Significance
Insulin-like growth factor 2 (IGF2) is an important growth factor with a critical role for fetal growth in mammals. The ZBED6 transcription factor is unique to placental mammals and has evolved from a domesticated DNA transposon. This study shows that ZBED6 and its interaction with the Igf2 locus play a prominent role in regulating postnatal growth of skeletal muscle and internal organs (kidney, liver, and heart) in placental mammals. This prominent role in mammalian biology provides a reasonable explanation why ZBED6 is highly conserved among all families of placental mammals and why 16 base pairs encompassing the ZBED6 binding site in an intron of Igf2 are conserved among the great majority of, if not all, placental mammals.
Keywords: ZBED6, IGF2, mouse, muscle growth, metabolic regulation
Abstract
A single nucleotide substitution in the third intron of insulin-like growth factor 2 (IGF2) is associated with increased muscle mass and reduced subcutaneous fat in domestic pigs. This mutation disrupts the binding of the ZBED6 transcription factor and leads to a threefold up-regulation of IGF2 expression in pig skeletal muscle. Here, we investigated the biological significance of ZBED6–IGF2 interaction in the growth of placental mammals using two mouse models, ZBED6 knock-out (Zbed6−/−) and Igf2 knock-in mice that carry the pig IGF2 mutation. These transgenic mice exhibit markedly higher serum IGF2 concentrations, higher growth rate, increased lean mass, and larger heart, kidney, and liver; no significant changes were observed for white adipose tissues. The changes in body and lean mass were most pronounced in female mice. The phenotypic changes were concomitant with a remarkable up-regulation of Igf2 expression in adult tissues. Transcriptome analysis of skeletal muscle identified differential expression of genes belonging to the extracellular region category. Expression analysis using fetal muscles indicated a minor role of ZBED6 in regulating Igf2 expression prenatally. Furthermore, transcriptome analysis of the adult skeletal muscle revealed that this elevated expression of Igf2 was derived from the P1 and P2 promoters. The results revealed very similar phenotypic effects in the Zbed6 knock-out mouse and in the Igf2 knock-in mouse, showing that the effect of ZBED6 on growth of muscle and internal organs is mediated through the binding site in the Igf2 gene. The results explain why this ZBED6 binding site is extremely well conserved among placental mammals.
Insulin-like growth factor 2 (IGF2) was discovered as a growth-promoting factor in the 1970s (1–3). Since that time, IGF2 has been subjected to intensive studies to understand its biological role in growth and development and to reveal how IGF2 expression is regulated in different tissues and developmental stages. IGF2 is a maternally imprinted gene normally expressed only from the paternal allele (4, 5), with the expression being derived from multiple promoters. For instance, human IGF2 is regulated by four promoters (P1–P4), whereas mouse Igf2 is under the control of three promoters (P1–P3) (6). During the development of human embryos, expression of IGF2 is mainly arising from the fetal-specific promoters P2–P4. After birth, expression of IGF2 is dramatically reduced, and in adult tissues, only low expression, mainly from the P1 promoter, is found (7).
ZBED6 was discovered as a transcriptional repressor of IGF2 subsequent to the discovery that a single base change in IGF2 intron 3 in pigs is causing a major quantitative trait locus affecting muscle growth, size of the heart, and fat deposition (8, 9). The mutation disrupts a ZBED6 binding site that is extremely well conserved among placental mammals, and results in a threefold up-regulation of IGF2 expression in pig skeletal muscle. A single-nucleotide substitution in the binding site (G > A) in domestic pigs was associated with threefold increase in IGF2 mRNA in skeletal muscle and resulted in increased muscle mass and heart size and reduced subcutaneous fat mass (8). The importance of this ZBED6 binding site for regulating IGF2 mRNA expression has been documented in several human and murine cell lines (9–12). Furthermore, ChIP-seq analysis has demonstrated that ZBED6 binds, in addition to the IGF2 site, thousands of sites in the human and murine genomes. In the present study, we established both a Zbed6 knock-out (Zbed6−/−) and an Igf2 knock-in (Igf2G/A) mouse, the latter carrying the same mutation as previously described in pigs, to investigate the physiological importance of the ZBED6–IGF2 axis. This experimental design allowed us to investigate which phenotypic effects caused by Zbed6 inactivation are mediated through its interaction with the binding site in the Igf2 gene.
Results
Establishment of Zbed6 Knock-Out and Igf2 Knock-In Transgenic Mice.
ZBED6 is encoded by an intronless gene located in the first intron of the Zc3h11a gene. The conditional Zbed6 knock-out mouse was generated by inserting loxP sites flanking the coding sequence of Zbed6, using homologous recombination (Fig. 1A). These mice were crossed with mice expressing Cre in germ-line (PGK-Cre), which resulted in deletion of Zbed6 in all tissues. The Igf2 allele (Igf2A) was generated by introducing a single-nucleotide substitution of G to A by homologous recombination at the ZBED6 binding site GGCTCG in the first intron of Igf2, which is identical to the regulatory mutation in the domestic pig (8) (Fig. 1B). The aim of the experimental design was to only compare wild-type Igf2G/G homozygotes with Igf2pA/mG heterozygotes (p = paternal, m = maternal), because it is well established that Igf2 is paternally expressed in mice. The intercross between Zbed6 knock-out and Igf2 knock-in mice was established by crossing male mice heterozygous at both Zbed6 and Igf2 (♂ Zbed6+/−, Igf2A/G) with females that were heterozygous at Zbed6 and homozygous wild-type at Igf2 (♀ Zbed6+/−, Igf2G/G) to obtain paternal expression of the Igf2 mutation in 50% of the offspring. Littermates representing four of the six genotype classes were used in the phenotypic characterization and downstream analysis; the Zbed6+/− mice were only used in the expression analysis of fetal tissue. The following abbreviations are used in the figures to represent the four genotypes: WT-G = Zbed6+/+, Igf2G/G; WT-G/A = Zbed6+/+, Igf2pA/mG; KO-G = Zbed6−/−, Igf2G/G; and KO-G/A = Zbed6−/−, Igf2pA/mG.
