above and monitored Ifnb1 expression over time. In all infections and time points examined, STING-deficient cells were capable of inducing Ifnb1 expression. In fact, in the absence of STING, virus-induced Ifnb1 expression was greater than what was observed in infected shCTRL cells (Fig. 1 E and G). Similar trends were observed when we examined the expression of the ISGs Rsad2 and Cxcl10 (Fig. S1 A and B).
We reasoned that the increased expression of IFN and ISGs in STING-deficient cells was due to increased virus replication in these cells, as viral replication should yield a greater abundance of RLR ligands. To test this hypothesis, we performed infections in MEFs that were depleted of STING or MAVS with siRNA, and Ifnb1 expression was monitored (Fig. S1C). Under all conditions examined, cells lacking MAVS were unable to induce Ifnb1 expression in response to RNA virus infection (Fig. S1D). In contrast, cells lacking STING were not defective for virus-induced Ifnb1 expression (Fig. S1D). These collective results indicate that STING does not control the induction of IFNs and ISGs during RNA virus infection, but this protein is required to restrict the replication of diverse RNA viruses.
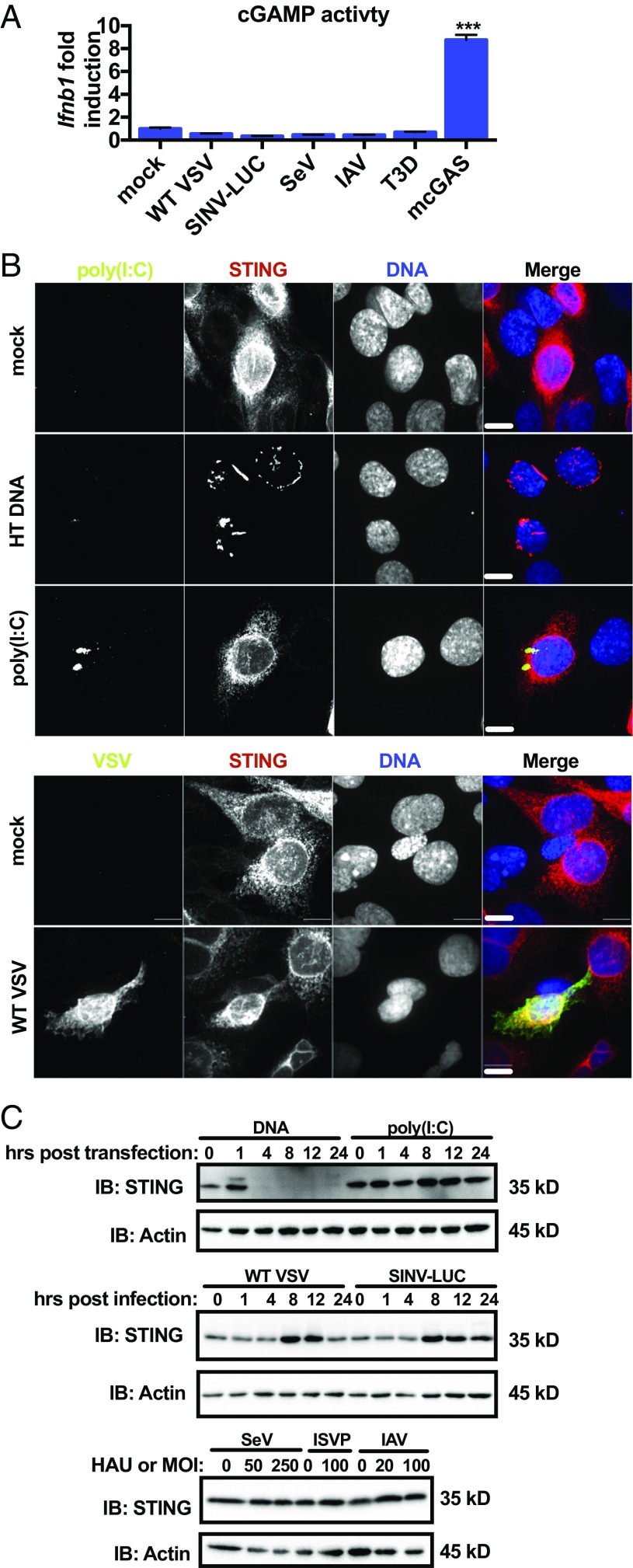
Despite the observations described above, it was possible that STING contributes to Ifnb1 expression during RNA virus infection but that MAVS signaling masked this activity. If this possibility is correct, then cells should display evidence of STING activation during RNA virus infection. To test this possibility, we monitored four markers of STING pathway activation: cGAMP production (10), STING trafficking from the ER (11), STING phosphorylation (14, 15), and degradation (14).
We detected cGAMP using an assay for biological activity of cGAMP in cell lysates. We detected inducible amounts of cGAMP activity from cells overexpressing cGAS but did not detect any cGAMP activity after 18 h of RNA virus infection (Fig. 2A). Next, we determined whether RNA ligands or virus infection induced the trafficking of STING from the ER. Using immunofluorescence, we determined the localization of HA-tagged STING after transfection with DNA, poly(I:C), or infection with VSV. Before transfection or infection, STING staining was consistent with the ER, in that the protein was scattered throughout the cell and concentrated at the nuclear envelope (Fig. 2B). After DNA transfection, STING localization shifted, and the protein concentrated in perinuclear vesicles that resemble the sites of signal transduction (Fig. 2B) (11). However, the subcellular localization of STING did not change after transfection with poly(I:C) or infection with VSV (Fig. 2B). Lastly, we assayed STING phosphorylation and degradation. One hour after DNA transfection, western analysis revealed a band corresponding to STING that exhibited a slower mobility (Fig. 2C), which is consistent with phosphorylation (14, 15). Degradation of STING was observed 4 h after DNA transfection (Fig. 2C). In contrast, no changes in STING electrophoretic mobility or stability were observed after poly(I:C) transfection or infection with RNA viruses (Fig. 2C). Taken together, these results indicate RNA virus infection does not induce any of the hallmarks of STING activation that are associated with an IFN response.
