Significance
The presynaptic active zone is important for precise and fast information transfer at central nervous synapses. Invertebrate studies have indicated that Liprin-α proteins are important for active zone assembly and function, but localization and function of vertebrate Liprin-α proteins have remained enigmatic, and pre- or postsynaptic localization and roles have been proposed. Here we used stimulated emission depletion microscopy to directly show that Liprin-α3, but not Liprin-α2, prominently localizes to the active zone. We then generated Liprin-α3–KO mice and found that active zone structure and synaptic vesicle docking and exocytosis are impaired at their synapses. These data establish important functions of Liprin-α3 at the active zone of vertebrate synapses.
Keywords: Liprin-α, active zone, synaptic vesicle exocytosis, vesicle docking, active zone assembly
Abstract
The presynaptic active zone provides sites for vesicle docking and release at central nervous synapses and is essential for speed and accuracy of synaptic transmission. Liprin-α binds to several active zone proteins, and loss-of-function studies in invertebrates established important roles for Liprin-α in neurodevelopment and active zone assembly. However, Liprin-α localization and functions in vertebrates have remained unclear. We used stimulated emission depletion superresolution microscopy to systematically determine the localization of Liprin-α2 and Liprin-α3, the two predominant Liprin-α proteins in the vertebrate brain, relative to other active-zone proteins. Both proteins were widely distributed in hippocampal nerve terminals, and Liprin-α3, but not Liprin-α2, had a prominent component that colocalized with the active-zone proteins Bassoon, RIM, Munc13, RIM-BP, and ELKS. To assess Liprin-α3 functions, we generated Liprin-α3–KO mice by using CRISPR/Cas9 gene editing. We found reduced synaptic vesicle tethering and docking in hippocampal neurons of Liprin-α3–KO mice, and synaptic vesicle exocytosis was impaired. Liprin-α3 KO also led to mild alterations in active zone structure, accompanied by translocation of Liprin-α2 to active zones. These findings establish important roles for Liprin-α3 in active-zone assembly and function, and suggest that interplay between various Liprin-α proteins controls their active-zone localization.
Exocytosis of synaptic vesicles in a nerve terminal is mediated by specialized sites called active zones. Active zones provide slots for synaptic vesicle docking and priming, tether Ca2+ channels, and target fusion to membrane domains opposed to postsynaptic receptors (1). At vertebrate synapses, active zones are thought to be composed of six protein families: RIM, ELKS, Munc13, RIM-BP, Piccolo/Bassoon, and Liprin-α (Fig. 1A). Many additional proteins that are not restricted to the active zone, including cytoskeletal proteins, SNAREs, Ca2+ channels, and cell adhesion proteins, are also present.
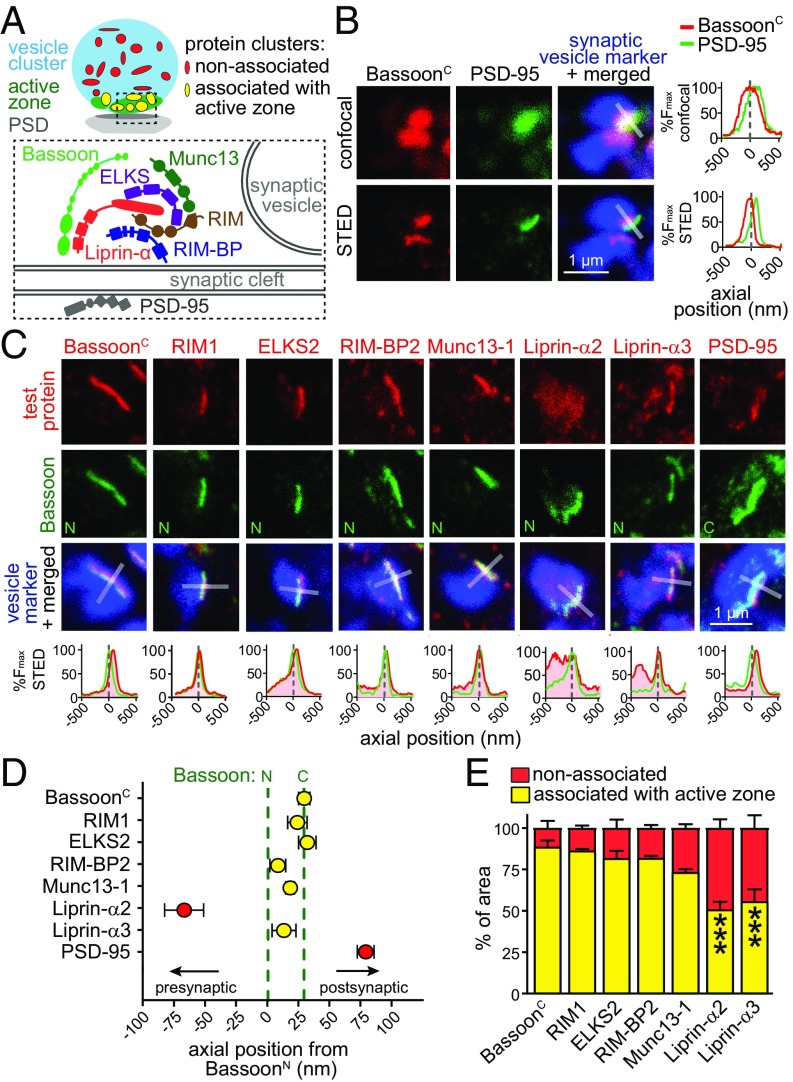
Fig. 1.
STED microscopy establishes active-zone localization of Liprin-α3 at excitatory hippocampal synapses. (A) Schematic illustration of a synapse. (B) Example images of hippocampal excitatory synapses in side view in confocal and STED microscopy stained for BassoonC and PSD-95. The synaptic vesicle cluster (labeled for synapsin) was imaged by confocal microscopy in all experiments. The intensity profile normalized to peak fluorescence is shown for the profile scan (transparent white rectangle) perpendicular through the center of Bassoon. (C) Representative STED images of individual excitatory synapses in side view stained for test proteins, Bassoon, and the vesicle marker vGlut1 (for PSD-95, synapsin antibodies were used due to limitations in antibody compatibility). Fluorescence intensity profiles are shown below. Dashed line at zero indicates the peak localization of BassoonN. Overview images and confocal scans are provided in Fig. S1. (D) Axial positions of test proteins relative to BassoonN. BassoonN, n = 920 synapses/8 independent cultures; BassoonC, n = 231/3; RIM1, n = 130/3; ELKS2, n = 141/3; RIM-BP2, n = 84/3; Munc13-1, n = 92/3; Liprin-α2, n = 105/3; Liprin-α3, n = 234/3; PSD-95, n = 79/3. (E) Percentage of test protein clusters associated (yellow) or not associated (red) with the active zone. BassoonC, n = 985 synapses/5 images/3 independent cultures; RIM1, n = 795/5/3; ELKS2, n = 845/6/3; RIM-BP2, n = 1,298/6/3; Munc13-1, n = 1,935/6/3; Liprin-α2, n = 1,296/6/3; Liprin-α3, n = 1,425/6/3. Statistical significance determined by two-way ANOVA (localization significant at P ≤ 0.001; test protein not significant, interaction significant at P ≤ 0.001) followed by Holm–Šídák posttests (reported in figure) comparing each distribution vs. that of BassoonC. All data are means ± SEM (***P ≤ 0.001).
