Significance
Understanding the functional diversity of specific microbial groups at the global scale is critical yet poorly developed. By combining the considerable knowledge accumulated through recent years on the molecular bases of photosynthetic pigment diversity in marine Synechococcus, a major phytoplanktonic organism, with the wealth of metagenomic data provided by the Tara Oceans expedition, we have been able to reliably quantify all known pigment types along its transect and provide a global distribution map. Unexpectedly, cells able to dynamically change their pigment content to match the ambient light color were ubiquitous and predominated in many environments. Altogether, our results unveiled the role of adaptation to light quality on niche partitioning in a key primary producer.
Keywords: marine cyanobacteria, metagenomics, light quality, phycobilisome, Tara Oceans
Abstract
Marine Synechococcus cyanobacteria are major contributors to global oceanic primary production and exhibit a unique diversity of photosynthetic pigments, allowing them to exploit a wide range of light niches. However, the relationship between pigment content and niche partitioning has remained largely undetermined due to the lack of a single-genetic marker resolving all pigment types (PTs). Here, we developed and employed a robust method based on three distinct marker genes (cpcBA, mpeBA, and mpeW) to estimate the relative abundance of all known Synechococcus PTs from metagenomes. Analysis of the Tara Oceans dataset allowed us to reveal the global distribution of Synechococcus PTs and to define their environmental niches. Green-light specialists (PT 3a) dominated in warm, green equatorial waters, whereas blue-light specialists (PT 3c) were particularly abundant in oligotrophic areas. Type IV chromatic acclimaters (CA4-A/B), which are able to dynamically modify their light absorption properties to maximally absorb green or blue light, were unexpectedly the most abundant PT in our dataset and predominated at depth and high latitudes. We also identified populations in which CA4 might be nonfunctional due to the lack of specific CA4 genes, notably in warm high-nutrient low-chlorophyll areas. Major ecotypes within clades I–IV and CRD1 were preferentially associated with a particular PT, while others exhibited a wide range of PTs. Altogether, this study provides important insights into the ecology of Synechococcus and highlights the complex interactions between vertical phylogeny, pigmentation, and environmental parameters that shape Synechococcus community structure and evolution.
Marine Synechococcus is the second most abundant phytoplankton group in the world’s oceans and constitutes a major contributor to global primary production and carbon cycling (1, 2). This genus displays a wide genetic diversity, and several studies have shown that, among the ∼20 clades defined based on various genetic markers, five (clades I–IV and CRD1) predominate in situ and can be broadly associated with distinct sets of physicochemical parameters (3–5). In a recent study, we further defined ecologically significant taxonomic units (ESTUs), that is, organisms belonging to the same clade and co-occurring in the field, and highlighted that the three main parameters affecting the in situ distribution of these ESTUs were temperature and availability of iron and phosphorus (6). However, marine Synechococcus also display a wide pigment diversity, suggesting that light could also influence their ecological distribution, both qualitatively and quantitatively (7, 8).
This pigment diversity comes from differences in the composition of their main light-harvesting antennae, called phycobilisomes (PBSs) (7–9). These water-soluble macromolecular complexes consist of a central core anchoring at least six radiating rods made of several distinct phycobiliproteins, that is, proteins to which specific enzymes (phycobilin lyases) covalently attach chromophores called phycobilins (7, 10). Although the PBS core is conserved in all marine Synechococcus, rods have a very variable composition, and three main pigment types (PTs) are usually distinguished (Fig. S1) (7, 11). In PT 1, PBS rods are solely made of phycocyanin (PC) (encoded by the cpcBA operon) and bear the red-light–absorbing phycocyanobilin (PCB) (Amax = 620 nm) as the sole chromophore. In PT 2, rods are made of PC and phycoerythrin I (PE-I) (encoded by cpeBA) and attach both PCB and the green-light–absorbing phycoerythrobilin (PEB) (Amax = 550 nm). All other marine Synechococcus belong to PT 3 and have rods made of PC, PE-I, and PE-II (encoded by mpeBA) that bind PCB, PEB, and the blue-light–absorbing phycourobilin (PUB) (Amax = 495 nm; Fig. S1). Several subtypes can be defined within PT 3, based on the fluorescence excitation ratio at 495 and 545 nm (hereafter, Ex495:545; Fig. S1), a proxy for the PUB:PEB ratio. This ratio is low (Ex495:545 < 0.6) in subtype 3a (green-light specialists), intermediate in subtype 3b (0.6 ≤ Ex495:545 < 1.6), and high (Ex495:545 ≥ 1.6) in subtype 3c (blue-light specialists) (7, 11). Additionally, strains of subtype 3d are able to change their PUB:PEB ratio depending on ambient light color, a process called type IV chromatic acclimation (hereafter CA4), allowing them to maximally absorb blue or green light (11–14). Comparative genomic analyses showed that genes involved in the synthesis and regulation of PBS rods are gathered into a dedicated genomic region, the content and organization of which correspond to the different PTs (7). Similarly, chromatic acclimation has been correlated with the presence of a small specific genomic island (CA4 genomic island) that exists in two distinct configurations (CA4-A and -B) (11). Both contain two regulators (fciA and fciB) and a phycobilin lyase (mpeZ in CA4-A or mpeW in CA4-B), thus defining two distinct CA4 genotypes: 3dA and 3dB, respectively (11, 14, 15). Finally, some strains possess a complete or partial CA4 genomic island but are not able to perform CA4, displaying a fixed Ex495:545 corresponding to 3a, 3b, or 3c phenotypes (11).
As there is no correspondence between pigmentation and core genome phylogeny (7, 16, 17), deciphering the relative abundance and niche partitioning of Synechococcus PTs in the environment requires specific approaches. In the past 30 y, studies have been based either on (i) proxies of the PUB:PEB ratio as assessed by flow cytometry (18–20), fluorescence excitation spectra (21–27), and epifluorescence microscopy (28); or (ii) phylogenetic analyses of cpcBA or cpeBA (17, 29–34). These studies showed that PT 1 is restricted to and dominates in low salinity surface waters and/or estuaries, which are characterized by a high turbidity resulting in a red-wavelengths–dominated light field (18, 22, 31–38), whereas PT 2 is found in coastal shelf waters or in the transition zones between brackish and oceanic environments with intermediate optical properties (18, 27, 34, 36–39). Finally, PT 3 with increasing PUB:PEB ratio are found over gradients from onshore mesotrophic waters, characterized by green-light dominance, to offshore oligotrophic waters, where blue light penetrates the deepest (19–24, 28, 36, 38, 40). Some authors reported an increase in the PUB:PEB ratio with depth (19, 21, 24), while others observed a constant ratio throughout the water column, a variability potentially linked to the location, water column features, and/or environmental parameters (22, 25, 28).
However, these analyses based on optical properties could only describe the distribution of high- and low-PUB populations without being able to differentiate green- (3a) or blue-light (3c) specialists from CA4 cells (3d) acclimated to green or blue light, while genetic analysis solely based on cpcBA and/or cpeBA could not differentiate all PTs. For instance, only two studies have reported CA4 populations in situ either in the western English Channel (17) or in subpolar waters of the western Pacific Ocean (29), but none of them was able to differentiate CA4-B from high-PUB (i.e., 3c) populations. As a consequence, the global relative abundance of the different Synechococcus PTs, particularly CA4, and the link between genetic and pigment diversity have remained largely unclear.
Here, we analyzed 109 metagenomic samples collected from all major oceanic basins during the 2.5-y Tara Oceans (2009–2011) expedition (41) using a bioinformatic pipeline combining a metagenomic read recruitment approach (6, 42) to recruit single reads from multiple PBS gene markers and placement of these reads in reference trees to assign them to a given PT. This pipeline allowed a description of the worldwide distribution of all known Synechococcus PTs, as well as of their realized environmental niches (sensu ref. 43). This study provides a synoptic view of how a major photosynthetic organism adapts to natural light color gradients in the ocean.