Fig. 2.
cGAS–STING pathway is not activated canonically during RNA virus infection. (A) WT MEFs were infected with VSV (MOI 10), SINV (MOI 10), SeV (250 HAU/mL), IAV (50 HAU/mL), or reovirus (MOI 100) for 18 h or transfected with a plasmid encoding mouse cGAS for 20 h. Virally infected or transfected cytoplasmic lysates were incubated with WT MEFs in the presence of reversible permeabilization buffer. Ifnb1 transcript was quantified by qRT-PCR. Data are displayed as fold induction of the indicated gene compared with uninfected. Data are represented as mean ± SEM. (B) WT MEFs expressing STING-HA were mock-transfected, transfected with herring testes DNA (5 μg/mL), poly(I:C) (2 μg/mL), mock-infected, or infected with VSV (MOI 1). MEFs transfected with DNA were fixed 1 h after transfection. Mock- and poly(I:C) transfected MEFs were fixed 4 h after transfection. VSV-infected and mock-infected MEFs were fixed 8 h after infection. MEFs were stained with antibodies detecting HA, dsRNA, or VSV M and stained to visualize nuclear DNA. Scale bar represents 10 μm. (C) WT MEFs were transfected with herring testes DNA (5 μg/mL), poly(I:C) (2 μg/mL), or infected with VSV (MOI 1) or SINV-LUC (MOI 1), SeV (0, 50, 250 HAU/mL), IAV (0, 20, 50 HAU/mL), or reovirus (MOI 100). SeV, IAV, and reovirus samples were collected 24 h postinfection. Lysates were separated by SDS/PAGE, and endogenous STING and actin were detected by western analysis. ***P < 0.0001 (Student’s t test).
The Abundance of Basal Antiviral Transcripts Does Not Influence RNA Virus Replication.
In uninfected cells, low levels of IFN are constitutively expressed and secreted (28). Since many IFN signaling components are regulated by IFN, low-level IFN secretion may prime cells to respond to infection (28). Similar to cells that lack cGAS (29), shSTING cells display lower levels of basal Ifnb1 transcripts than shCTRL counterparts (Fig. 1B).
To determine if the abundance of basal antiviral transcripts influences RNA virus replication, we experimentally altered the levels of ISGs using the pan-Janus Kinase (JAK) inhibitor pyridone 6 (P6). This treatment inhibits signaling from all IFN receptors and thus prevents activities of constitutive IFNs. In the absence of P6, the abundance of the ISG Rsad2 was lower in STING-deficient cells than in shCTRL cells (Fig. S2A). Twenty-four hours of P6 treatment lowered basal Rsad2 transcripts in shCTRL cells to levels comparable to what was observed in shSTING cells (Fig. S2A). This approach therefore allowed us to examine the influence of basal antiviral factors on RNA virus infection. We infected shCTRL and shSTING cells in the presence or absence of P6 with VSV-LUC. Viral replication was monitored via luciferase activity and plaque assay over 24 h. Whereas P6 strongly suppressed basal ISG expression, these changes had a minor influence of viral replication (Fig. S2B). Specifically, we observed no difference in viral replication when comparing P6-treated cells to DMSO-treated cells (Fig. S2B). In contrast, there was a strong difference in viral replication in the presence or absence of STING, regardless of the levels of basal ISGs. These results suggest that the abundance of ISG products in a cell before infection does not strongly influence the rate of VSV replication and therefore cannot explain the antiviral actions of STING.
Antiviral STING Activities Are Not Mediated by Autophagy, Mitochondrial DNA, or an Inducible Transcriptional Response.
We reasoned that if the sole antiviral activities of STING are via transcription, then blocking host cell gene expression should render WT cells more permissive to viral replication. To address this possibility, we blocked transcription by RNA polymerase II using the inhibitor actinomycin D. Treatments with actinomycin D strongly reduced the abundance of Tbp transcripts, which encode TATA-binding protein (Fig. S2C), confirming host cell transcription was inhibited. Cells were pretreated with actinomycin D and then infected with VSV-LUC in the presence of inhibitor. Viral replication was monitored over time. ShCTRL cells were not more permissive for VSV replication in the presence of actinomycin D but were rather slightly less permissive (Fig. S2D). A similar trend was observed upon actinomycin D treatment of shSTING cells, in that the absence of inducible gene expression did not promote more effective viral replication. These results suggest that transcriptional responses are not the primary strategy used by infected cells to restrict VSV replication. The antiviral activities of STING must therefore involve a transcription-independent cellular process.
One candidate antiviral activity that occurs independent of transcription is autophagy. Autophagy is a process by which intracellular microbes are enveloped in membranes and delivered to lysosomes for degradation (30). STING-induced autophagy restricts intracellular M. tuberculosis (31). To determine if STING mediates autophagy during RNA virus infection, we transduced shCTRL and shSTING cells with a retroviral vector expressing an LC3-GFP fusion protein. LC3 is incorporated into autophagosomes and is retained on these organelles as they are delivered to lysosomes. Once in lysosomes, LC3 is degraded (30). We monitored the release of the GFP epitope tag present on the LC3 transgene as a readout of autophagosome delivery to lysosomes during viral infection (32). Infection with VSV, but not SeV, led to the appearance of free GFP (indicating cleavage) in shCTRL and STING-deficient cells (Fig. S2E). As SeV replication is restricted by STING (Fig. 1F) but does not induce detectable changes in LC3 cleavage (Fig. S2E), autophagy induction does not correlate with STING-dependent antiviral activities. Moreover, in the case of VSV infection, we observed no defect in LC3 processing in the absence of STING (Fig. S2E).
To complement this marker-based analysis, we examined the requirement of autophagy for the restriction of viral replication. Phosphoinositide 3-kinase (PI3K) regulates the induction of autophagy that occurs during viral infection. We treated cells with the PI3K inhibitors wortmannin (WMN) and 3-methyladenine (3-MA) and examined their effect on VSV replication. We did not observe higher virus replication in cells treated with either inhibitor (Fig. S2F). Thus, autophagy induction during viral infection does not depend on STING, and preventing autophagy does not enhance VSV infection.
RNA virus restriction may occur due to sensing of mitochondrial DNA by cGAS, which then signals through STING (33). While we have not found that IFN induction is regulated by STING during RNA virus replication, we sought to fully assess the requirement for mitochondrial DNA during infection. We treated shCTRL and shSTING cells with low-dose ethidium bromide for 2 wk. This procedure generates ρ0 cells, which are depleted of mitochondrial DNA (34). We confirmed mitochondrial DNA depletion by monitoring the abundance of mitochondrial genomes (Fig. S2G). Mock-treated and ρ0 cells were infected with VSV-LUC, and replication was monitored over time. ShCTRL cells retained the ability to restrict VSV replication, by a STING-dependent process, even in ρ0 cells (Fig. S2H). These data suggest that the release of mitochondrial DNA does not promote the STING-dependent control of RNA virus replication.
The cGAS–STING pathway also regulates cellular senescence (35, 36). We addressed whether differences in viral control could be associated with senescence. We observed equal incorporation of BrdU in shCTRL and shSTING MEFs and observed no senescence-associated β-galactosidase activity in either cell population (Fig. S2 I and J). In contrast, high passage primary WT MEFs displayed β-galactosidase activity (Fig. S2J). These results eliminate the possible contribution of senescence to the STING-dependent phenotypes we have observed.