Although mouse genetic experiments have led to insights into functions of most vertebrate active-zone proteins, such experiments have not been pursued for Liprin-α. Liprin-α was identified through the binding to the phosphotyrosine phosphatase LAR (2) and was hypothesized to be a vertebrate active-zone protein based on biochemical interactions with RIM and ELKS (3, 4). Invertebrate data support broad presynaptic functions for Liprin-α. At the Caenorhabditis elegans neuromuscular junction, deletion of the Liprin-α homolog syd-2 alters active-zone structure and disrupts synaptic vesicle accumulation in nerve terminals (5–7). The fly homolog Dliprin-α regulates presynaptic morphogenesis of the neuromuscular junction and is required for target selection in the retina (8–10). These effects are consistent with the many biochemical activities of Liprin-α. In addition to interacting with LAR, RIM, and ELKS (2–4), Liprin-α binds to itself and β-liprins, GIT, CASK, RSY-1, mDiaphenous, and KIF1A (1, 2, 7, 11, 12). These interactions predict broad cellular functions.
Vertebrates express four genes, Ppfia1–Ppfia4, encoding Liprin-α1–Liprin-α4. Liprin-α1 is expressed within and outside the nervous system, and, like Liprin-α4, brain expression is low and restricted to a few areas. In contrast, Liprin-α2 and Liprin-α3 are strongly and specifically expressed in the brain (2, 13–15). Their subcellular localization and function, however, remained uncertain. Confocal microscopy partially supported synaptic localization of Liprin-α2 and Liprin-α3 (14, 15), and postsynaptic localization was favored by immunogold labeling experiments with Liprin-α2 antibodies (16). A recent study, however, found that knockdown of Liprin-α2 impaired synaptic transmission, and effects were more consistent with presynaptic than with postsynaptic roles (17).
Here, we set out to determine localization and function of Liprin-α in the brain. We found that Liprin-α3 and Liprin-α2 are widely distributed in hippocampal nerve terminals. In addition, Liprin-α3, but not Liprin-α2, had a prominent component that strongly colocalized with active-zone proteins. Liprin-α3–KO mice, which we generated for this study, displayed impairments in synaptic vesicle docking, tethering, and exocytosis, establishing presynaptic roles for Liprin-α3 in synaptic transmission.
Results
Localization of Liprin-α and Active-Zone Proteins Using Stimulated Emission Depletion Microscopy.
We set out to evaluate Liprin-α2 and Liprin-α3 localization at hippocampal synapses. Similar to the approach used at the fly neuromuscular junction (18), we performed stimulated emission depletion (STED) superresolution microscopy because diffraction-limited microscopy is not sufficient to determine whether a protein is localized to the active zone, to synaptic vesicles inside a nerve terminal, or to the postsynaptic density (PSD). We first imaged cultured hippocampal neurons stained with antibodies against the Bassoon N terminus (BassoonN) or C terminus (BassoonC), the excitatory PSD marker PSD-95, and a synaptic vesicle protein to identify synapses. We identified synapses in side view (Fig. 1B and Fig. S1A) by identifying a cluster of synaptic vesicles with an active zone marked by Bassoon appearing as a narrow band (531 ± 25 nm long; n = 45 synapses) at one edge of the vesicle cloud. At these synapses, PSD-95 was opposed to Bassoon and could be reliably separated from Bassoon in STED but not confocal microscopy.
We next analyzed pairwise stainings of synapses in side view for all known active-zone proteins, including Liprin-α2 and Liprin-α3, with Bassoon (Fig. 1 C and D and Fig. S1). We expressed test protein localization relative to BassoonN by identifying the peak intensity in line profiles (1 μm long and 250 nm wide) perpendicular through the center of the active zone, and calculated the average peak position from the individual peaks. In this analysis, negative values are proximal to BassoonN inside the nerve terminal, whereas positive values are distal toward the presynaptic plasma membrane, the synaptic cleft, and the postsynaptic compartment. As observed before with stochastic optical resolution microscopy (STORM) (19), the ∼450-kDa protein Bassoon had an orientation at the active zone with BassoonC being closer to the plasma membrane than BassoonN (Fig. 1 C and D), and the peaks were separated by +30 ± 4 nm. The peak signal of PSD-95, indicating the position of the PSD, was +80 ± 7 nm distal from BassoonN. The peak fluorescent signals of Liprin-α3, RIM1, Munc13-1, ELKS2, and RIM-BP2, but not of Liprin-α2, were between the peaks of BassoonN and BassoonC. These data establish close colocalization of these proteins at the active zone, with the exception of Liprin-α2.
Bassoon, RIM-BP2, RIM1, ELKS2, and Munc13-1 all showed relatively sharp peaks at the active zone. Interestingly, Liprin-α3 had an additional widespread component (Fig. 1C) that was localized inside the nerve terminal. This component was pronounced for Liprin-α2 (Fig. 1C) with no sharp peak at the active zone, which led to a shift of the average Liprin-α2 peak position toward the inside of the nerve terminal at −67 ± 15 nm relative to BassoonN (Fig. 1D).
To assess the localization of each protein within and outside the active zone, we generated two masks (Fig. 1A): one that outlined the nerve terminal (vGlut1 or Synapsin; Fig. 1A, blue), and one that defined the active zone (BassoonN; Fig. 1A, green). We then quantified the area associated with the active zone for each protein (Fig. 1A, yellow) and compared it vs. the area of the same protein outside the active zone (Fig. 1A, red), normalized to the sum of the area of active zone and nonactive zone components (Fig. 1E). This provides a measure of signal distribution within a nerve terminal without accounting for the signal intensity within the area. The majority of the area stained for BassoonC, RIM1, ELKS2, RIM-BP2, and Munc13-1 was at the active zone (73–90%; Fig. 1E). Liprin-α2 and Liprin-α3 had ∼50% of positively stained area outside the active zone. Thus, both proteins are widespread in a presynaptic nerve terminal. The key difference between Liprin-α2 and -α3 is that the peak intensity for Liprin-α3 is within the active zone (Fig. 1D), whereas the peak of Liprin-α2 is away from other active-zone proteins shifted toward the synaptic vesicle cloud, which arises as an average from its widespread distribution throughout the nerve terminal.