Results
A Robust Approach for Estimating Pigment Type Abundance from Metagenomes.
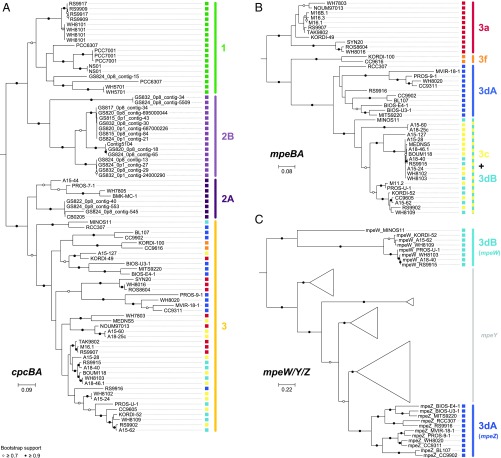
We developed a multimarker approach combining phylogenetic information retrieved from three different genes or operons (cpcBA, mpeBA, and mpeW; Fig. 1 and Datasets S1 and S2) to overcome the issue of fully resolving the whole range of PTs. While cpcBA discriminated PT 1, 2, and 3 (Fig. 1A), only the mpeBA operon, a PT 3-specific marker, was able to distinguish the different PT 3 subtypes (Fig. 1B), although as for cpeBA it could not differentiate PT 3dB (CA4-B) from PT 3c (i.e., blue light specialists) (11, 29). The mpeW marker was thus selected to specifically target PT 3dB and, by subtraction, enumerate PT 3c (Fig. 1C). Using the cpcBA marker, members of PT 2 were split into two well-defined clusters, 2A and 2B (Fig. 1A), the latter corresponding to a purely environmental PT identified from assembled metagenomes of the Baltic Sea (38). Strains KORDI-100 and CC9616 also clustered apart from other strains in the mpeBA phylogeny, suggesting that they have a divergent evolutionary history from other PT 3 members (Fig. 1B). This is supported by the diverged gene content and order of their PBS rod genomic region, and these strains were recently referred to as PT 3f, even though they have a similar phenotype as PT 3c (Ex495:545 ratio ≥ 1.6) (30). To investigate the phylogenetic resolution of small fragments of these three markers, sequences were removed one at a time from the reference database, and simulated reads (150 bp long compared with 164 bp in average for Tara Oceans cleaned/merged reads) generated from this sequence were assigned using our bioinformatic pipeline against a database comprising the remaining sequences. Inferred and known PTs were then compared. The percentage of simulated reads assigned to the correct PT was between 93.2% and 97.0% for all three markers, with less than 2.1–5.6% of reads that could not be classified and an error rate below 2%, showing that all three markers display a sufficient resolution to reliably assign the different PTs (Fig. S2 B, D, and F).
Fig. 1.
Maximum-likelihood phylogenetic trees of (A) cpcBA operon, (B) mpeBA operon, and (C) the mpeW/Y/Z gene family. The cpcBA tree includes both strains with characterized pigment type (PT) and environmental sequences (prefixed with GS) assembled from metagenomes of the Baltic Sea (38). Circles at nodes indicate bootstrap support (black, >90%; white, >70%). Note that, for PT 2B clade, only environmental sequences are available. The PT associated with each sequence is indicated as a colored square. The scale bars represent the number of substitutions per nucleotide position. Please note that there are two to four copies of the cpcBA operon in PT 1, which are either identical or closely related.
To ensure that the different markers could be quantitatively compared in a real dataset, we examined the correlations between estimates of PT abundances using the different markers in the 109 metagenomes analyzed in this study. Total cpcBA counts were highly correlated (R2 = 0.994, n = 109; Fig. S3A) with total Synechococcus counts obtained with the petB gene, which was previously used to study the phylogeography of marine picocyanobacteria (6), and the correlation slope was not significantly different from 1 (slope, 1.040; Wilcoxon’s paired difference test P value = 0.356). cpcBA is thus as good as petB at capturing the total population of Synechococcus reads. Moreover, counts of cpcBA reads assigned to PT 3 and total mpeBA counts (specific for PT 3) were also strongly correlated (R2 = 0.996, n = 109; Fig. S3B), and not skewed from 1 (slope of 0.991; Wilcoxon’s P value = 0.607). Although no redundant information for PT 3dB is available with the three selected markers, another marker targeting 3dB (fciAB) was tested and produced results similar to mpeW (Fig. S3C). These results demonstrate that our multimarker approach can be used to reliably and quantitatively infer the different Synechococcus PTs from short metagenomic reads, with PT 1, 2A, and 2B abundances being assessed by cpcBA normalized counts; PT 3a, 3f, and 3dA by mpeBA normalized counts; PT 3dB by mpeW normalized counts; and PT 3c by the difference between mpeBA normalized counts for 3c + 3dB and mpeW normalized counts. We thus used this approach on the Tara Oceans metagenomes, generated from 109 samples collected at 65 stations located in the major oceanic basins (Fig. 2).
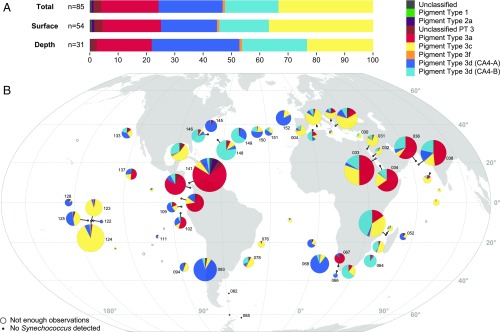
Fig. 2.
Distribution of Synechococcus pigment types (PTs). (A) Relative abundance of each PT in the whole dataset (Total), in surface, and at the deep chlorophyll maximum (DCM). (B) Map showing the global distribution of all Synechococcus PTs in surface waters along the Tara Oceans transect. Diameters of pies are proportional to the number of cpcBA reads normalized by the sequencing effort. Stations with less than 30 cpcBA or mpeBA reads are indicated by open circles, and those with no cpcBA reads by black dots. Numbers next to pies correspond to Tara Oceans stations.
CA4 Populations Are Widespread and Predominate at Depth and High Latitudes.
The latitudinal distribution of Synechococcus inferred from cpcBA counts is globally consistent with previous studies (2, 6, 44), with Synechococcus being present in most oceanic waters, but quasi-absent (<20 cpcBA counts) beyond 60°S (Southern Ocean stations TARA_082 to TARA_085; Fig. 2B). Overall, the number of recruited cpcBA reads per station was between 0 and 8,151 (n = 63; median, 449; mean, 924; SD, 1,478) for surface and 0 and 3,200 (n = 46; median, 170; mean, 446; SD, 664) for deep chlorophyll maximum (DCM) samples, respectively. Stations with less than 30 cpcBA reads were excluded from further analysis.
PT 1 and 2, both of which are known to be mostly found and abundant in coastal waters (29, 36, 38, 45), were expectedly almost absent from this dataset (total of 15 and 513 cpcBA reads, respectively; Fig. 2) since the Tara cruise sampling was principally performed in oceanic waters. While PT 2A was mostly found at the surface at one station off Panama (TARA_141, 417 out of 6,637 reads at this station; Fig. 2B), PT 2B was virtually absent (total of 3 cpcBA reads) from our dataset and might thus be confined to the Baltic Sea (38). This low abundance of PT 1 and 2B precluded the correlation analysis between their distribution and physicochemical parameters. PT 3 was by far the most abundant along the Tara Oceans transect, accounting for 99.1 ± 1.4% (mean ± SD) of cpcBA reads at stations with ≥30 cpcBA read counts. Interestingly, several PT 3 subtypes often co-occurred at a given station.