The Ability to Productively Infect Cells Is Mediated by STING, Independent of the Viral Entry Mechanism.
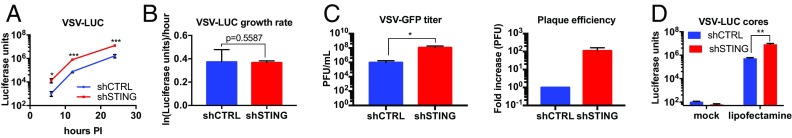
We have observed that viral replication was higher in the absence of STING (Fig. 1 D and F). One of two possible models could explain this observation: (i) The rate of viral replication is higher in STING-deficient cells, or (ii) the establishment of infection is more efficient in the absence of STING. To distinguish between these possibilities, we infected shCTRL or shSTING cells with VSV-LUC and modeled the rate of viral replication or determined the efficiency of plaque formation.
We first determined the rate of VSV replication in shCTRL and shSTING MEFs. This was accomplished by infecting cells with VSV-LUC and tracking viral luciferase expression over time (Fig. 3A). Since virus replication is an exponential process, we modeled the viral curves as exponential growth equations and extracted the growth rate. This analysis indicated that the growth rate was similar in the presence and absence of STING (Fig. 3B). This result indicates that the increased viral replication observed in shSTING cells was not due to faster virus replication rates. Given this finding, we hypothesized that the probability that a cell becomes infected is increased in shSTING cells.
Fig. 3.
STING decreases infection probability. (A) The MEFs indicated were infected with VSV-LUC at MOI 1.0. Infection was monitored by luciferase assay at the indicated time points. (B) Virus growth rates [ln((Luciferase units)/hour] were determined from the exponential phase of the data in A by linear regression and are shown as the best fit values with 95% confidence intervals. Growth rates were compared using the Mann–Whitney U test. (C) Plaque forming assays were performed on the MEFs indicated. Plaques were counted by fluorescence microscopy, and a viral titer (pfu/mL) was calculated. Relative plaque efficiency was calculated from the fold increase in titer calculated on shCTRL and shSTING cells. Data are represented as mean ± SEM. (D) The MEFs indicated were transfected in the presence of lipofectamine 2000 or mock-transfected with purified VSV-LUC cores. Luciferase was quantified 5 h after transfection by luciferase assay. Data are represented as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.0001 (Student’s t test).
If STING controls the infection probability, then a fixed concentration of virus will yield a greater amount of pfus when titered on STING-deficient cells, compared with shCTRL cells. Consistent with this model, the titer of VSV-GFP on shCTRL cells was calculated to be 9.4 × 105 pfu/mL, and the titer of the same inoculum on shSTING cells was 1.1 × 108 pfu/mL (Fig. 3C). The relative probability of plaque formation was therefore ∼100-fold higher in shSTING cells than in shCTRL cells (Fig. 3C). Thus, with a fixed input of virus, more productive infections occur in STING-deficient cells than in shCTRL counterparts. These data demonstrate that in the absence of STING, the probability that a cell will become infected is increased.
To determine if the increased probability of infection in the absence of STING depends on a specific entry mechanism, we bypassed the natural route of VSV entry by transfecting cells with the viral core of VSV-LUC. This strategy has been used to understand post-entry aspects of VSV replication (37, 38). Under these conditions, VSV replication (as assessed by luciferase production) was higher in shSTING cells than in shCTRL cells (Fig. 3D). Thus, STING-dependent antiviral activities are evident even when the natural entry route is bypassed. These results suggest that STING restricts viral replication at a stage after entry into the cells.
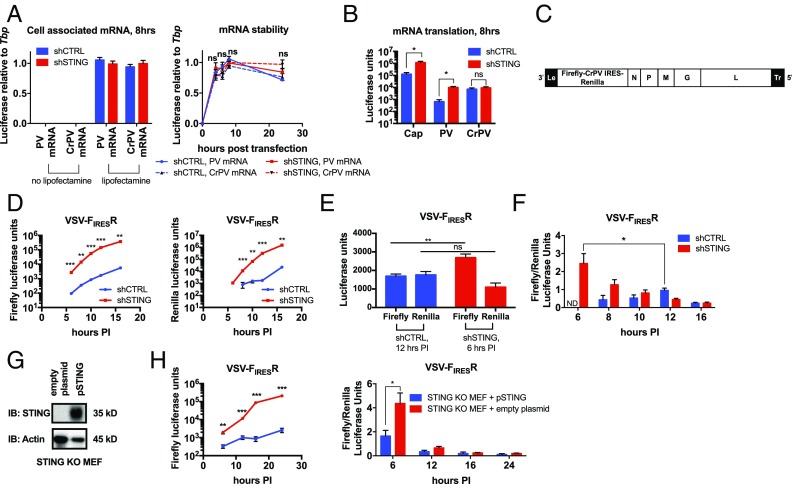
STING Regulates the Translation of Virus and Host mRNAs.
Previous work on RNA viruses demonstrated that infectivity potential can be determined by the efficiency of viral mRNA translation (39). It was therefore possible that STING regulates translation. We first tested this model by determining if STING controlled the amount of protein produced from an in vitro transcribed mRNA. We used bicistronic mRNAs that express firefly luciferase via cap-dependent translation with renilla luciferase expressed downstream of either a poliovirus IRES (PV mRNA) or the intergenic cricket paralysis virus IRES (CrPV mRNA). We transfected shCTRL and shSTING cells and used qPCR to ensure that an equal number of transcripts were delivered to the cells (Fig. 4A). We also monitored the presence of the transfected mRNA within cells over 24 h and found that the stability of the transcript was similar in shCTRL and shSTING cells (Fig. 4A). We then tested the amount of protein produced by cap-, PV IRES-, and CrPV IRES-dependent mechanisms. Approximately 10-fold higher luciferase signal was detected from cap-dependent translation in shSTING cells, compared with shCTRL cells (Fig. 4B). Additionally, more luciferase was produced from the PV IRES in the absence of STING (Fig. 4B). However, the CrPV IRES displayed no increase in translation in the absence of STING (Fig. 4B). These findings indicate that STING is necessary to limit translation of exogenous mRNAs. As CrPV does not require any host translation initiation factors to begin protein synthesis (40), STING likely regulates the step of translation initiation.
Fig. 4.