Finally, we tested whether localization of Liprin-α3 depends on other active-zone proteins (Fig. S2). We generated KO neurons for RIM1, RIM2, ELKS1, and ELKS2, which leads to strong disruption of the active zone (20). In this mutant, however, Liprin-α3 synaptic fluorescence and peak localization were not affected, and the line profile peak intensity was only mildly impaired, indicating that active-zone localization of Liprin-α3 is largely independent of RIM and ELKS.
Generation of Liprin-α3–KO Mice.
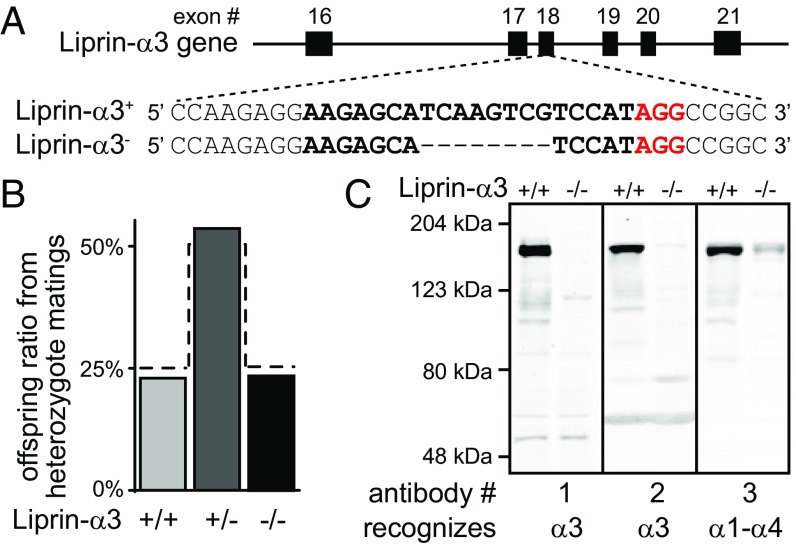
The localization of Liprin-α3 at the active zone suggests that Liprin-α3 may play a role in active-zone function. To test this hypothesis, we generated Liprin-α3–KO mice by using CRISPR/Cas9 gene editing in single-cell zygotes. We selected an allele that had an 8-bp deletion (Fig. 2A), inducing a frame-shift mutation leading to nonsense-mediated decay. Heterozygote matings produced offsprings with a Mendelian distribution of homozygote KO (Liprin-α3−/−), heterozygote (Liprin-α3+/−), and WT (Liprin-α3+/+) sibling mice (Fig. 2B). Western blotting of brain homogenates revealed that Liprin-α3 was removed, and we did not detect bands that would establish the presence of protein fragments (Fig. 2C). Liprin-α3−/− mice were viable and fertile, but had a significantly increased distance traveled and were less spatially confined in an open-field assessment of locomotion (Fig. S3 A–D), suggesting alterations in brain function. No changes beyond removal of Liprin-α3 were detected when we assessed morphology and protein composition in hippocampi of Liprin-α3−/− mice (Fig. S3 E–H).
Fig. 2.
Generation of Liprin-α3–KO mice. (A) CRISPR/Cas9-mediated genome editing in single-cell zygotes. The sgRNA-targeting sequence is shown in bold, the protospacer-adjacent motif (PAM) is red, and the dashed line indicates the deletion. (B) Survival analysis of offspring of heterozygous matings. The dashed line represents a Mendelian distribution (n = 418 animals/54 litters). (C) Liprin-α3 expression in hippocampi analyzed by fluorescent Western blotting (quantification shown in Fig. S3 G and H). The Liprin-α3 antibody 3 cross-reacts with Liprin-α1–α4.
Impaired Synaptic Vesicle Exocytosis in Liprin-α3−/− Mice.
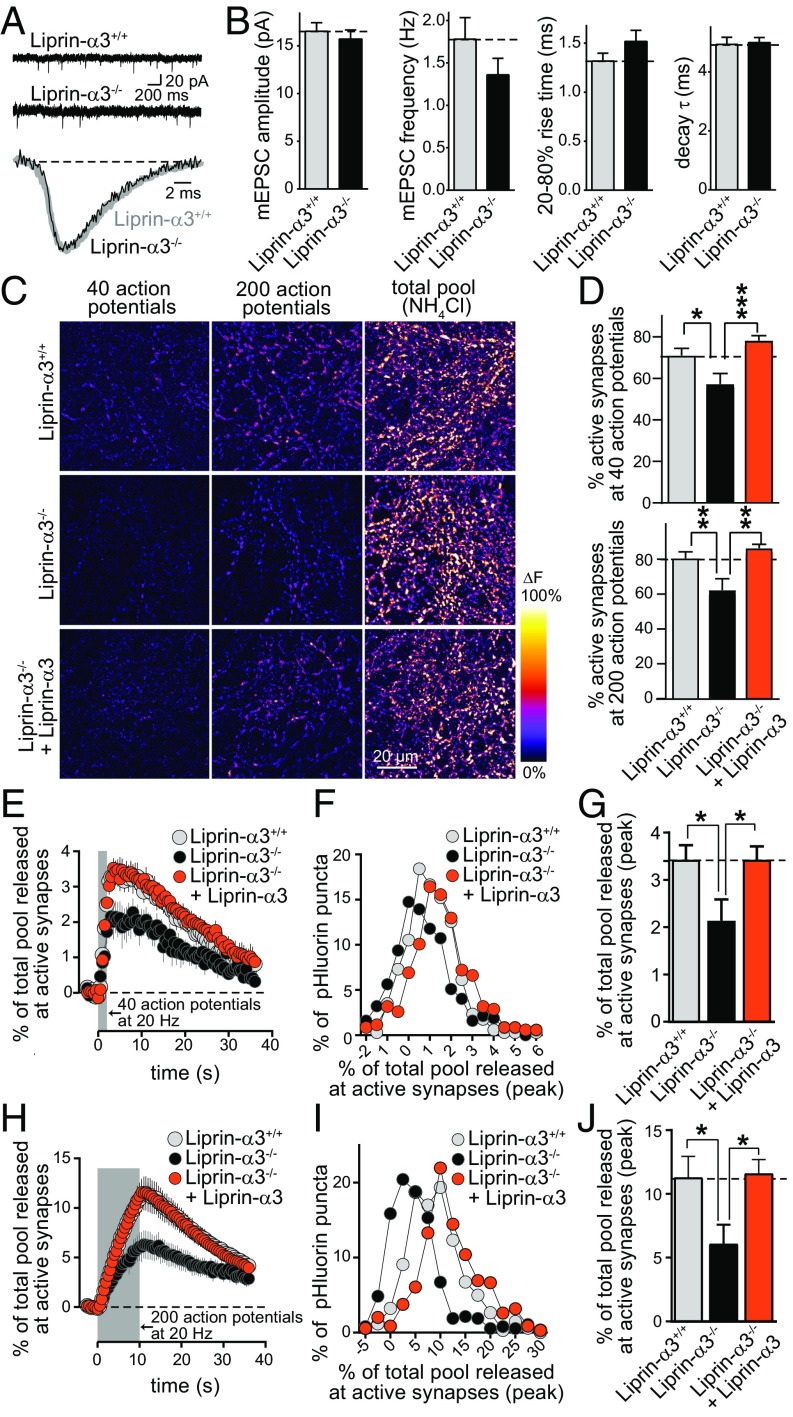
We assessed synaptic transmission in Liprin-α3−/− and Liprin-α3+/+ neurons cultured from hippocampi of littermate newborn mice. We first measured AMPA receptor-mediated miniature excitatory postsynaptic currents (mEPSCs; Fig. 3 A and B). The mEPSC amplitudes and kinetics were indistinguishable between the two genotypes. There was a ∼20% trend toward a reduction in mEPSC frequency, which was not statistically significant. These results indicate that postsynaptic AMPA receptor numbers are not strongly affected by Liprin-α3 KO. Given that previous studies found developmental and postsynaptic roles for Liprin-α, we decided to measure exocytosis of synaptic vesicles directly. We used imaging of synaptic-vesicle cycling upon lentiviral expression of synaptophysin-pHluorin (sypHy; Fig. 3 C–J). We compared exocytosis of synaptic vesicles, as indicated by an increase in pHluorin fluorescence, in Liprin-α3+/+ neurons, Liprin-α3−/− neurons, and Liprin-α3−/− neurons transduced with a lentivirus that expressed HA-tagged Liprin-α3 from a neuron-specific human synapsin promoter. In hippocampal neurons, Liprin-α3 is expressed as early as day in vitro (DIV) 1 (Fig. S4A). To mimic this early expression, we transduced Liprin-α3−/− neurons for rescue with a Liprin-α3–expressing lentivirus at DIV 1. Rescue Liprin-α3 was localized normally as assessed by STED microscopy (Fig. S4 B–D). Neurons were stimulated with 40 or 200 action potentials at 20 Hz, followed by application of NH4Cl to unquench pHluorin fluorescence. No differences in the number of pHluorin-positive or NH4Cl-responsive puncta was observed across conditions (Fig. S5), indicating that removing or reexpressing Liprin-α3 did not change synapse numbers.