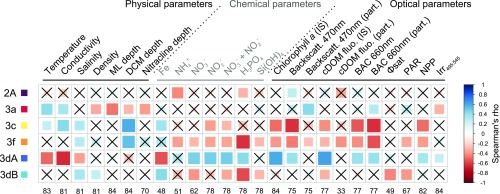
PT 3a (green-light specialists) totaled 20.3% of read counts, with similar abundance between surface (20.5%) and DCM (19.4%) samples, and was particularly abundant in intertropical oceanic borders and regional seas, including the Red Sea, the Arabian Sea, and the Panama/Gulf of Mexico area (Fig. 2B). Correlation analyses show that this PT is consistently associated with high temperatures but also with greenish (as estimated from a low blue to green downwelling irradiance ratio, Irr495:545), particle-rich waters (high particle backscattering at 470 nm and beam attenuation coefficient at 660 nm; Fig. 3). Still, in contrast with previous studies that reported the distribution of low-PUB populations (19, 21, 23, 24, 26, 27), this PT does not seem to be restricted to coastal waters, explaining its absence of correlation with chlorophyll concentration and colored dissolved organic matter (cDOM).
Fig. 3.
Correlation analysis between Synechococcus pigment types (PTs) and environmental parameters measured along the Tara Oceans transect for all sampled depths. The scale shows the degree of correlation (blue) or anticorrelation (red) between two variables. Nonsignificant correlations (adjusted P value > 0.05) are indicated by crosses. Number of observations for each environmental parameter is indicated at Bottom. Abbreviations: BAC, beam attenuation coefficient; Backscatt., backscattering; cDOM fluo., colored dissolved organic matter fluorescence; DCM, deep chlorophyll maximum; Irr495:545, ratio of downwelling irradiance at 495 and 545 nm; IS, in situ; MLD, mixed layer depth; NPP, net primary production; PAR, photosynthetically active radiation; part., particulate; Φsat, satellite-based nonphotochemical quenching (NPQ)-corrected quantum yield of fluorescence (proxy for iron limitation) (6).
Blue light specialists (PT 3c) appear to be globally widespread, with the exception of high-latitude North Atlantic waters, and accounted for 33.4% of reads, with a higher relative abundance at the surface (36.8%) than at the DCM (23.3%; Fig. 2A). This PT is dominant in transparent, oligotrophic, iron-replete areas such as the Mediterranean Sea as well as South Atlantic and Indian Ocean gyres (Figs. 2B and 4C). In the South Pacific, PT 3c was also found to be predominant in the Marquesas Islands area (TARA_123 and 124), where the coast proximity induced a local iron enrichment (6). Consistently, PT 3c was found to be positively associated with iron concentration, high temperature, and DCM depth, and anticorrelated with chlorophyll fluorescence, nitrogen concentrations, net primary production (NPP), as well as other related optical parameters, such as backscattering at 470 nm and beam attenuation coefficient at 660 nm (Fig. 3). Despite its rarity, PT 3f seems to thrive in a similar environment, with the highest relative abundances in the Indian Ocean and Mediterranean Sea (Figs. 2B and 4C). Its occurrence in the latter area might explain its strong anticorrelation with phosphorus availability.
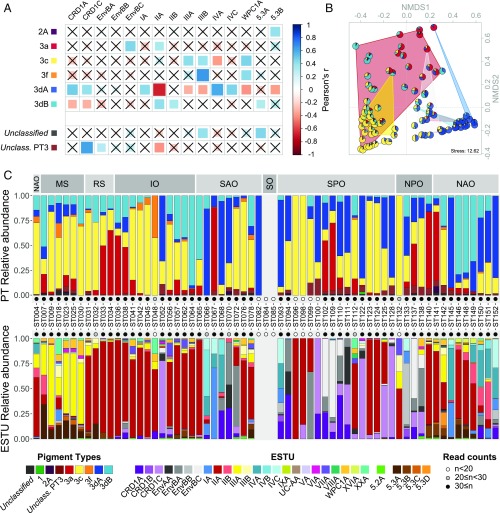
Fig. 4.
Relationship between Synechococcus pigment types (PTs) and ecologically significant taxonomic units (ESTUs) (as defined in ref. 6). (A) Correlation analysis between Synechococcus PTs and the most abundant ESTUs (>1% relative abundance) for all sampled depths (the complete dataset is shown in Fig. S5). Nonsignificant correlations (adjusted P value > 0.05) are indicated by crosses. The surface of station TARA_067, identified as an outlier (Fig. S7), was removed for this analysis. (B) NMDS analysis of stations according to Bray–Curtis distance between PT assemblages. Samples that belong to the same ESTU assemblage have been contoured with a background color according to the color code used in ref. 6, namely, red, assemblage 1 dominated by ESTU IIA; yellow, assemblage 2 dominated by ESTU IIIA; dark blue, assemblage 4 dominated by ESTUs IA and IVA/B; pink, assemblage 5 codominated by ESTUs IIB and IVA/B; gray, assemblage 6 codominated by ESTUs CRD1C and EnvBC; light blue, assemblage 8 codominated by ESTUs IVA/B, EnvBB, and CRD1A/B. (C) PT and ESTU relative abundance at each surface station along the Tara Oceans transect. Oceanic provinces are indicated in the Top gray panels. IO, Indian Ocean; MS, Mediterranean Sea; NAO, North Atlantic Ocean; NPO, North Pacific Ocean; RS, Red Sea; SAO, South Atlantic Ocean; SO, Southern Ocean; SPO, South Pacific Ocean.
Both CA4 types, 3dA and 3dB, which represented 22.6% and 18.9% of reads, respectively, were unexpectedly widespread and could locally account for up to 95% of the total Synechococcus population (Figs. 2 and 4C and Fig. S4). In contrast to blue- and green-light specialists, both CA4 types were proportionally less abundant at the surface (19.8% and 17.5%, for 3dA and 3dB, respectively) than at depth (30.9% and 22.9%). Interestingly, PT 3dA and 3dB generally displayed complementary distributions along the Tara Oceans transect (Fig. 2B). PT 3dA was predominant at high latitude in the Northern Hemisphere as well as in other vertically mixed waters such as in the Chilean upwelling (TARA_093) or in the Agulhas current (TARA_066 and 68; Fig. 2B). Accordingly, PT 3dA distribution seems to be driven by low-temperature, high-nutrient, and highly productive waters (high NPP, chlorophyll a, and optical parameters), a combination of physicochemical parameters almost opposite to those observed for blue-light specialists (PT 3c; Fig. 3). In contrast, PT 3dB shares a number of characteristics with PT 3c, including the anticorrelation with nitrogen concentration and association with iron availability (as indicated by both a positive correlation with [Fe] and negative correlation with the iron limitation proxy Φsat; Fig. 3), consistent with their widespread occurrence in iron-replete oceanic areas. Also noteworthy, PT 3dB was one of the sole PTs (with 3f) to be associated with low photosynthetically available radiation (PAR).
Niche Partitioning of Synechococcus Populations Relies on a Subtle Combination of ESTU and PT Niches.
We previously showed that temperature and iron and phosphorus availability constituted major factors influencing the diversification and niche partitioning of Synechococcus ESTUs (i.e., genetically related subgroups within clades that co-occur in the field) (6). However, these results cannot be extended to PTs since the pigment content does not follow the vertical phylogeny (7). To decipher the respective roles of genetic and pigment diversity in Synechococcus community structure, we examined the relationships between ESTUs and PTs in situ abundances through correlation and nonmetric multidimensional scaling (NMDS) analyses (Fig. 4 A and B) and compared their respective distributions (Fig. 4C and Fig. S4).