STING restricts translation of exogenous and viral mRNAs. (A) The indicated mRNA was transfected into the MEFs indicated with or without lipofectamine 2000. RNA was isolated at the indicated times, and luciferase transcript levels were analyzed by qRT-PCR. Data are represented as mean ± SEM. (B) The indicated mRNAs were transfected into the MEFs indicated. Luciferase was quantified by dual luciferase assay 8 h after transfection. Data are represented as mean ± SEM. (C) Schematic of VSV-FIRESR genome; “le” denotes the leader region, and “tr” denotes the trailer. (D) The MEFs indicated were infected with VSV-FIRESR at MOI 1.0. Infections were monitored by dual luciferase assay at the indicated time postinfection (hours PI). Firefly and renilla luciferase units are represented at the indicated time. (E) Firefly and renilla luciferase units from the indicated cell line and time postinfection from D. (F) The ratio between the firefly and renilla signal from the VSV-FIRESR infections in D at the indicated time. Data are represented as mean ± SEM. (G) Lysates from STING KO primary MEFs transfected with empty plasmid or plasmid encoding STING were separated by SDS/PAGE. STING and actin were detected by western analysis. (H) Primary STING KO MEFs transfected with empty plasmid or plasmid encoding STING (pSTING) were infected with VSV-FIRESR at MOI 1.0. Infections were monitored by dual luciferase assay at the indicated time postinfection (hours PI). Left, firefly luciferase units from the indicated cells and time postinfection are plotted. Right, the ratio between the firefly and renilla signal from the VSV-FIRESR infections at the indicated time. Data are represented as mean ± SEM. ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.0001 (Student’s t test).
As in vitro transcribed mRNAs had a translation advantage in the absence of STING, it was possible that viral mRNAs also had a translation advantage. We utilized the observation that the CrPV IRES was translated similarly in the presence and absence of STING and created a recombinant VSV that expresses a bicistronic mRNA that has firefly luciferase driven by a capped VSV 5′UTR and a renilla luciferase after the CrPV IRES (VSV-FIRESR) (Fig. 4C). This viral strain allowed us to estimate the number of proteins produced per viral transcript by calculating the ratio between STING-sensitive translation (firefly luciferase signal) and STING-insensitive translation (renilla luciferase signal). We infected shCTRL and shSTING cells and monitored STING-sensitive firefly luciferase expression over time. As we observed using other VSV strains, the VSV-FIRESR strain replicated more robustly in the absence of STING (Fig. 4D). When we compared a time point where approximately equal amounts of STING-insensitive renilla were expressed in shCTRL and shSTING cells, we observe approximately twice the amount of firefly signal in shSTING cells (Fig. 4E). This result indicates that more viral translation is occurring per transcript in shSTING cells compared with the shCTRL cells. We quantified the viral translation efficiency (firefly to renilla ratio) over the course of infection. We observed a maximum translation efficiency in shSTING cells at 6 h, whereas the maximum in shCTRL cells was reached later at 12 h (Fig. 4F). At these time points, the efficiency in the shSTING cells was significantly higher, together indicating that infection in shSTING cells leads to more efficient translation earlier in infection. We confirmed these findings in primary STING KO cells (Fig. 4 G and H). Thus, similar to transfected mRNAs, translation of viral mRNAs is more efficient in the absence of STING.
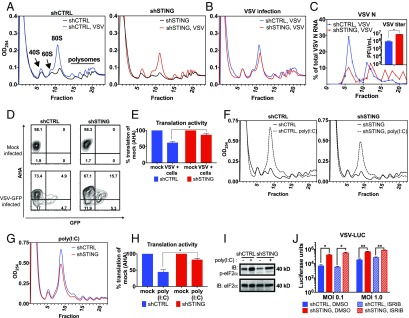
Translational efficiency often coincides with the distribution of a given mRNA in more translationally active polysomes. To determine if the presence of viral transcripts in polysomes is influenced by STING, polysome profiling was performed. The UV absorbance spectra (OD254) of the polysome sucrose gradients demonstrated a higher 80S monosome accumulation in infected shCTRL cells compared with uninfected shCTRL cells (Fig. 5A). This finding is consistent with the cessation of robust protein synthesis and the partial collapse of polysomes into monosomes. Interestingly, 80S accumulation after viral infection was not as robust in shSTING cells as in shCTRL cells (Fig. 5 A and B), suggesting that maximal translation shutdown depended on STING function. Quantification of VSV N mRNA was performed in each fraction, and we found that a larger proportion of viral mRNAs were present within polysomes from shSTING cells than in polysomes isolated from shCTRL cells (Fig. 5C). These results are consistent with the idea that translation of exogenous (transfected or viral) mRNA is more efficient in the absence of STING.
Fig. 5.
Translation is enhanced in virally infected and poly(I:C)-transfected cells lacking STING. (A) UV absorbance spectra of polysome sedimentation from shCTRL and shSTING cells infected at MOI 2, 12 h postinfection. (B) Overlay of VSV-infected shCTRL and shSTING profiles from A. (C) qRT-PCR analysis of VSV N mRNA from each fraction. Inset is virus titer calculated by pfu assays of supernatant virus at time of RNA isolation. Data are represented as mean ± SEM. (D) The MEFs indicated were infected or mock-infected with VSV-GFP for 10 h at MOI 5. Cells were depleted of methionine and labeled with AHA. Cells were fixed and a fluorophore was attached to AHA. Infected and mock-infected cells were analyzed by flow cytometry. VSV-infected cells were gated by GFP signal. (E) AHA MFI of all populations was determined from the data in D. Data are shown as percent MFI of mock-infected cells. Data are represented as mean ± SEM. (F) UV absorbance spectra of polysome sedimentation from shCTRL and shSTING MEFs 6 h after transfection with 2 μg/mL of poly(I:C) or mock-transfected. (G) Overlay of poly(I:C)-transfected shCTRL and shSTING profiles from F. (H) The MEFs indicated were transfected with 2 μg/mL of poly(I:C) and then labeled with AHA. Six hours after transfection, cells were fixed and AHA incorporation was measured with flow cytometry. AHA MFI of all populations was determined. Data are shown as percent MFI of mock-transfected cells. Data are represented as mean ± SEM. (I) MEFs were transfected with 2 μg/mL of poly(I:C) or mock-transfected. Lysates were separated by SDS/PAGE, and endogenous phospho-eIF2α and total eIF2α were detected by western analysis. (J) The MEFs indicated were incubated with 1 μM ISRIB for 10 min before infection with VSV-LUC at the indicated MOI, in the presence of ISRIB. Viral gene expression was determined by luciferase activity assay 12 h postinfection. *P < 0.05; **P < 0.01; ***P < 0.0001 (Student’s t test).
The larger proportion of viral mRNAs found in the polysomes suggests that there is more protein synthesis occurring in VSV-infected shSTING cells. To address this possibility directly, we used metabolic labeling to quantify the amount of translation occurring within VSV-infected cells. We infected (or mock-infected) shCTRL and shSTING cells with VSV-GFP. Ten hours postinfection, cells were depleted of methionine and labeled with the methionine homolog, l-azidohomoalanine (AHA). Cells were then fixed and click chemistry was used to attach a fluorophore to AHA. This procedure creates a correlation between cell-associated fluorescence and protein synthesis (41). Flow cytometry was then used to quantify translational activity (Fig. 5D).