Fig. 3.
Synaptic vesicle exocytosis is impaired upon Liprin-α3 KO. (A and B) Example traces (A) and quantification (B) of mEPSCs. Liprin-α3+/+, n = 47 cells/4 independent cultures; Liprin-α3−/−, n = 48/4. (C) Pseudocolored images of peak fluorescence change in sypHy-expressing Liprin-α3+/+ neurons, Liprin-α3−/− neurons, and Liprin-α3−/− neurons rescued with lentiviral expression of Liprin-α3 transduced at DIV 1. Neurons were stimulated with 40 or 200 action potentials and dequenched with NH4Cl. (D) Quantification of the percentage of sypHy puncta responsive to 40 (Top) or 200 (Bottom) action potentials. (E–G) Quantification of sypHy mean fluorescence changes at active synapses stimulated with 40 action potentials expressed as percentage of the NH4Cl-responsive vesicle pool (E), and the frequency distribution (F) and peak response (G) of the same data. Liprin-α3+/+, n = 2,365 NH4Cl-responsive synapses/1,338 active synapses/11 coverslips/6 independent cultures; Liprin-α3−/− 2,371/1,093/11/6; Liprin-α3−/− + Liprin-α3, n = 2,438/1,463/11/6. (H–J) Quantification as in E–G but for neurons stimulated with 200 action potentials. Liprin-α3+/+, n = 2,353/1,485/11/6; Liprin-α3−/−, n = 2,346/1,164/11/6; Liprin-α3−/− + Liprin-α3, n = 2,427/1,598/11/6. All data are means ± SEM [*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001, Student’s t tests (B; all P > 0.05) or one-way ANOVA (D, G, and J) followed by Holm–Šídák multiple comparisons test comparing each condition vs. Liprin-α3−/−]. The number of coverslips was used as a basis for statistics in sypHy imaging.
Removal of Liprin-α3 led to a decrease in the number of synapses responsive to action potentials (Fig. 3 C and D). At responsive synapses, KO of Liprin-α3 was accompanied by a 38% or 46% decrease in pHluorin fluorescence after stimulation with 40 or 200 action potentials, respectively (Fig. 3 E–J). Upon reexpression of Liprin-α3, release recovered to levels that were indistinguishable from WT littermate neurons. Efficient Liprin-α3 expression and rescue of its function required viral transduction early after plating (Fig. S6). These experiments establish a defect in synaptic vesicle exocytosis upon loss of Liprin-α3. The full rescue indicates that the impairment in exocytosis is caused by loss of Liprin-α3 in the mutant mice, and the reversibility of the impairment establishes that postnatal, neuronal expression of Liprin-α3 is sufficient to mediate its role in exocytosis. Together with the presynaptic localization observed with STED microscopy, these experiments establish a presynaptic function of Liprin-α3 to enhance synaptic strength. Field recordings in acute brain slices confirmed a defect in synaptic strength in excitatory Schaffer collateral to CA1 synapses of the hippocampus (Fig. S7).
Synaptic Ultrastructure in Liprin-α3–KO Neurons.
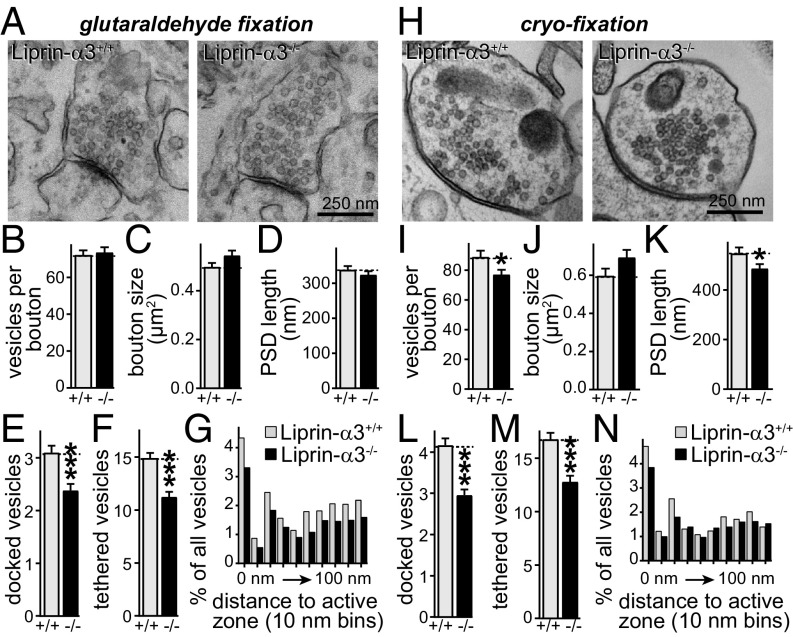
To determine whether Liprin-α3 has a structural role in vertebrate presynaptic morphogenesis, we pursued EM analysis of synapse structure in cultured hippocampal neurons after chemical fixation (Fig. 4 A–G). We observed a 23% loss of vesicles docked to the presynaptic plasma membrane upon KO of Liprin-α3, accompanied by a 25% loss of vesicles within 100 nm of the target membrane, referred to as tethered vesicles (Fig. 4 E and F). Bouton size, PSD length, and the number of vesicles per bouton were not changed.