Interestingly, all PTs are either preferentially associated with or excluded from a subset of ESTUs. PT 2A is found at low abundance at a few stations along the Tara Oceans transect, and, when present, it is seemingly associated with the rare ESTU 5.3B (Fig. 4A), an unusual PT/ESTU combination so far only observed in metagenomes from freshwater reservoirs (46). PT 3a is associated with ESTUs EnvBC (occurring in low-iron areas) and IIA, the major ESTU in the global ocean (Fig. 4A), a result consistent with NMDS analysis, which shows that PT 3a is found in assemblages dominated by these two ESTUs (indicated by red and gray backgrounds in Fig. 4B), as well as with independent observations on cultured strains (Dataset S3). PT 3c is associated with ESTU IIIA (the dominant ESTU in P-depleted areas), as observed on many isolates (Dataset S3), and is also linked, like PT 3f, with ESTUs IIIB and WPC1A, both present at lower abundance than IIIA in P-poor waters (Fig. 4A). PT 3f is also associated with the newly described and low-abundance ESTU XXA (previously EnvC; Fig. S5) (4, 6). Both PT 3f and ESTU XXA were rare in our dataset but systematically co-occurred, in agreement with the fact that the only culture representative of the latter clade belongs to PT 3f (Dataset S3).
PT 3dA appears to be associated with all ESTUs from clades CRD1 (specific to iron-depleted areas) as well as with those representative of coastal and cold waters (IA, IVA, IVC) but is anticorrelated with most other major ESTUs (IIA, IIIA and -B, WPC1A, and 5.3B; Fig. 4A). This pattern is opposite to PT 3dB, which is preferentially found associated with ESTU IIA, IIB, and 5.3A, but not in CRD1A or -C (Fig. 4A). Thus, it seems that the two types of CA4 are found in distinct and complementary sets of ESTUs. Interestingly, our analysis might suggest the occurrence of additional PTs not isolated so far, since a number of reads (0.7% and 2.7% of cpcBA and mpeBA counts, respectively; Fig. 2A) could not be assigned to any known PTs. For instance, while most CRD1C seem preferentially associated with PT 3dA, a fraction of the population could only be assigned at the PT 3 level (Fig. 4A). Similarly, a number of reads could not be assigned to any known PT in stations rich in ESTU 5.3A and XXA, although one cannot exclude that this observation might be due to a low number of representative strains, and thus PT reference sequences, for these ESTUs.
The preferred association of PTs with specific ESTUs is also well illustrated by some concomitant shifts of PTs and ESTU assemblages. For instance, in the wintertime North Atlantic Ocean, the shift from 3dB-dominated stations on the western side (TARA_142 and TARA_146 to 149) to 3dA-dominated stations near European coasts (TARA_150 to 152) and North of Gulf Stream (TARA_145) is probably related to the shift in ESTU assemblages occurring along this transect, with ESTU IIA being gradually replaced by ESTU IVA (Fig. 4C) (6). Similarly, the takeover of CRD1C by IIA in the Marquesas Island area (TARA_123 to 125), which is iron-enriched with regard to surrounding high-nutrient low-chlorophyll (HNLC) waters (TARA_122 and 128), perfectly matched the corresponding replacement of PT 3dA by 3c. However, in several other cases, PT shifts were not associated with a concomitant ESTU shift or vice versa. One of clearest examples of these dissociations is the transect from the Mediterranean Sea to the Indian Ocean, where the entry in the northern Red Sea through the Suez Canal triggered a sharp shift from a IIIA- to a IIA-dominated community (TARA_030 and 031), which was not accompanied by any obvious change in PTs. Conversely, a sharp rise in the relative abundance of PT 3a was observed in the southern Red Sea/northeastern Indian Ocean (TARA_033 to 038) without changes in the large dominance of ESTU IIA. Altogether, this strongly suggests that a subtle combination of ESTUs and PTs respective niche occupancy is responsible for the observed niche partitioning of Synechococcus populations.
Deficient Chromatic Acclimaters Are Dominant in HNLC Areas.
Although our results clearly indicate that CA4 cells represent a large proportion of the Synechococcus community in a wide range of ecological niches, this must be somewhat tempered by the fact that, in culture, about 30% of the strains possessing a CA4-A or -B genomic island are not able to chromatically acclimate (Dataset S3) (11). Some of these natural mutants have an incomplete CA4 genomic island (Fig. S6K). For example, strains WH8016 (ESTU IA) and KORDI-49 (WPC1A) both lack the CA4-A–specific lyase-isomerase MpeZ, an enzyme shown to bind a PUB molecule on PE-II (14), and display a green-light specialist phenotype (PT 3a, Ex495:545 ∼ 0.4) whatever the ambient light color (11). However, since they possess a PT 3a mpeBA allele, reads from field WH8016- or KORDI-49–like cells are adequately counted as PT 3a (Fig. S6K). Another CA4-deficient strain, BIOS-E4-1 (ESTU CRD1C), possesses mpeZ and a 3dA mpeBA allele but lacks the CA4 regulators FciA and FciB as well as the putative lyase MpeY and exhibits a fixed blue-light specialist phenotype (PT 3c, Ex495:545 ∼ 1.7; Fig. S6K) (11, 15). Thus, reads from such natural Synechococcus CA4-incapable mutants in the field are counted as 3dA using the mpeBA marker. Last, the strain MVIR-18-1 possesses a complete CA4-A island and a 3dA mpeBA allele but lacks mpeU, a gene necessary for blue-light acclimation (Fig. S6K) (47). While MVIR-18-1 displays a fixed green-light phenotype, reads from such Synechococcus are also erroneously counted as 3dA.
To assess the significance of these genotypes in the field, we compared the normalized read counts obtained for 3dA with mpeBA, fciAB, mpeZ, mpeU, and mpeY (Fig. S6 A–J). Overall, this analysis revealed a high consistency between these different markers (0.860 < R2 < 0.986), indicating that most mpeZ-containing populations also contained 3dA alleles for fciAB, mpeY, mpeU, and mpeBA and are therefore likely able to perform CA4. However, a number of stations, all located in HNLC areas (TARA_094, 111, and 122–128 in the Pacific Ocean and TARA_052 located northwest of Madagascar; Fig. 2B), displayed more than 10-fold higher mpeBA, mpeU, and mpeZ counts than fciAB and mpeY counts (Fig. S6 A, B, E, F, H, and I). This indicates that a large proportion or even the whole population (TARA_122 and 124) of 3dA in these HNLC areas is probably lacking the FciA/B regulators and MpeY and, like strain BIOS-E4-1 (Fig. S6K), might thus be stuck in the blue-light specialist phenotype (PT 3c) (11). Conversely, station TARA_067 exhibited consistently more than twice the fciAB and mpeZ counts compared with mpeBA, mpeY, or mpeU (Fig. S6 B–E, G, and H) and was a clear outlier when comparing pigment type and clade composition (Fig. S7). This suggests that the proportion of PT 3dA might have been underestimated at this station, as a significant proportion of this population probably corresponds to PT 3a genotypes that have acquired a CA4-A island by lateral gene transfer, as is seemingly the case for strains WH8016 and KORDI-49. Finally, no station exhibited markedly lower mpeU counts compared with all other genes, indicating that the genotype of strain MVIR-18-1 is probably rare in the oceans.
It must be noted that two out of the six sequenced CA4-B strains (WH8103 and WH8109) also have a deficient CA4 phenotype and display a constant, intermediate Ex495:545 ratio (0.7 and 1, respectively), despite any obvious PBS- or CA4-related gene deletion (11). Accordingly, the plot of 3dB normalized read counts obtained with mpeW vs. fciAB shows no clear outlier (Fig. S3C).
Discussion
Marine Synechococcus display a large pigment diversity, with different PTs preferentially harvesting distinct regions of the light spectrum. Previous studies based on optical properties or on a single genetic marker could not differentiate all PTs (17, 29–31), and thus neither assess their respective realized environmental niches (43) nor the role of light quality on the relative abundance of each PT. Here, we showed that a metagenomic read recruitment approach combining three genetic markers can be used to reliably predict all major PTs. Applied to the extensive Tara Oceans dataset, this original approach, which avoids PCR amplification and cloning biases, allowed us to describe the distribution of the different Synechococcus PTs at the global scale and to refine our understanding of their ecology.