We gated on VSV-infected cells, as determined by the virally encoded GFP signal, and quantified the mean fluorescence intensity (MFI) of AHA staining in the population. We normalized the MFI of AHA staining in the infected cell population to the MFI of mock-infected cells, thereby providing a percent translation activity, compared with mock infected. Consistent with the polysome analysis performed, VSV-infected shCTRL cells displayed lower translation activity after infection than VSV-infected shSTING cells (Fig. 5 D and E). These collective results demonstrate that STING regulates the translational activity of infected cells.
Role of the Initiation Factor eIF2α in STING-Dependent Translational Control.
We sought to determine if STING regulates the translation of exogenous mRNAs (transfected and viral) specifically or if STING regulates translation globally during RNA virus infection. However, because viruses utilize and modify host translation machinery, it is difficult to deconvolute host translation from viral translation. We therefore used a model for RNA virus infection, poly(I:C) transfection.
We transfected shCTRL and shSTING cells with poly(I:C) and then subjected the cells to polysome analysis. Poly(I:C) transfection led to the expected increase in the abundance of 80S monosomes, as poly(I:C) is recognized to diminish the translation activity of cells (Fig. 5 F and G).
Similar to what we observed during VSV infections, poly(I:C) transfection led to a greater amount of 80S monosome accumulation in shCTRL cells than cells lacking STING (Fig. 5G). To corroborate these findings, we labeled shCTRL and shSTING cells with AHA after poly(I:C) transfection and monitored the extent of protein synthesis directly. In agreement with our results using VSV infection, more protein synthesis occurred after poly(I:C) transfection in the absence of STING than in the presence of STING (Fig. 5H). These results verify that during RNA virus infection [or poly(I:C) transfection], STING regulates translational activity.
To determine how STING restricts translation, we considered the mechanism that regulates translation inhibition after poly(I:C) transfection. This process involves the phosphorylation of the initiation factor eIF2α by the dsRNA sensor PKR. Phosphorylated eIF2α (phospho-eIF2α) binds and inhibits the GEF activity of another initiation factor, eIF2B, which prevents eIF2α from being recycled after one round of translation and initiating further rounds of translation (42). Similar to poly(I:C) transfection, RNA virus replication activates PKR and phospho-eIF2α accumulates during VSV replication in MEFs (43). To determine if STING regulates eIF2α phosphorylation, we analyzed phospho-eIF2α in shCTRL and shSTING cells after poly(I:C) transfection. No STING-dependent changes in eIF2α phosphorylation were observed, as shCTRL and shSTING cells contained similar amounts of phospho-eIF2α (Fig. 5I).
When eIF2α is phosphorylated, it binds and inhibits eIF2B (44). However, if eIF2B GEF activity is restored, translation proceeds in the presence of phospho-eIF2α (42). We reasoned that if depletion of STING leads to an enhancement of eIF2B GEF activity, then we would observe increased translation in STING-deficient cells, even in the presence of phospho-eIF2α. A drug that maintains GEF activity when eIF2α is phosphorylated should therefore phenocopy STING deficiency during virus infection of shCTRL cells. We tested this hypothesis by treating shCTRL and shSTING cells with the drug ISRIB, which maintains eIF2B GEF activity in the presence of phospho-eIF2α (45). To confirm the activity of the drug, we treated shCTRL and shSTING cells with thapsigargin, which induced eIF2α phosphorylation via the kinase PERK. Five minutes before metabolically labeling the cells with puromycin, we treated thapsigargin-treated cells with ISRIB. As observed by western analysis for puromycin labeling, ISRIB restored protein production quickly in the presence of phospho-eIF2α and thapsigargin (Fig. S3A). The puromycin signal was quantified and normalized to actin (Fig. S3B). We then assayed viral gene expression in shCTRL and shSTING cells treated with ISRIB during the infection. After infection with VSV-LUC, viral gene expression did not significantly increase in the shCTRL cells (Fig. 5J). This finding suggests that the STING-dependent translation restriction is not mediated through eIF2B. Based on these findings, we propose that an eIF2-independent mechanism underlies the ability of STING to control protein synthesis within cells infected with RNA viruses.
Genetic Pathway Analysis of STING-Dependent Translation Regulation.
In all our experiments, STING restriction of translation occurs in the presence of an RNA ligand that stimulates the RLR–MAVS pathway. Thus, we hypothesized that STING restriction depends on RIG-I, melanoma differentiation-associated gene 5 (MDA5), and/or MAVS. To test this hypothesis, we first determined if STING regulates translation in response to a stimulus that does not involve exogenous RNAs. The ER stress response induced by thapsigargin provides such a stimulus. The kinase PERK is activated by thapsigargin to phosphorylate eIF2α and diminish translation (46). We treated cells with thapsigargin for 6 h and then labeled cells with puromycin and quantified the amount of puromycin incorporation by western analysis. We normalized the puromycin signal to actin and then normalized the thapsigargin-treated sample to the amount of puromycin in the DMSO-treated cells. From this analysis, we observed no significant difference in translation activity between shCTRL and shSTING cells (Fig. S3C). This result indicates that STING does not broadly regulate all pathways that lead to translation inhibition but specifically controls pathways activated by exogenous RNA. Consistent with this idea, we found that STING does not regulate translation in response to cytosolic DNA. Translation, as measured by AHA incorporation, was not significantly different between shCTRL and shSTING cells transfected with DNA (Fig. S3D).
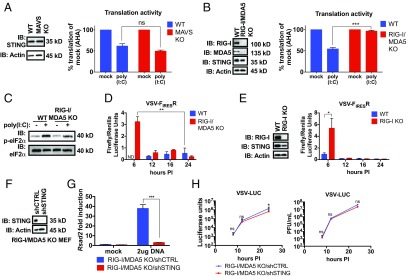
To examine the role of the RLR–MAVS pathway in translation regulation, we determined if MAVS was required to restrict translation. We assayed translation activity within WT or MAVS KO MEFs after transfection with poly(I:C) using metabolic labeling with AHA. After transfection, translation was inhibited in WT and MAVS KO MEFs (Fig. 6A). These data indicate that STING-dependent translation restriction does not depend on MAVS.
Fig. 6.