Fig. 4.
Impaired synaptic vesicle docking and tethering after Liprin-α3 KO. (A) Representative EM images of synapses of cultured hippocampal neurons fixed with glutaraldehyde. (B–F) Quantification of vesicles per bouton (B), bouton size (C), PSD length (D), docked vesicles (E), and vesicles tethered within 100 nm (F). Liprin-α3+/+, n = 150 synapses/3 independent cultures; Liprin-α3−/−, n = 147/3. (G) Histogram of the vesicle distribution in the first 100 nm of the active zone in 10-nm bins expressed as a percentage of all synaptic vesicles. Numbers of synapses and cultures are as in B–F. (H–N) Same as A–G, but for neurons cryofixed by high-pressure freezing. Liprin-α3+/+, n = 104/3; Liprin-α3−/−, n = 121/3. Data are means ± SEM unless stated otherwise (*P ≤ 0.05 and ***P ≤ 0.001, unpaired Student’s t test).
Chemical fixation leads to collapse of intra- and extracellular space, potentially confounding the analysis of synaptic ultrastructure. We therefore repeated this experiment with the use of high-pressure freezing followed by freeze substitution, directly comparing the two methods and the Liprin-α3–KO phenotypes (Fig. 4 H–M). Liprin-α3–KO phenotypes were essentially the same, with a 30% and 24% loss of docked and tethered vesicles, respectively. In addition, we observed a small decrease in bouton size and PSD length upon Liprin-α3 KO, perhaps suggesting mild structural roles for Liprin-α3 in bouton formation.
Deficits in vesicle docking are often accompanied by accumulation of nondocked vesicles within 10–20 nm of the presynaptic plasma membrane, for example in Munc13 and SNARE mutants (21). The decrease in the number of tethered vesicles (Fig. 4 F and M) suggested that this is not the case in Liprin-α3–KO neurons. To better analyze this, we plotted histograms of the distribution of vesicles within 100 nm of the presynaptic membrane in 10-nm bins (Fig. 4 G and N). We did not observe an accumulation of undocked vesicles near the plasma membrane, but, instead, mild reductions of vesicles were present across the first 100 nm, establishing a synaptic vesicle tethering defect.
Altered Active-Zone Structure upon Deletion of Liprin-α3.
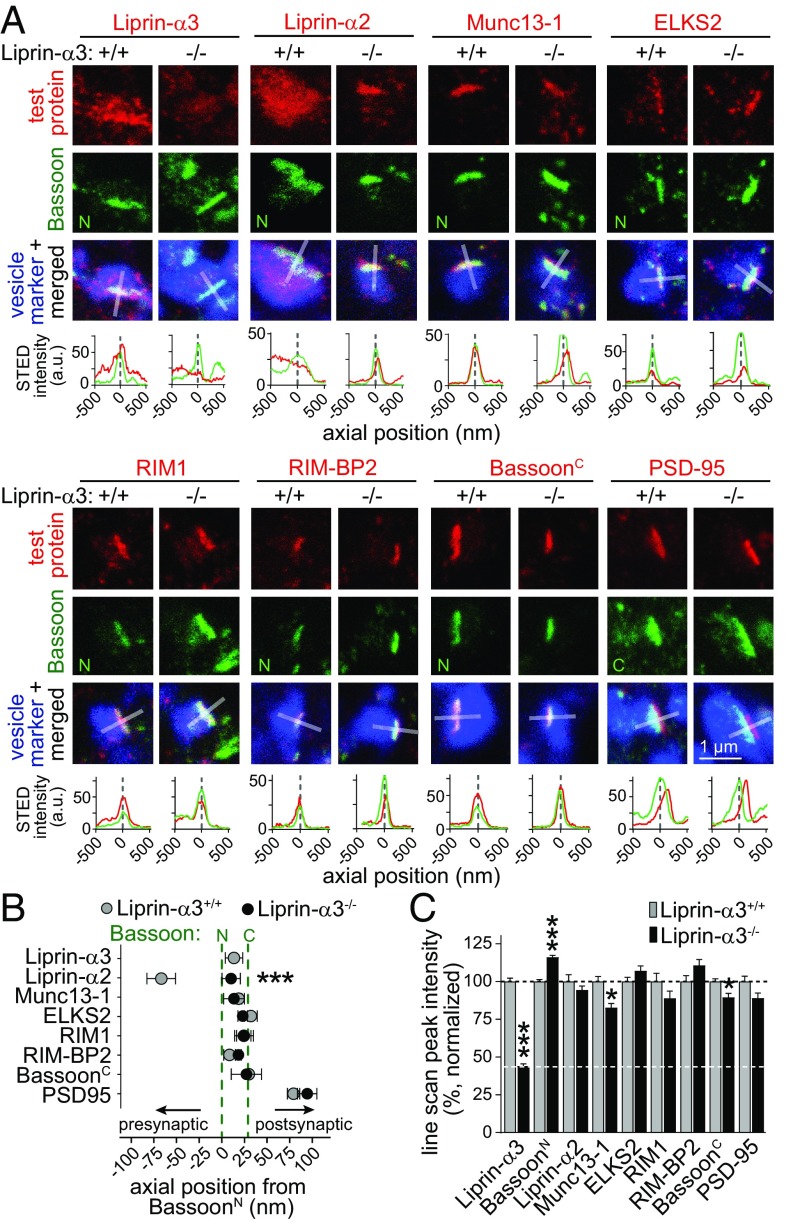
To assess whether Liprin-α3 KO induced structural changes in the active zone, we compared Liprin-α3+/+ with Liprin-α3−/− synapses by STED microscopy (Fig. 5 and Fig. S8). Compellingly, at Liprin-α3−/− synapses, the peak of Liprin-α2 staining moved to a position between the Bassoon N and C termini, akin to the localization of Liprin-α3 (Fig. 5B). Hence, only upon deletion of Liprin-α3, Liprin-α2 is strongly localized to the active zone. Liprin-α2, however, cannot functionally compensate for the loss of Liprin-α3 because vesicle docking, tethering, and release are impaired at Liprin-α3−/− synapses despite translocation of Liprin-α2 to the active zone. Effects on other active-zone proteins were mild. For each active-zone protein, the peak localization was not affected in Liprin-α3–KO mice (Fig. 5B). Nevertheless, we observed modest changes in Bassoon and Munc13-1 labeling (Fig. 5C and Figs. S8 and S9). These changes likely reflect rearrangements of the release-site structure that contribute to the impairment in synaptic vesicle tethering, docking, and exocytosis.