PT 3 was found to be largely dominant over PT 1 and 2 along the oceanic Tara Oceans transect, in agreement with the coastal-restricted distribution of the latter PTs (18, 22, 27, 31–34, 37–39). Biogeography and correlation analyses with environmental parameters provided several important insights concerning niche partitioning of PT 3 subtypes. Green- (PT 3a) and blue-light (PT 3c) specialists were both shown to dominate in warm areas but display clearly distinct niches, with 3a dominating in Synechococcus-rich stations located on oceanic borders, while 3c predominated in purely oceanic areas where the global abundance of Synechococcus is low. These results are in agreement with the prevailing view of an increase in the PUB:PEB ratio from green onshore mesotrophic waters to blue offshore oligotrophic waters (19–24, 26–29, 40, 48). Similarly, we showed that PT 3dB, which could not be distinguished from PT 3c in previous studies (17, 29–31), prevails in more coastal and/or mixed temperate waters than do 3c populations. The realized environmental niche of the second type of CA4 (PT 3dA) is the best defined of all PTs as it is clearly associated with nutrient-rich waters and with the coldest stations of our dataset, occurring at high latitude, at depth, and/or in vertically mixed waters (e.g., TARA_068, 093, and 133). This result is consistent with a recent study demonstrating the dominance of 3dA in subarctic waters of the Northwest Pacific Ocean (29), suggesting that the prevalence of 3dA at high latitude can be generalized. The decrease of PT 3c (blue-light specialists) with depth is unexpected given previous reports of a constant (22, 25, 28, 49) or increasing (19, 21, 24) PUB:PEB ratio throughout the water column. However, the high abundance of CA4 can reconcile these observations with the decreased abundance of PT 3c, as cells capable of CA4 likely have a blue-light phenotype at depth. Altogether, while little was previously known about the abundance and distribution of CA4 populations in the field, here we show that they are ubiquitous, dominate in a wide range of niches, are present both in coastal and oceanic mixed waters, and overall are the most abundant Synechococcus PT.
The relationship between ESTUs and PTs shows that some ESTUs are preferentially associated with only one PT, while others present a much larger pigment diversity. ESTU IIA, the most abundant and ubiquitous ESTU in the field (5, 6), displays the widest PT diversity (Fig. 4B), a finding confirmed by clade II isolates spanning the largest diversity of pigment content, with representative strains of PT 2A, 3a, 3c, and 3dB within this clade (Dataset S3) (7, 11, 50–52). This suggests that this ESTU can colonize all light color niches, an ability that might be partially responsible for its global ecological success. Our current results do not support the previously observed correlation between clade III and PT 3a (29) since the two ESTUs defined within this clade (IIIA and B) were associated with PT 3c and/or 3f. This discrepancy could be due either to the different methods used in these studies or to the occurrence of genetically distinct clade III populations in coastal areas of the northwestern Pacific Ocean and along the Tara Oceans transect. However, the pigment phenotype of strains isolated to date is more consistent with our findings (Dataset S3) (16, 36).
In contrast to most other PTs, the association between PT 3dA and ESTUs was found to be nearly exclusive in the field, as ESTUs from clades I, IV, CRD1, and EnvA were not associated with any other PT, and reciprocally PT 3dA is only associated with these clades (Fig. 4A). An interesting exception to this general rule was observed in the Benguela upwelling (TARA_ 067), where the dominant ESTU IA population both displays a 3a mpeBA allele and possesses fciA/B and mpeZ genes (Figs. S6K and S7), suggesting that cells, which were initially green-light specialists (PT 3a), have inherited a complete CA4-A island through lateral gene transfer at this station. Interestingly, among the seven clade I strains sequenced to date, three possess a 3a mpeBA allele, among which WH8016 also has a CA4-A island but only partial (lacking mpeZ) and therefore not functional (11). It is thus difficult to conclude whether the lateral transfer of this island, likely a rare event since it was only observed in populations of the Benguela upwelling, has conferred these populations the ability to perform CA4.
Another important result of this study was the unsuspected importance of populations that have likely lost the ability to chromatically acclimate, specifically in warm HNLC areas, which cover wide expanses of the South Pacific Ocean (53). Interestingly, populations living in these ultra-oligotrophic environments have a different genetic basis for their consistently elevated PUB phenotype than do typical blue-light specialists (i.e., PT 3c), since they have lost the CA4 regulators fciA/B and accumulated mutations in mpeY, a yet-uncharacterized member of the phycobilin lyase family, as observed in strain BIOS-E4-1 (Fig. S6K) (11). This finding, consistent with the previous observation that the south Pacific is dominated by high-PUB Synechococcus (22), is further supported by the recent sequencing of three isolates from the Equatorial Pacific, strains MITS9504, MITS9509 (both CRD1C), and MITS9508 (CRD1A) (54), all of which contain, like BIOS-E4-1, a 3dA mpeBA allele, a CA4-A island lacking fciA/B, and a partial (MITS9508) or highly degenerated (two other MIT strains) mpeY gene sequence (Fig. S6K). Thus, these natural CA4-A mutants seem to have adapted to blue, ultra-oligotrophic waters by inactivating a likely energetically costly acclimation mechanism (positive selection), although we cannot exclude that it might be a consequence of the lower selection efficiency associated to the reduced effective population size of Synechococcus in such an extreme environment (genetic drift). If, as we hypothesize, all Synechococcus cells counted as 3dA at these stations are CA4-deficient, these natural mutants would represent about 15% of the total 3dA population. In contrast, CRD1-A populations of the eastern border of the Pacific Ocean (TARA_102, 109–110, 137) are likely true CA4 populations as they possess all CA4 genes (Fig. S6K).
In conclusion, our study provided insights into the distribution, ecology, and adaptive value of all known Synechococcus PTs. Unexpectedly, the sum of 3dA and 3dB constituted about 40% of the total Synechococcus counts in the Tara Oceans dataset, making chromatic acclimaters (PT 3d) the most globally abundant PT, even when taking into account potential CA4-deficient natural mutants. In addition, this PT made up 95% of the Synechococcus population at high latitudes and was present in every one of the five major clades in the field (I, II, III, IV, and CRD1). This suggests that chromatic acclimation likely confers a strong adaptive advantage compared with strains with a fixed pigmentation, particularly in vertically mixed environments and at depth at stations with a stratified water column. The occurrence of natural CA4 mutants and evidence for lateral transfer of the CA4 genomic island further support previous hypotheses that not only temperature and nutrient availability (3, 5, 6) but also light quality (7, 52) coexert selective pressures affecting marine Synechococcus evolution. Thus, changes in pigment diversity could occur in response to changes in light niches by acquisition or loss of specific PBS synthesis and/or regulation genes, as previously observed for phosphorus and nitrogen transport genes in Prochlorococcus (55–57). Still, the complex interactions between PTs, vertical phylogeny, and environmental parameters remain unclear and more work is needed to refine our understanding of the balance between the forces shaping community composition and Synechococcus evolution. At the boundaries of Synechococcus environmental niche(s), where the harshest conditions are encountered, both pigment and clade diversity are drastically reduced, and this concomitant reduction tends to support a coselection by light quality and other environmental parameters. On the contrary, the diverse PTs occurring within some clades, as well as the co-occurrence of different PTs at most stations compared with more clearcut clade shifts (e.g., in the Red Sea/Indian Ocean) might indicate that light quality is not the strongest selective force or that light changes are too transient to allow the dominance and fixation of a particular PT in a population. Future experimental work exploring the fitness of distinct ESTU/PT combinations under different controlled environmental conditions (including temperature, nutrients, and light) might help clarifying the respective effects of these parameters on the diversification of this ecologically important photosynthetic organism.