RIG-I and STING form a genetic pathway to control RNA virus translation and replication. (A, Left) Lysates were separated by SDS/PAGE, and endogenous STING and actin were detected by western analysis. (A, Right) Translation activity of WT and MAVS KO cells quantified as in Fig. 5H. (B, Left) Lysates were separated by SDS/PAGE, and the endogenous proteins indicated were detected by western analysis. (B, Right) Translation activity of WT and RIG-I/MDA5 KO cells quantified as in Fig. 5H. (C) Lysates were separated by SDS/PAGE and the endogenous proteins indicated were detected by western analysis. (D) The MEFs indicated were infected with VSV-FIRESR. Infections were monitored by dual luciferase assay at the indicated time postinfection (hours PI). Data are the ratio between the firefly and renilla signal at the indicated time. Data are represented as mean ± SEM. (E, Left) Lysates were separated by SDS/PAGE, and endogenous actin, RIG-I, and STING were detected by western analysis. Right, WT and RIG-I KO MEFs were infected with VSV-FIRESR. Infections were monitored by dual luciferase assay at the indicated time postinfection (hours PI). Data are the ratio between the firefly and renilla signal at the indicated time. Data are represented as mean ± SEM. (F) Lysates were separated by SDS/PAGE. Endogenous actin and STING were detected by western analysis. (G) The indicated cell lines were mock-transfected or transfected with 2 μg of herring testes DNA for 8 h. RNA was isolated and Rsad2 transcript was analyzed by qRT-PCR. Data are represented as mean ± SEM. (H) The MEFs indicated were infected with VSV-LUC at MOI 0.1. Infections were monitored by luciferase assay at the indicated time postinfection (hours PI). Left shows firefly luciferase units at the indicated time. Right shows viral titer as determined by pfu assays. Data are represented as mean ± SEM. ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.0001 (Student’s t test).
Next, we tested the requirement for RIG-I and MDA5. Using metabolic labeling with AHA, we assayed the amount of translation that occurs after poly(I:C) transfection in WT MEFs and MEFs that lack RIG-I and MDA5 (RIG-I/MDA5 KO). In WT MEFs, translation activity was decreased to ∼50% of mock after poly(I:C) transfection. However, in MEFs lacking RIG-I and MDA5, translation was maintained at 96% of mock (Fig. 6B). We also detected a slight decrease in eIF2α phosphorylation in the absence of RIG-I and MDA5 (Fig. 6C). In contrast (and similar to STING-deficient cells), RIG-I/MDA5 KO cells displayed no defects in thapsigargin-induced translation inhibition or eIF2α phosphorylation (Fig. S3E). Translation regulation in response to exogenous RNA was therefore dependent on RLRs.
To determine if RIG-I and/or MDA5 influenced translation of viral mRNAs, we infected WT and RIG-I/MDA5 KOs with VSV-FIRESR. We quantified the firefly and renilla signal over time. Similar to what was observed in shSTING and STING KO cells, we observed an early enhancement of viral translation (6 h) in the absence of RIG-I/MDA5 (Fig. 6D).
Because detection of VSV RNA ligands is dependent on RIG-I (47), we hypothesized that RIG-I is necessary to control VSV translation. We infected WT or RIG-I KO primary MEFs with VSV-FIRESR. We again observed an early enhancement of viral translation in the absence of RIG-I (Fig. 6E). Therefore, RIG-I (and not MAVS) is necessary to inhibit VSV mRNA translation.
Given that viral translation is regulated in a similar way by STING and RIG-I, we reasoned that these proteins form a pathway to control RNA virus replication. If STING is downstream of RNA sensing by RLRs, then RIG-I/MDA5 KO cells should control virus replication to the same extent as cells lacking all three of these regulators. However, if STING controls RNA virus replication independently of RIG-I and MDA5, we would expect to see an enhancement of RNA virus replication when RIG-I, MDA5, and STING are all missing from a cell. Using our shRNA that targets STING, we depleted RIG-I/MDA5 KO cells of STING (RIG-I/MDA5/shSTING) or transduced them with the nontargeted control (RIG-I/MDA5/shCTRL). We confirmed STING depletion by western analysis (Fig. 6F). After STING depletion, cells no longer induced Rsad2 in response to transfected DNA, confirming that the depletion was functional (Fig. 6G). We infected both cell lines with VSV-LUC and monitored the infection by luciferase activity assay and viral titer over time. We observed no difference in viral replication between RIG-I/MDA5/shCTRL cells and RIG-I/MDA5/shSTING cells (Fig. 6H). This result supports a model whereby RLRs have distinct effector responses during RNA virus infection. One response is mediated by MAVS to induce IFN, and a distinct response is mediated by STING to diminish translation.
Discussion
In this study, we explored the role of STING in the establishment of a functional antiviral state within murine fibroblasts. While some reports have indicated a role for STING in the induction of IFNs and ISGs during RNA virus infection, substantial evidence supports a model whereby STING specifically controls IFN expression when DNA viruses enter cells (11, 13, 16). Our studies support this latter conclusion, in that STING-deficient cells retained the ability to induce IFNs during infections with multiple phylogenetically diverse RNA viruses. Despite this ability to induce IFNs and ISGs, STING-deficient cells were unable to restrict the replication of RNA viruses. These results indicate that IFN expression is not sufficient to restrict viral replication and that other antiviral mechanisms are important to fully protect a cell from infection. Consistent with this finding are studies demonstrating that STAT1-deficient cells (which are defective for IFN signaling) are not more permissive for VSV replication than WT cells (22, 23). We have verified these results in this study with the finding that the inhibitor P6, which blocks signaling by JAKs, does not result in an increase in VSV replication. Viral replication in the absence of STING unmasked an IFN-independent mechanism of viral control.
We have explored several possible mechanisms by which STING could restrict RNA virus replication, including the role of basal ISGs, mitochondrial DNA, and autophagy. We found that VSV replication was largely insensitive to experimental alterations in basal ISG abundance, removal of mitochondrial DNA, or prevention of autophagy. These findings prompted an examination of the most primordial antiviral activity known—the control of mRNA translation. Diverse host strategies prevent viral mRNA from gaining access to the translational machinery that is critical for pathogen propagation. These strategies include the use of restriction enzymes to hydrolyze bacteriophage genomes, which prevents transcription (48), the use of RNA interference to degrade viral mRNAs within insects (2), and the use of PKR to globally diminish translation in mammalian cells (26). Our findings add STING to the list of factors used by host cells to restrict translation of viral mRNAs, as STING is necessary for global translation inhibition during RNA virus infections. Whether STING-mediated translation control operates in cell types other than fibroblasts is unclear. Several evolutionary questions also remain unanswered. In particular, it is unknown if STING proteins present in nonmammals display similar activities to those described herein.