Fig. 5.
STED analysis of Liprin-α3–KO synapses. (A) Representative STED images of side-view synapses and line profiles [as in Fig. 1 but showing arbitrary units (a.u.)]. (B) Axial positions of the test proteins relative to Bassoon: BassoonN (+/+, n = 841/8; −/−, n = 817/8); Liprin-α3 (+/+, n = 234/3); Liprin-α2 (+/+, n = 105/3; −/−, n = 120/3); BassoonC (+/+, n = 55/3; −/−, n = 28/3); Munc13-1 (+/+, n = 92/3; −/−, n = 77/3); ELKS2 (+/+, n = 141/3; −/−, n = 196/3); RIM1 (+/+, n = 130/3; −/−, n = 114/3); RIM-BP2 (+/+, n = 84/3; −/−, n = 107/3); and PSD-95 (+/+, n = 79/3; −/−, n = 116/3). Statistical significance determined by two-way ANOVA (genotype significant at P ≤ 0.05; test protein significant at P ≤ 0.001; interaction significant at P ≤ 0.001) followed by Holm–Šídák multiple-comparisons posttests (reported in figure). (C) Peak intensity of line profiles normalized to the average of Liprin-α3+/+ synapses: Liprin-α3 (+/+, n = 234/3; −/−, n = 175/3); BassoonN (+/+, n = 772/8; −/−, n = 805/8); Liprin-α2 (+/+, n = 105/3; −/−, n = 120/3); BassoonC (+/+, n = 231/3; −/−, n = 169/3); Munc13-1 (+/+, n = 92/3; −/−, n = 77/3); ELKS2 (+/+, n = 141/3; −/−, n = 175/3); RIM1 (+/+, n = 130/3; −/−, n = 114/3); RIM-BP2 (+/+, n = 84/3; −/−, n = 107/3); and PSD-95 (+/+, n = 116/3; −/−, n = 158/3). Statistical significance determined by two-way ANOVA (genotype not significant; test protein significant at P ≤ 0.001; interaction significant at P ≤ 0.001) followed by Holm–Šídák multiple-comparisons posttests (reported in figure). All data are means ± SEM (*P ≤ 0.05 and ***P ≤ 0.001). The data were acquired simultaneously with identical settings in a blind experiment in Liprin-α3+/+ control and Liprin-α3−/− neurons, and the Liprin-α3+/+ data are the same data that are shown in Fig. 1. A second, independent assessment of active-zone protein localization is shown in Fig. S2. Overview images and additional analyses are in Figs. S8 and S9.
Discussion
The localization and function of vertebrate Liprin-α has been uncertain, and pre-, post-, and extrasynaptic functions have been proposed. We establish here that localization of Liprin-α3, but not Liprin-α2, peaks at the active zone (Fig. 1). We find that Liprin-α3 is important for synaptic vesicle docking and release (Figs. 2–4), functions that are likely mediated by a role for Liprin-α3 in determining active-zone composition (Fig. 5). Hence, Liprin-α3 functions at the presynaptic active zone to enhance synaptic strength.
Analysis of Active-Zone Protein Localization at Vertebrate Synapses Using STED Microscopy.
A large body of literature suggests that Piccolo/Bassoon, RIM, Munc13, ELKS, RIM-BP, and Liprin-α define the active zone at vertebrate synapses, with biochemical and morphological evidence in support of active-zone association. Although localization was typically established by qualitative immuno-EM, colocalization with other active-zone proteins at a nanoscale has not been systematically studied. Recent advances in superresolution microscopy provide new tools to tackle this limitation. A first study employed STORM microscopy and found colocalization of Bassoon/Piccolo and RIM (19). Two recent papers provided additional evidence for active-zone localization of RIM-BP, Bassoon, and Munc13 by using STED or STORM microscopy (22, 23). Here we extend these studies by systematic mapping of active-zone protein localization, including vertebrate ELKS and Liprin-α, proteins that have not been studied by using superresolution microscopy to our knowledge. We find that members of each protein family localize to the active zone and are opposed to the PSD at excitatory hippocampal synapses. Notably, the peaks of RIM-BP2, Liprin-α3, Munc13-1, RIM1, and ELKS2 all fall within the 25 nm between the Bassoon N and C termini, strongly supporting that these proteins are associated with one another at active zones.
Our work relates to comprehensive studies at the fly neuromuscular junction, where STED microscopy has revealed that Brp, a partial homolog of ELKS, forms prominent umbrella-shaped T-bars that reach 200–300 nm into the nerve terminal. The fly homologs of RIM-BP, Liprin-α, and Munc13 each form small, distinct clusters surrounding the base of these T-bars (1, 18, 24). Our data at hippocampal synapses suggest finer subclustering of active-zone proteins, resulting in relatively homogenous appearance of the active zone in STED microscopy. A contributing factor could be that hippocampal synapses are more diverse and likely less stereotyped than the fly neuromuscular junction.
Liprin-α3 as an Active-Zone Scaffold.
Our data firmly establish presynaptic localization and function of Liprin-α3. However, several observations set Liprin-α3 apart from other active-zone proteins. First, Liprin-α3 has a broader distribution than other active-zone proteins, with antibody staining spread throughout the nerve terminal (Fig. 1). Second, at synapses that lack RIM and ELKS, Liprin-α3 localization is more mildly affected than that of other active-zone proteins (20) (Fig. S2). The most parsimonious model is that interactions with LAR-PTPs and Liprin-α self-association play roles in Liprin-α targeting and presynaptic assembly (2, 11). Studies of active-zone assembly in C. elegans suggested that Liprin-α may act early in active-zone assembly (6, 7, 25), and fit with our finding that Liprin-α3 is recruited to the presynaptic plasma membrane independent of RIM and ELKS. In invertebrates, syd-1 is required upstream of Liprin-α/syd-2, and overexpression of Liprin-α/syd-2 bypasses the need for syd-1 (26, 27). This suggested that syd-1 may recruit Liprin-α/syd-2 to synapses. Vertebrates express only distal orthologs of syd-1, called SYD1A/syde1 and SYD1B/syde2, with a Rho-GAP activity that is not present in invertebrate syd-1 but essential for cytoskeletal remodeling in vertebrates (28, 29), and localization of the vertebrate orthologs is not known. Future studies are necessary to address potential roles of LAR-RPTPs and SYD1A/B for targeting Liprin-α3 to vertebrate active zones.