Materials and Methods
Metagenomic Samples.
This study focused on 109 metagenomic samples corresponding to 65 stations from the worldwide oceans collected during the 2.5-y Tara Oceans circumnavigation (2009–2011). Water sample and sequence processing are the same as in ref. 6. Dataset S4 describes all metagenomic samples with location and sequencing effort. Sequencing depths ranged from 16 × 106 to 258 × 106 reads per sample after quality control and paired-reads merging, and corresponding fragments lengths averaged 164 ± 20 bp (median, 168 bp).
Databases: Reference and Outgroup Sequences.
A reference database comprising the full-length gene or operon nucleotide sequences was generated for each marker used in this study (cpcBA, mpeBA, and mpeW) based on culture isolates with characterized pigment type (Dataset S1). These databases comprised 83 cpcBA sequences (64 unique), including 18 PT 1, 5 PT 2A, 19 PT 2B, and 39 PT 3; 41 mpeBA sequences (all unique), including 11 PT 3a, 2 PT 3f, 11 PT 3dA, and 17 PT 3dB; and 5 unique mpeW sequences. For each marker, a reference alignment was generated with MAFFT L-INS-i, version 6.953b (58), and a reference phylogenetic tree was inferred with PhyML, version 20120412 (GTR + I + G, 10 random starting trees, best of SPR and NNI moves, 500 bootstraps) (59); and drawn using the ETE Toolkit (60).
A database of outgroups was also built, comprising paralogous sequences from marine Synechococcus or Prochlorococcus as well as orthologous sequences from other marine and freshwater organisms retrieved from public databases. For cpcBA and mpeBA, the outgroup databases comprised apcA, apcB, apcD, apcF, and cpeBA from marine Synechococcus, ppeBA from Prochlorococcus, cpcBA and cpeBA from other nonpicocyanobacterial organisms as well as either mpeBA or cpcBA from marine Synechococcus, respectively (Datasets S1 and S2). For mpeW, the outgroup database was made of paralogous genes (mpeZ, mpeY, and cpeY) from marine Synechococcus or Prochlorococcus, as no ortholog could be identified in public databases. Similarly, for mpeY and mpeZ, the outgroup database comprised cpeY, mpeW, as well as mpeZ or mpeY, respectively. The outgroup database for mpeU comprised cpeF paralogous sequences from marine Synechococcus and Prochlorococcus. No outgroup database was used for fciAB, as no paralogs or other distantly related sequences were found either in marine Synechococcus and Prochlorococcus or in public databases.
Read Assignation and Estimation of PT Abundance.
Reads were preselected using BLAST+ (61) with relaxed parameters (blastn, maximum E-value of 1e-5, minimum percent identity of 60%, minimum 75% of read length aligned), using reference sequences as subjects; the selection was then refined by a second BLAST+ round against databases of outgroups: reads with a best hit to outgroup sequences were excluded from downstream analysis. Selected reads were then aligned to the marker reference alignment with MAFFT, version 7.299b (–addfragments –adjustdirectionaccurately), and placed in the marker reference phylogenetic tree with pplacer (62). For each read, pplacer returns a list of possible positions (referred to as placements) at which it can be placed in the tree and their associated “likelihood weight ratio” (LWR) (proxy for the probability of the placement; see pplacer publication and documentation for more details). Reads were then assigned to a pigment type using a custom classifier written in Python. Briefly, internal nodes of the reference tree were assigned a pigment type based on the pigmentation of descending nodes (PT of child reference sequences if the same for all of them, “unclassified” otherwise). For each read, placements were assigned to their nearest ascending or descending node based on their relative position on the edge, and the lowest common ancestor (LCA) of the set of nodes for which the cumulated LWR was greater than 0.95 (LCA of possible placements at 95% probability) was then computed. Finally, the read was assigned to the pigment type of this LCA. Different combinations of read assignment parameters (LCA at 90%, 95%, or 100%; assignation of placements to the ascending, descending, or nearest node) were also assessed, and resulted either in higher rates of unassigned reads or of wrongly assigned reads (Fig. S2).
Read counts were normalized by adjusted marker length: for each marker and each sequence file, counts were normalized by (L − ℓ + 1), with L, the length of the marker gene (cpcBA mean length: 1,053.7 bp; mpeBA mean length: 1,054.6 bp; mpeW mean length: 1,193.3 bp), and ℓ, the mean length of reads in the sequence file. Finally, the abundance of PT 1, 2A, and 2B was defined as the normalized cpcBA read counts of these PTs, the abundance of PT 3a, 3f, and 3dA as the normalized mpeBA read counts of these PTs, 3dB as the normalized mpeW count and 3c as the difference between the normalized mpeBA (3c + 3dB) read count and the PT 3dB count assessed with mpeW. The abundance of unclassified sequences was also taken into account. Detailed petB counts for clade and ESTU abundances were obtained from ref. 6.
Read Assignment Simulations.
For each marker, simulated reads were generated from one reference sequence at a time using a sliding window of 100, 125, or 150 bp (Tara Oceans mean read length, 164.2 bp; median, 169 bp) and steps of 5 bp. Simulated reads were then assigned to a pigment type with the aforementioned bioinformatic pipeline, using all reference sequences except the one used to simulate reads (“leave-one-out” cross-validation scheme). Inferred PTs of simulated fragments were then compared with known PTs of reference sequences.
Statistical Analyses.
All environmental parameters used for statistical analyses are the same as in ref. 6, except the blue to green irradiance ratio that was modeled as described in SI Materials and Methods. Hierarchical clustering and NMDS analyses of stations were performed using R (63) packages cluster, version 1.14.4 (64) and MASS, version 7.3-29 (65), respectively. PT contingency tables were filtered by considering only stations with more than 30 cpcBA reads and 30 mpeBA reads, and only PT appearing in at least two stations and with more than 150 reads in the whole dataset. Contingency tables were normalized using Hellinger transformation that gives lower weights to rare PT. The Bray–Curtis distance was then used for ordination (isoMDS function; maxit, 100; k, 2). Correlations were performed with R package Hmisc_3.17–4 with Benjamini and Hochberg multiple-comparison adjusted P value (66).
Supplementary Material
Acknowledgments
We warmly thank Dr. Annick Bricaud for fruitful discussions on biooptics, members of the Analysis and Bioinformatics for Marine Science (ABiMS) platform (Roscoff) for providing us an efficient storage and computing facility for our bioinformatics analyses, as well as the Natural Environment Research Council Biomolecular Analysis Facility (Centre for Genomic Research, University of Liverpool) for sequencing some Synechococcus genomes used in this study. We also thank the support and commitment of the Tara Oceans coordinators and consortium, Agnès b. and E. Bourgois, the Veolia Environment Foundation, Région Bretagne, Lorient Agglomeration, World Courier, Illumina, the Electricité de France (EDF) Foundation, Fondation pour la Recherche sur la Biodiversité, the Prince Albert II de Monaco Foundation, and the Tara schooner and its captains and crew. Tara Oceans would not exist without continuous support from 23 institutes (https://oceans.taraexpeditions.org/en/). This work was supported by the French “Agence Nationale de la Recherche” Programs SAMOSA (Synechococcus as a model genus for studying adaptation of marine phytoplankton to environmental changes) (Grant ANR-13-ADAP-0010) and France Génomique (Grant ANR-10-INBS-09), the French Government “Investissements d’Avenir” programs World Ocean Bioressources, Biotechnologies and Earth-System Services (OCEANOMICS) (Grant ANR-11-BTBR-0008), the European Union’s Seventh Framework Programs FP7 MicroB3 (Grant Agreement 287589), and Marine Microorganisms: Cultivation Methods for Improving Their Biotechnological Applications (Macumba; Grant Agreement 311975), UK Natural Environment Research Council Grant NE/I00985X/1, and the Spanish Ministry of Science and Innovation Grant MicroOcean PANGENOMICS (GL2011-26848/BOS). This article is contribution number 69 of Tara Oceans.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequencing data and accession numbers are provided in Dataset S1.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717069115/-/DCSupplemental.