While the precise mechanism by which STING influences translation is unclear, our analysis suggests that this process is distinct from that mediated by PKR. Indeed, eIF2α phosphorylation, which is the target of PKR, was not affected by STING deficiency, and a broader range of viruses are restricted by STING than are capable of inducing eIF2α phosphorylation. For example, SINV is resistant to eIF2α translation control and VSV-M51R does not activate eIF2α kinases (43, 49), but these viruses replicate to higher amounts in the absence of STING. STING was observed to associate with a subunit of the translocon complex (12). Given this observation, we considered the possibility that STING influences translation via the regulation of protein translocation. However, translation of mRNAs that do not utilize the translocon (luciferase and VSV N) was suppressed by STING, suggesting that the mechanism of translation inhibition is not related to the translocon. Some other strategy must therefore exist within cells that is mediated by STING to influence translation inhibition, and this process is necessary to suppress RNA virus replication.
Our findings support the idea that STING has distinct and context-dependent antiviral effector mechanisms. There are few other examples of innate immune regulators that exhibit distinct functional activities. Of those that do exhibit diverse downstream effector functions, such as TLR4 and TLR9, these functions are induced from distinct organelles (6). For example, TLR4 induces cytokine expression from the plasma membrane and IFN expression from endosomes, and TLR9 induces cytokine and IFN expression from distinct endosomal populations (6). Caspase-11 also can induce either pyroptosis or cell hyperactivation in response to different upstream stimuli, independent of subcellular localization (50). Our findings suggest that STING is regulated by a process similar to TLRs and caspase-11. For example, like caspase-11, STING induces different responses when different upstream stimuli (cGAMP or RLR activation) are encountered. Our studies of STING subcellular trafficking also suggest similarities with the TLRs, as STING must leave the ER to stimulate IFN expression. In contrast, STING-mediated translation inhibition was not associated with changes in its subcellular positioning. STING may therefore have the ability to induce translation inhibition from the ER.
The final notable aspect of this study came from our genetic analysis, which identified a pathway that links RLRs to STING, independent of MAVS. These data expand the functions of RLRs in infected cells beyond the transcriptional induction of antiviral genes. Our results support a model where RLRs detect a common upstream ligand (viral RNA) and then engage distinct sets of downstream factors to induce IFN expression or translation inhibition (Fig. S4). We propose that while IFN expression is critical to protect uninfected cells from a future infection, translation inhibition is important to protect during the earliest stages of an infection, in a cell-intrinsic manner. The findings reported herein therefore provide a mandate to explore whether other innate immune signaling pathways coordinate diverse antiviral mechanisms to maximize the ability to restrict infection.
Materials and Methods
Virus Stocks, Infections, and Virus Detection.
Reovirus Type 3 Dearing Cashdollar (T3D) was propagated in L929 cells and plaque purified as described (51). SeV was obtained from Charles River Laboratories. Influenza virus (A/Puerto Rico/8/34, H1N1) was propagated in Vero cells as described (52). VSV (Indiana), VSV-LUC, VSV-FIRESR, and VSV-GFP was propagated and purified as described (53). VSV-FIRESR was generated by cloning the CrPV IRES Renilla sequence from pFR_CrPV_xb directly downstream of the firefly luciferase gene in the VSV-LUC backbone. The virus was rescued using a previously published protocol (54). SINV was propagated and purified as described (55).
Cells were seeded 16 h before infection. On the day of infection, cell monolayers were incubated with the indicated amounts of virus in serum-free DMEM for 1 h at 37 °C, washed, and incubated in normal media. Luciferase activity readings were performed using Bright-Glo Luciferase Assay System or Dual-Glo Luciferase Assay System (Promega) according to the manufacturer’s instructions. VSV titers were determined by pfu assay on Vero cells as described (55). VSV-LUC ribonucleoproteins were isolated and transfected into cells as described (56).
Generation of ρ0 MEFs.
ρ0 MEFs were generated as described (34). Depletion of mitochondria DNA was assessed by qPCR for mitochondrial genomes using the primers and procedure previously described (57). DNA was analyzed on a CFX384 real-time cycler (Bio-Rad) using iTaq Universal SYBR Green Supermix.
cGAMP Activity Assay.
cGAMP activity was measured essentially as described (10). Full details can be found in SI Materials and Methods.
Metabolic Labeling, Flow Cytometry.
Cell labeling with 50 μM AHA was performed via the manufacturer’s protocol (Life Technologies). AHA was stained with 100 µM of APC-phosphine according to the manufacturer’s protocol (Life Technologies). Cells labeled with puromycin were labeled and analyzed by western analysis as described (58). For AHA experiments, populations were gated based on cells treated with cycloheximide (10 μg/mL) before AHA labeling. All data for flow cytometry experiments were acquired on a BD FACSCanto II (Becton Dickinson) and analyzed using FlowJo v10 (FlowJo; LLC).
Polysome Profiling and mRNA Detection.
MEFs were mock-infected or infected at an MOI of 2 with VSV for 12 h. Cells were incubated in complete DMEM with 100 μg/mL cycloheximide for 10 min at 37 °C. Cells were washed with cold PBS and 100 μg/mL cycloheximide. Cells were collected after trypsinization, resuspended in 500 μL of PEB (20 mM Tris⋅HCl, pH 7.5, 50 mM KCl, 10 mM MgCl2, 1 mM DTT, 100 μg/mL cycloheximide, 200 μg/mL heparin) with 1% Triton-X, and incubated on ice for 15 min. Lysate was spun for 15 min at 4 °C at 12,000 × g, and supernatant was sedimented on a 10–50% (wt/vol) sucrose gradient by centrifugation at 38,000 rpm at 4 °C for 2 h in a Beckman SW40 Ti rotor. Fractions were collected from the top of the gradient using a Gradient Station (BioComp). RNA was isopropanol precipitated overnight, pelleted by centrifugation at 4 °C at 12,000 × g, and resuspended in lysis buffer. RNA was isolated using PureLink RNA Mini Kit (Life Technologies). cDNA was synthesized from RNA using iScript cDNA synthesis kit (BioRad). VSV N mRNA expression was analyzed on a CFX384 real-time cycler (Bio-Rad) using iTaq Universal SYBR Green Supermix with primers described (56).
RNA was isolated from cell culture using PureLink RNA Mini Kit (Life Technologies). Gene expression was analyzed on a CFX384 real-time cycler (Bio-Rad) using TaqMan RNA-to-CT 1-Step Kit (Applied Biosystems) with probes purchased from Life Technologies.
Supplementary Material
Acknowledgments
We thank members of the J.C.K. laboratory for helpful discussions. J.C.K. is supported by NIH Grants AI093589, AI116550, and P30 DK34854 and an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. K.M.F. is supported by the Harvard Herchel Smith Graduate Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716937115/-/DCSupplemental.