Our functional and morphological data support a scaffolding role for Liprin-α3. We observe a loss of tethered and docked synaptic vesicles that is well matched with a reduction in exocytosis. Although recent studies at the fly neuromuscular junction support roles for Liprin-α in targeting of a specific Munc13 isoform (24), there is currently no support for a direct interaction between Liprin-α3 and Munc13. Because the reduction of Munc13 levels at Liprin-α3−/− synapses is mild, and because Liprin-α3−/− synapses have a loss in docked and tethered vesicles [tethered vesicles are increased in Munc13-KO synapses (21)], it is unlikely that the secretory deficit in Liprin-α3−/− synapses is the result of an isolated loss of Munc13. Instead, the exocytotic defect is probably a result of a combination of active-zone defects caused by a loss of scaffolding by Liprin-α3.
Redundant and Diverse Roles for Vertebrate Liprin-α in Brain Development and Function.
Although our studies establish localization and function of Liprin-α3 at the presynaptic active zone, they do not exclude other roles for Liprin-α3, for example, in cellular transport or morphogenesis, as generally suggested for Liprin-αs from biochemical studies and invertebrate genetics (1, 9, 10). Our data, however, suggest that these roles are not essential or are compensated for by other Liprin-αs, because Liprin-α3–KO mice survive and develop mostly normally (Fig. 2 and Fig. S3).
Diverse roles for Liprin-α are also strongly supported by the variable localization of the various Liprin-α proteins within neurons and by studies in nonneuronal cells. Within neurons, Liprin-α1 is mostly localized dendritically, and knockdown and overexpression experiments support roles in dendrite morphogenesis (14, 15, 30). These functions are consistent with studies in nonneuronal cells, where Liprin-α1 controls edge dynamics in motile cancer cells through focal adhesion (31). Liprin-α2 localization appears more restricted to synapses, but, so far, it was unclear whether it is pre- or postsynaptically localized (14–16). A functional study using knockdown of Liprin-α2 suggested a presynaptic function in regulating the localization and turnover of other presynaptic proteins to control release (17). Together with the widespread presynaptic localization established here, these data suggest that one function of Liprin-α2 may be the trafficking and/or capture of presynaptic components, consistent with roles of Dliprin-α in flies (9).
Our data reveal insight into the interplay between Liprin-α2 and Liprin-α3. Compellingly, when Liprin-α3 was removed, Liprin-α2 translocated to the active zone, mimicking the localization of Liprin-α3. There are several potential alternative explanations. First, it is possible that a limited number of Liprin-α binding sites at the active zone accounts for this interplay, and Liprin-α3 outcompetes Liprin-α2 for localization because it is expressed higher or has a higher affinity for key interacting partners. Similar mechanisms were observed for presynaptic Ca2+ channels, which compete for a limited number of slots (32). Alternatively, Liprin-α3 could have an active role in suppressing active-zone localization of Liprin-α2, for example, by recruiting proteins that inhibit Liprin-α2 active-zone localization. Finally, Liprin-α3 expression may suppress Liprin-α2 expression and therefore its active-zone levels. This is unlikely because there is no strong increase in Liprin-α2 expression in Liprin-α3–KO animals. Regardless of the mechanism, our data indicate that the functions of Liprin-α3 are not identical to those of Liprin-α2 because Liprin-α3−/− neurons have a secretory impairment that is not compensated for by Liprin-α2. A previous study complements this point: Liprin-α3 cannot fully compensate for knockdown of Liprin-α2 (17). Future studies are needed to generate KO mice for Liprin-α2 and other Liprin-αs and to systematically compare single- and double-KO phenotypes. Together, this and previous studies support diverse roles for Liprin-α in morphogenesis, cellular trafficking, and synaptic transmission. These roles are shared by a single gene in invertebrates, they have likely diverged across several Liprin-α genes in vertebrates.
Methods
Mice.
The constitutive Liprin-α3–KO mice were generated by CRISPR/Cas9 gene editing in one-cell zygotes. All animal experiments were performed according to institutional regulations at Harvard University.
Neuronal Cultures.
Primary mouse hippocampal cultures were generated from newborns as previously described (20), and all analyses were performed at DIV 14–17.
Morphology.
Confocal and STED experiments were performed with a Leica SP8 Confocal/STED 3× microscope, and detailed information is outlined in SI Methods. For transmission EM, neuronal cultures were fixed with glutaraldehyde or frozen with a Leica EM ICE high-pressure freezer. Tissue processing, imaging, and data analyses were performed as described previously (20). All experiments were done by an experimenter blind to the genotype.
Electrophysiology and Imaging.
mEPSC recordings and sypHy imaging experiments were performed in cultured neurons at DIV 14–17. SypHy was delivered by lentiviral infection. All experiments were done by an experimenter blind to the genotype.
Supplementary Material
Acknowledgments
We thank L. Bickford, J. Wang, E. Atwater, and M. Sanghvi for technical assistance; Dr. A. deJong, Dr. W. Regehr, and R. Held for comments; Drs. T. Südhof and S. Schoch for antibodies; and Drs. M. Verhage and J. Broeke for a MATLAB program to analyze EM images. This work was supported by National Institutes of Health (NIH) Grants R01NS083898 and R01MH113349 (to P.S.K.); the Brain Research Foundation (P.S.K.); the Harvard Brain Initiative (P.S.K.); the Armenise Harvard Foundation (P.S.K.); fellowships from the Croucher Foundation (to M.Y.W.) and the Lefler Foundation (to M.Y.W.); the Brooks, Fix and Gordon Fellowship Funds (to C.L.); and National Science Foundation graduate research fellowship DGE1144152 (to S.S.H.W.). The following Harvard Medical School core facilities supported this project: Neurobiology Imaging (supported by NIH Grant P30NS072030), Conventional Electron Microscopy, the Neurodiscovery Center, and the Transgenic Cores at Brigham Women’s and Boston Children’s Hospital (Intellectual and Developmental Disabilities Research Center, supported by NIH Grant P30HD18655).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719012115/-/DCSupplemental.