References
- 1.Guidi L, et al. Tara Oceans Coordinators Plankton networks driving carbon export in the oligotrophic ocean. Nature. 2016;532:465–470. doi: 10.1038/nature16942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flombaum P, et al. Present and future global distributions of the marine cyanobacteria Prochlorococcus and Synechococcus. Proc Natl Acad Sci USA. 2013;110:9824–9829. doi: 10.1073/pnas.1307701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zwirglmaier K, et al. Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ Microbiol. 2008;10:147–161. doi: 10.1111/j.1462-2920.2007.01440.x. [DOI] [PubMed] [Google Scholar]
- 4.Mazard S, Ostrowski M, Partensky F, Scanlan DJ. Multi-locus sequence analysis, taxonomic resolution and biogeography of marine Synechococcus. Environ Microbiol. 2012;14:372–386. doi: 10.1111/j.1462-2920.2011.02514.x. [DOI] [PubMed] [Google Scholar]
- 5.Sohm JA, et al. Co-occurring Synechococcus ecotypes occupy four major oceanic regimes defined by temperature, macronutrients and iron. ISME J. 2016;10:333–345. doi: 10.1038/ismej.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farrant GK, et al. Delineating ecologically significant taxonomic units from global patterns of marine picocyanobacteria. Proc Natl Acad Sci USA. 2016;113:E3365–E3374. doi: 10.1073/pnas.1524865113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Six C, et al. Diversity and evolution of phycobilisomes in marine Synechococcus spp.: A comparative genomics study. Genome Biol. 2007;8:R259. doi: 10.1186/gb-2007-8-12-r259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alberte RS, Wood AM, Kursar TA, Guillard RRL. Novel phycoerythrins in marine Synechococcus spp.: Characterization and evolutionary and ecological implications. Plant Physiol. 1984;75:732–739. doi: 10.1104/pp.75.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ong LJ, Glazer AN. Phycoerythrins of marine unicellular cyanobacteria. I. Bilin types and locations and energy transfer pathways in Synechococcus spp. phycoerythrins. J Biol Chem. 1991;266:9515–9527. [PubMed] [Google Scholar]
- 10.Sidler WA. 1994. Phycobilisome and phycobiliprotein structures. The Molecular Biology of Cyanobacteria, Advances in Photosynthesis (Springer, Dordrecht, The Netherlands), pp 139–216.
- 11.Humily F, et al. A gene island with two possible configurations is involved in chromatic acclimation in marine Synechococcus. PLoS One. 2013;8:e84459. doi: 10.1371/journal.pone.0084459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palenik B. Chromatic adaptation in marine Synechococcus strains. Appl Environ Microbiol. 2001;67:991–994. doi: 10.1128/AEM.67.2.991-994.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everroad C, et al. Biochemical bases of type IV chromatic adaptation in marine Synechococcus spp. J Bacteriol. 2006;188:3345–3356. doi: 10.1128/JB.188.9.3345-3356.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shukla A, et al. Phycoerythrin-specific bilin lyase-isomerase controls blue-green chromatic acclimation in marine Synechococcus. Proc Natl Acad Sci USA. 2012;109:20136–20141. doi: 10.1073/pnas.1211777109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanfilippo JE, et al. Self-regulating genomic island encoding tandem regulators confers chromatic acclimation to marine Synechococcus. Proc Natl Acad Sci USA. 2016;113:6077–6082. doi: 10.1073/pnas.1600625113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toledo G, Palenik B, Brahamsha B. Swimming marine Synechococcus strains with widely different photosynthetic pigment ratios form a monophyletic group. Appl Environ Microbiol. 1999;65:5247–5251. doi: 10.1128/aem.65.12.5247-5251.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Humily F, et al. Development of a targeted metagenomic approach to study a genomic region involved in light harvesting in marine Synechococcus. FEMS Microbiol Ecol. 2014;88:231–249. doi: 10.1111/1574-6941.12285. [DOI] [PubMed] [Google Scholar]
- 18.Jiang T, et al. Temporal and spatial variations of abundance of phycocyanin- and phycoerythrin-rich Synechococcus in Pearl River Estuary and adjacent coastal area. J Ocean Univ China. 2016;15:897–904. [Google Scholar]
- 19.Olson RJ, Chisholm SW, Zettler ER, Armbrust EV. Pigments, size, and distributions of Synechococcus in the North Atlantic and Pacific Oceans. Limnol Oceanogr. 1990;35:45–58. [Google Scholar]
- 20.Sherry ND, Wood AM. Phycoerythrin-containing picocyanobacteria in the Arabian Sea in February 1995: Diel patterns, spatial variability, and growth rates. Deep Sea Res Part II Top Stud Oceanogr. 2001;48:1263–1283. [Google Scholar]
- 21.Lantoine F, Neveux J. Spatial and seasonal variations in abundance and spectral characteristics of phycoerythrins in the tropical northeastern Atlantic Ocean. Deep Sea Res Part I Oceanogr Res Pap. 1997;44:223–246. [Google Scholar]
- 22.Neveux J, Lantoine F, Vaulot D, Marie D, Blanchot J. Phycoerythrins in the southern tropical and equatorial Pacific Ocean: Evidence for new cyanobacterial types. J Geophys Res Oceans. 1999;104:3311–3321. [Google Scholar]
- 23.Campbell L, et al. Response of microbial community structure to environmental forcing in the Arabian Sea. Deep Sea Res Part II Top Stud Oceanogr. 1998;45:2301–2325. [Google Scholar]
- 24.Wood AM, Lipsen M, Coble P. Fluorescence-based characterization of phycoerythrin-containing cyanobacterial communities in the Arabian Sea during the Northeast and early Southwest Monsoon (1994–1995) Deep Sea Res Part II Top Stud Oceanogr. 1999;46:1769–1790. [Google Scholar]
- 25.Yona D, Park MO, Oh SJ, Shin WC. Distribution of Synechococcus and its phycoerythrin pigment in relation to environmental factors in the East Sea, Korea. Ocean Sci J. 2014;49:367–382. [Google Scholar]
- 26.Hoge FE, Wright CW, Kana TM, Swift RN, Yungel JK. Spatial variability of oceanic phycoerythrin spectral types derived from airborne laser-induced fluorescence emissions. Appl Opt. 1998;37:4744–4749. doi: 10.1364/ao.37.004744. [DOI] [PubMed] [Google Scholar]
- 27.Wood AM, Phinney DA, Yentsch CS. Water column transparency and the distribution of spectrally distinct forms of phycoerythrin-containing organisms. Mar Ecol Prog Ser. 1998;162:25–31. [Google Scholar]
- 28.Campbell L, Iturriaga R. Identification of Synechococcus spp. in the Sargasso Sea by immunofluorescence and fluorescence excitation spectroscopy performed on individual cells. Limnol Oceanogr. 1988;33:1196–1201. [Google Scholar]
- 29.Xia X, et al. Phylogeography and pigment type diversity of Synechococcus cyanobacteria in surface waters of the northwestern pacific ocean. Environ Microbiol. 2017;19:142–158. doi: 10.1111/1462-2920.13541. [DOI] [PubMed] [Google Scholar]
- 30.Xia X, Liu H, Choi D, Noh JH. Variation of Synechococcus pigment genetic diversity along two turbidity gradients in the China Seas. Microb Ecol. 2017;75:10–21. doi: 10.1007/s00248-017-1021-z. [DOI] [PubMed] [Google Scholar]
- 31.Xia X, Guo W, Tan S, Liu H. Synechococcus assemblages across the salinity gradient in a salt wedge estuary. Front Microbiol. 2017;8:1254. doi: 10.3389/fmicb.2017.01254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu H, Jing H, Wong THC, Chen B. Co-occurrence of phycocyanin- and phycoerythrin-rich Synechococcus in subtropical estuarine and coastal waters of Hong Kong. Environ Microbiol Rep. 2014;6:90–99. doi: 10.1111/1758-2229.12111. [DOI] [PubMed] [Google Scholar]