References
- 1.Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding SW. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R. Approaching the asymptote: 20 years later. Immunity. 2009;30:766–775. doi: 10.1016/j.immuni.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate immune pattern recognition: A cell biological perspective. Annu Rev Immunol. 2015;33:257–290. doi: 10.1146/annurev-immunol-032414-112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma DY, Suthar MS. Mechanisms of innate immune evasion in re-emerging RNA viruses. Curr Opin Virol. 2015;12:26–37. doi: 10.1016/j.coviro.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samanta M, Iwakiri D, Kanda T, Imaizumi T, Takada K. EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN. EMBO J. 2006;25:4207–4214. doi: 10.1038/sj.emboj.7601314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu J, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li XD, et al. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155:688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 16.Aguirre S, et al. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 2012;8:e1002934. doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu CY, et al. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog. 2012;8:e1002780. doi: 10.1371/journal.ppat.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xing Y, et al. The papain-like protease of porcine epidemic diarrhea virus negatively regulates type I interferon pathway by acting as a viral deubiquitinase. J Gen Virol. 2013;94:1554–1567. doi: 10.1099/vir.0.051169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding Q, et al. Hepatitis C virus NS4B blocks the interaction of STING and TBK1 to evade host innate immunity. J Hepatol. 2013;59:52–58. doi: 10.1016/j.jhep.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 20.Holm CK, et al. Influenza A virus targets a cGAS-independent STING pathway that controls enveloped RNA viruses. Nat Commun. 2016;7:10680. doi: 10.1038/ncomms10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, et al. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell. 2014;5:369–381. doi: 10.1007/s13238-014-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Obuchi M, Fernandez M, Barber GN. Development of recombinant vesicular stomatitis viruses that exploit defects in host defense to augment specific oncolytic activity. J Virol. 2003;77:8843–8856. doi: 10.1128/JVI.77.16.8843-8856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katzenell S, Chen Y, Parker ZM, Leib DA. The differential interferon responses of two strains of Stat1-deficient mice do not alter susceptibility to HSV-1 and VSV in vivo. Virology. 2014;450-451:350–354. doi: 10.1016/j.virol.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McNab F, Mayer-Barber K, Sher A, Wack A, O’Garra A. Type I interferons in infectious disease. Nat Rev Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levin D, London IM. Regulation of protein synthesis: Activation by double-stranded RNA of a protein kinase that phosphorylates eukaryotic initiation factor 2. Proc Natl Acad Sci USA. 1978;75:1121–1125. doi: 10.1073/pnas.75.3.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmed M, et al. Ability of the matrix protein of vesicular stomatitis virus to suppress beta interferon gene expression is genetically correlated with the inhibition of host RNA and protein synthesis. J Virol. 2003;77:4646–4657. doi: 10.1128/JVI.77.8.4646-4657.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taniguchi T, Takaoka A. A weak signal for strong responses: Interferon-alpha/beta revisited. Nat Rev Mol Cell Biol. 2001;2:378–386. doi: 10.1038/35073080. [DOI] [PubMed] [Google Scholar]
- 29.Schoggins JW, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe. 2012;11:469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun B, et al. Dengue virus activates cGAS through the release of mitochondrial DNA. Sci Rep. 2017;7:3594. doi: 10.1038/s41598-017-03932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.King MP, Attardi G. Isolation of human cell lines lacking mitochondrial DNA. Methods Enzymol. 1996;264:304–313. doi: 10.1016/s0076-6879(96)64029-4. [DOI] [PubMed] [Google Scholar]
- 35.Yang H, Wang H, Ren J, Chen Q, Chen ZJ. cGAS is essential for cellular senescence. Proc Natl Acad Sci USA. 2017;114:E4612–E4620. doi: 10.1073/pnas.1705499114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glück S, et al. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat Cell Biol. 2017;19:1061–1070. doi: 10.1038/ncb3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szilágyi JF, Uryvayev L. Isolation of an infectious ribonucleoprotein from vesicular stomatitis virus containing an active RNA transcriptase. J Virol. 1973;11:279–286. doi: 10.1128/jvi.11.2.279-286.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee AS, Burdeinick-Kerr R, Whelan SP. A genome-wide small interfering RNA screen identifies host factors required for vesicular stomatitis virus infection. J Virol. 2014;88:8355–8360. doi: 10.1128/JVI.00642-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edgil D, Diamond MS, Holden KL, Paranjape SM, Harris E. Translation efficiency determines differences in cellular infection among dengue virus type 2 strains. Virology. 2003;317:275–290. doi: 10.1016/j.virol.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 40.Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 41.Asrat S, Dugan AS, Isberg RR. The frustrated host response to Legionella pneumophila is bypassed by MyD88-dependent translation of pro-inflammatory cytokines. PLoS Pathog. 2014;10:e1004229. doi: 10.1371/journal.ppat.1004229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sidrauski C, et al. Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the integrated stress response. eLife. 2015;4:e07314. doi: 10.7554/eLife.07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Connor JH, Lyles DS. Inhibition of host and viral translation during vesicular stomatitis virus infection. eIF2 is responsible for the inhibition of viral but not host translation. J Biol Chem. 2005;280:13512–13519. doi: 10.1074/jbc.M501156200. [DOI] [PubMed] [Google Scholar]
- 44.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sidrauski C, McGeachy AM, Ingolia NT, Walter P. The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. eLife. 2015;4:e05033. doi: 10.7554/eLife.05033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 47.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 48.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 49.Ventoso I, et al. Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: A strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev. 2006;20:87–100. doi: 10.1101/gad.357006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zanoni I, et al. An endogenous caspase-11 ligand elicits interleukin-1 release from living dendritic cells. Science. 2016;352:1232–1236. doi: 10.1126/science.aaf3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Furlong DB, Nibert ML, Fields BN. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J Virol. 1988;62:246–256. doi: 10.1128/jvi.62.1.246-256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.García-Sastre A, et al. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 53.Cureton DK, Massol RH, Saffarian S, Kirchhausen TL, Whelan SP. Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLoS Pathog. 2009;5:e1000394. doi: 10.1371/journal.ppat.1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whelan SP, Ball LA, Barr JN, Wertz GT. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc Natl Acad Sci USA. 1995;92:8388–8392. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chow J, Marka Z, Bartos I, Marka S, Kagan JC. Environmental stress causes lethal neuro-trauma during asymptomatic viral infections. Cell Host Microbe. 2017;22:48–60.e45. doi: 10.1016/j.chom.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee AS, Burdeinick-Kerr R, Whelan SP. A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc Natl Acad Sci USA. 2013;110:324–329. doi: 10.1073/pnas.1216454109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakahira K, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 59.Tan Z, et al. A new panel of NS1 antibodies for easy detection and titration of influenza A virus. J Med Virol. 2010;82:467–475. doi: 10.1002/jmv.21709. [DOI] [PubMed] [Google Scholar]
- 60.Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang GP, Bushman FD. A statistical method for comparing viral growth curves. J Virol Methods. 2006;135:118–123. doi: 10.1016/j.jviromet.2006.02.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.