References
- 1.Südhof TC. The presynaptic active zone. Neuron. 2012;75:11–25. doi: 10.1016/j.neuron.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serra-Pagès C, Medley QG, Tang M, Hart A, Streuli M. Liprins, a family of LAR transmembrane protein-tyrosine phosphatase-interacting proteins. J Biol Chem. 1998;273:15611–15620. doi: 10.1074/jbc.273.25.15611. [DOI] [PubMed] [Google Scholar]
- 3.Schoch S, et al. RIM1alpha forms a protein scaffold for regulating neurotransmitter release at the active zone. Nature. 2002;415:321–326. doi: 10.1038/415321a. [DOI] [PubMed] [Google Scholar]
- 4.Ko J, Na M, Kim S, Lee JR, Kim E. Interaction of the ERC family of RIM-binding proteins with the Liprin-alpha family of multidomain proteins. J Biol Chem. 2003;278:42377–42385. doi: 10.1074/jbc.M307561200. [DOI] [PubMed] [Google Scholar]
- 5.Zhen M, Jin Y. The Liprin protein SYD-2 regulates the differentiation of presynaptic termini in C. elegans. Nature. 1999;401:371–375. doi: 10.1038/43886. [DOI] [PubMed] [Google Scholar]
- 6.Dai Y, et al. SYD-2 Liprin-alpha organizes presynaptic active zone formation through ELKS. Nat Neurosci. 2006;9:1479–1487. doi: 10.1038/nn1808. [DOI] [PubMed] [Google Scholar]
- 7.Chia PH, Patel MR, Wagner OI, Klopfenstein DR, Shen K. Intramolecular regulation of presynaptic scaffold protein SYD-2/Liprin-α. Mol Cell Neurosci. 2013;56:76–84. doi: 10.1016/j.mcn.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Astigarraga S, Hofmeyer K, Farajian R, Treisman JE. Three Drosophila Liprins interact to control synapse formation. J Neurosci. 2010;30:15358–15368. doi: 10.1523/JNEUROSCI.1862-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller KE, et al. Direct observation demonstrates that Liprin-alpha is required for trafficking of synaptic vesicles. Curr Biol. 2005;15:684–689. doi: 10.1016/j.cub.2005.02.061. [DOI] [PubMed] [Google Scholar]
- 10.Choe K-M, Prakash S, Bright A, Clandinin TR. Liprin-alpha is required for photoreceptor target selection in Drosophila. Proc Natl Acad Sci USA. 2006;103:11601–11606. doi: 10.1073/pnas.0601185103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taru H, Jin Y. The Liprin homology domain is essential for the homomeric interaction of SYD-2/Liprin-α protein in presynaptic assembly. J Neurosci. 2011;31:16261–16268. doi: 10.1523/JNEUROSCI.0002-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel MR, Shen K. RSY-1 is a local inhibitor of presynaptic assembly in C. elegans. Science. 2009;323:1500–1503. doi: 10.1126/science.1169025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zürner M, Schoch S. The mouse and human Liprin-alpha family of scaffolding proteins: Genomic organization, expression profiling and regulation by alternative splicing. Genomics. 2009;93:243–253. doi: 10.1016/j.ygeno.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Zürner M, Mittelstaedt T, tom Dieck S, Becker A, Schoch S. Analyses of the spatiotemporal expression and subcellular localization of Liprin-α proteins. J Comp Neurol. 2011;519:3019–3039. doi: 10.1002/cne.22664. [DOI] [PubMed] [Google Scholar]
- 15.Spangler SA, et al. Differential expression of Liprin-α family proteins in the brain suggests functional diversification. J Comp Neurol. 2011;519:3040–3060. doi: 10.1002/cne.22665. [DOI] [PubMed] [Google Scholar]
- 16.Wyszynski M, et al. Interaction between GRIP and Liprin-alpha/SYD2 is required for AMPA receptor targeting. Neuron. 2002;34:39–52. doi: 10.1016/s0896-6273(02)00640-2. [DOI] [PubMed] [Google Scholar]
- 17.Spangler SA, et al. Liprin-α2 promotes the presynaptic recruitment and turnover of RIM1/CASK to facilitate synaptic transmission. J Cell Biol. 2013;201:915–928. doi: 10.1083/jcb.201301011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fouquet W, et al. Maturation of active zone assembly by Drosophila Bruchpilot. J Cell Biol. 2009;186:129–145. doi: 10.1083/jcb.200812150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dani A, Huang B, Bergan J, Dulac C, Zhuang X. Superresolution imaging of chemical synapses in the brain. Neuron. 2010;68:843–856. doi: 10.1016/j.neuron.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang SSH, et al. Fusion competent synaptic vesicles persist upon active zone disruption and loss of vesicle docking. Neuron. 2016;91:777–791. doi: 10.1016/j.neuron.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imig C, et al. The morphological and molecular nature of synaptic vesicle priming at presynaptic active zones. Neuron. 2014;84:416–431. doi: 10.1016/j.neuron.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 22.Tang A-H, et al. A trans-synaptic nanocolumn aligns neurotransmitter release to receptors. Nature. 2016;536:210–214. doi: 10.1038/nature19058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grauel MK, et al. RIM-binding protein 2 regulates release probability by fine-tuning calcium channel localization at murine hippocampal synapses. Proc Natl Acad Sci USA. 2016;113:11615–11620. doi: 10.1073/pnas.1605256113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Böhme MA, et al. Active zone scaffolds differentially accumulate Unc13 isoforms to tune Ca(2+) channel-vesicle coupling. Nat Neurosci. 2016;19:1311–1320. doi: 10.1038/nn.4364. [DOI] [PubMed] [Google Scholar]
- 25.Ackley BD, et al. The two isoforms of the Caenorhabditis elegans leukocyte-common antigen related receptor tyrosine phosphatase PTP-3 function independently in axon guidance and synapse formation. J Neurosci. 2005;25:7517–7528. doi: 10.1523/JNEUROSCI.2010-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Owald D, et al. A Syd-1 homologue regulates pre- and postsynaptic maturation in Drosophila. J Cell Biol. 2010;188:565–579. doi: 10.1083/jcb.200908055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel MR, et al. Hierarchical assembly of presynaptic components in defined C. elegans synapses. Nat Neurosci. 2006;9:1488–1498. doi: 10.1038/nn1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wentzel C, et al. mSYD1A, a mammalian synapse-defective-1 protein, regulates synaptogenic signaling and vesicle docking. Neuron. 2013;78:1012–1023. doi: 10.1016/j.neuron.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lo HF, et al. Association of dysfunctional synapse defective 1 (SYDE1) with restricted fetal growth - SYDE1 regulates placental cell migration and invasion. J Pathol. 2017;241:324–336. doi: 10.1002/path.4835. [DOI] [PubMed] [Google Scholar]
- 30.Hoogenraad CC, et al. Liprinalpha1 degradation by calcium/calmodulin-dependent protein kinase II regulates LAR receptor tyrosine phosphatase distribution and dendrite development. Dev Cell. 2007;12:587–602. doi: 10.1016/j.devcel.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 31.Astro V, et al. Liprin-α1 and ERC1 control cell edge dynamics by promoting focal adhesion turnover. Sci Rep. 2016;6:33653. doi: 10.1038/srep33653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao YQ, et al. Presynaptic Ca2+ channels compete for channel type-preferring slots in altered neurotransmission arising from Ca2+ channelopathy. Neuron. 2004;43:387–400. doi: 10.1016/j.neuron.2004.07.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.