- 33.Chung C-C, Gong G-C, Huang C-Y, Lin J-Y, Lin Y-C. Changes in the Synechococcus assemblage composition at the surface of the East China Sea due to flooding of the Changjiang river. Microb Ecol. 2015;70:677–688. doi: 10.1007/s00248-015-0608-5. [DOI] [PubMed] [Google Scholar]
- 34.Haverkamp T, et al. Diversity and phylogeny of Baltic Sea picocyanobacteria inferred from their ITS and phycobiliprotein operons. Environ Microbiol. 2008;10:174–188. doi: 10.1111/j.1462-2920.2007.01442.x. [DOI] [PubMed] [Google Scholar]
- 35.Stomp M, et al. Colourful coexistence of red and green picocyanobacteria in lakes and seas. Ecol Lett. 2007;10:290–298. doi: 10.1111/j.1461-0248.2007.01026.x. [DOI] [PubMed] [Google Scholar]
- 36.Hunter-Cevera KR, Post AF, Peacock EE, Sosik HM. Diversity of Synechococcus at the Martha’s Vineyard coastal observatory: Insights from culture isolations, clone libraries, and flow cytometry. Microb Ecol. 2016;71:276–289. doi: 10.1007/s00248-015-0644-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fuller NJ, et al. Clade-specific 16S ribosomal DNA oligonucleotides reveal the predominance of a single marine Synechococcus clade throughout a stratified water column in the Red Sea. Appl Environ Microbiol. 2003;69:2430–2443. doi: 10.1128/AEM.69.5.2430-2443.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larsson J, et al. Picocyanobacteria containing a novel pigment gene cluster dominate the brackish water Baltic Sea. ISME J. 2014;8:1892–1903. doi: 10.1038/ismej.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen F, et al. Phylogenetic diversity of Synechococcus in the Chesapeake Bay revealed by ribulose-1,5-bisphosphate carboxylase-oxygenase (RuBisCO) large subunit gene (rbcL) sequences. Aquat Microb Ecol. 2004;36:153–164. [Google Scholar]
- 40.Choi DH, Noh JH. Phylogenetic diversity of Synechococcus strains isolated from the East China Sea and the East Sea. FEMS Microbiol Ecol. 2009;69:439–448. doi: 10.1111/j.1574-6941.2009.00729.x. [DOI] [PubMed] [Google Scholar]
- 41.Sunagawa S, et al. Tara Oceans Coordinators Ocean plankton. Structure and function of the global ocean microbiome. Science. 2015;348:1261359. doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- 42.Logares R, et al. Metagenomic 16S rDNA Illumina tags are a powerful alternative to amplicon sequencing to explore diversity and structure of microbial communities. Environ Microbiol. 2014;16:2659–2671. doi: 10.1111/1462-2920.12250. [DOI] [PubMed] [Google Scholar]
- 43.Pearman PB, Guisan A, Broennimann O, Randin CF. Niche dynamics in space and time. Trends Ecol Evol. 2008;23:149–158. doi: 10.1016/j.tree.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 44.Paulsen ML, et al. Synechococcus in the Atlantic gateway to the Arctic Ocean. Front Mar Sci. 2016;3:191. [Google Scholar]
- 45.Haverkamp THA, et al. Colorful microdiversity of Synechococcus strains (picocyanobacteria) isolated from the Baltic Sea. ISME J. 2009;3:397–408. doi: 10.1038/ismej.2008.118. [DOI] [PubMed] [Google Scholar]
- 46.Cabello-Yeves PJ, et al. Novel Synechococcus genomes reconstructed from freshwater reservoirs. Front Microbiol. 2017;8:1151. doi: 10.3389/fmicb.2017.01151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahmoud RM, et al. Adaptation to blue light in marine Synechococcus requires MpeU, an enzyme with similarity to phycoerythrobilin lyase isomerases. Front Microbiol. 2017;8:243. doi: 10.3389/fmicb.2017.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Veldhuis MJW, Kraay GW. Cell abundance and fluorescence of picoplankton in relation to growth irradiance and nitrogen availability in the Red Sea. Neth J Sea Res. 1993;31:135–145. [Google Scholar]
- 49.Katano T, Nakano S. Growth rates of Synechococcus types with different phycoerythrin composition estimated by dual-laser flow cytometry in relationship to the light environment in the Uwa Sea. J Sea Res. 2006;55:182–190. [Google Scholar]
- 50.Ahlgren NA, Rocap G. Culture isolation and culture-independent clone libraries reveal new marine Synechococcus ecotypes with distinctive light and N physiologies. Appl Environ Microbiol. 2006;72:7193–7204. doi: 10.1128/AEM.00358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bemal S, Anil AC. Genetic and ecophysiological traits of Synechococcus strains isolated from coastal and open ocean waters of the Arabian Sea. FEMS Microbiol Ecol. 2016;92:fiw162. doi: 10.1093/femsec/fiw162. [DOI] [PubMed] [Google Scholar]
- 52.Everroad RC, Wood AM. Phycoerythrin evolution and diversification of spectral phenotype in marine Synechococcus and related picocyanobacteria. Mol Phylogenet Evol. 2012;64:381–392. doi: 10.1016/j.ympev.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 53.Morel A, et al. Optical properties of the “clearest” natural waters. Limnol Oceanogr. 2007;52:217–229. [Google Scholar]
- 54.Cubillos-Ruiz A, Berta-Thompson JW, Becker JW, van der Donk WA, Chisholm SW. Evolutionary radiation of lanthipeptides in marine cyanobacteria. Proc Natl Acad Sci USA. 2017;114:E5424–E5433. doi: 10.1073/pnas.1700990114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martiny AC, Huang Y, Li W. Occurrence of phosphate acquisition genes in Prochlorococcus cells from different ocean regions. Environ Microbiol. 2009;11:1340–1347. doi: 10.1111/j.1462-2920.2009.01860.x. [DOI] [PubMed] [Google Scholar]
- 56.Martiny AC, Coleman ML, Chisholm SW. Phosphate acquisition genes in Prochlorococcus ecotypes: Evidence for genome-wide adaptation. Proc Natl Acad Sci USA. 2006;103:12552–12557. doi: 10.1073/pnas.0601301103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martiny AC, Kathuria S, Berube PM. Widespread metabolic potential for nitrite and nitrate assimilation among Prochlorococcus ecotypes. Proc Natl Acad Sci USA. 2009;106:10787–10792. doi: 10.1073/pnas.0902532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 60.Huerta-Cepas J, Serra F, Bork P. ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol. 2016;33:1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Camacho C, et al. BLAST+: Architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matsen FA, Kodner RB, Armbrust EV. pplacer: Linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics. 2010;11:538. doi: 10.1186/1471-2105-11-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.R Core Team 2014 R: A language and environment for statistical computing (R Found Stat Comput, Vienna). Available at www.R-project.org/
- 64.Maechler M, Rousseeuw P, Struyf A, Hubert M, Hornik K. 2017 cluster: Cluster analysis basics and extensions. R package, version 2.0.6. Available at: http://CRAN.R-project.org/package=cluster. Accessed April 18, 2016.
- 65.Venables WN, Ripley BD. Modern Applied Statistics with S. 4th Ed Springer; New York: 2002. [Google Scholar]
- 66.Harrell FE. 2016 Hmisc: Harrell Miscellaneous. Available at CRAN.R-project.org/package=Hmisc. Accessed April 18, 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.