Fig. 1.
Development of Zbed6−/− and Igf2 mutant mouse models. (A) Two homology arms were used to insert loxP sites flanking the Zbed6 coding sequence. A single FRT site remained after the Neo cassette had been removed by FlpE-mediated recombination. The conditional knock-out mice were crossed with mice expressing Cre in germ line to eliminate the sequences between the loxP sites resulting in the elimination of the entire Zbed6 coding sequence. (B) The Igf2 mutant mouse model was generated using homologous recombination and resulted in the insertion of a single nucleotide substitution G to A at position 6 in the ZBED6 binding site (GGCTCG) in the first intron of Igf2. Two loxP sites and a single FRT remained in the transgene used in the present study. Blue arrows indicate Igf2 promoters (P1, P2 and P3) (6). (C) Serum concentrations of IGF2 were determined in male (Left) and female (Right) mice of different genotypes. Results are means ± SEM ***P < 0.001.
Disruption of ZBED6–IGF2 Interaction Resulted in Increased Circulating Levels of IGF2 and Increased Growth.
Serum IGF2 levels were about eightfold higher in Zbed6 knock-out and Igf2 knock-in mice compared with wild-type littermates (Fig. 1C). In wild-type mice, levels ranged between 1.2 and 3.2 ng/mL, whereas levels were >10 ng/mL in all three mutant genotype groups. There was no significant difference in IGF2 serum levels among the three mutant genotypes.
Both the Zbed6 knock-out and Igf2 knock-in mice showed increased body weight, around 20% in Igf2pA/mG and 15% in Zbed6−/− females and 14% in Igf2pA/mG and 4% in Zbed6−/− males at the age of 20 wk (Fig. 2A). Thus, the increased growth rate was most pronounced in the Zbed6 wild-type, Igf2 mutant mice (Fig. 2A). After 20 wk of age, the mice were transferred to a facility in which quantitative measurements of whole-body composition were performed using an EchoMRI system at 23 wk of age. At this age, the difference in body weight between wild-type and mutant mice was only significant in females (Fig. 2B). In fact, there appears to be a trend in males in which higher body weights in Zbed6−/− mice at early age (Fig. 2A) disappear over time and are absent at 23 wk of age (Fig. 2B). EchoMRI revealed an increased lean body mass in female Igf2pA/mG and Zbed6−/− mice (Fig. 2B). Inactivating Zbed6 or paternal expression of the mutant Igf2 allele had very similar effects on lean body mass (Fig. 2B).
Fig. 2.
Increased body weight and lean mass in Igf2pA/mG and Zbed6−/− mice. (A) Body weight measurements of females (Left) and males (Right) of different genotypes starting from 6 wk until 20 wk of age; lines represent the average for each group. Results are means ± SEM. (B) Body weight and results of body composition analysis using an EchoMRI system that were used to quantify fat mass and lean mass in females (Top) and males (Bottom) at 23 wk of age. **P < 0.01.
We further characterized the effects of the ZBED6–Igf2 interaction on lean body mass by individual dissection of four skeletal muscle types (gastro-gastrocnemius, soleus, tibialis anterior, and extensor digitorum longus). All muscle types were significantly increased in size in females for all three mutant genotype groups (Fig. 3A), whereas in males, the difference between genotypes was minor and only reached statistical significance in Zbed6 wild-type-Igf2 knock-in males (Fig. 3B). In contrast, quantitative PCR (qPCR) analysis of cDNA from tibialis anterior muscle showed a very similar marked up-regulation of Igf2 mRNA in males and females and across the three mutant genotypes (Fig. 3C). The up-regulation of Igf2 expression in the Igf2pA/mG Zbed6−/− double-mutant mice was very similar to that observed in the Igf2pA/mG or Zbed6−/− single-mutant mice. This indicates that there is no additive effect in Igf2 up-regulation by disrupting Zbed6 and by deleting the ZBED6 binding site in Igf2.
Fig. 3.
Increased muscle growth in Igf2pA/mG and Zbed6−/− mice. (A and B) Weight of dissected gastrocnemius (gastro), soleus, tibialis anterior, and extensor digitorum longus (EDL) muscles in the indicated genotypes for females (A) and males (B). (C) qPCR analysis of Igf2 mRNA in tibialis anterior muscle. (D) Respiratory exchange ratio (RER) during day and night in single-housed female and male mice with ad libitum access to food. (E and F) Blood glucose after i.p. administration of a glucose bolus and plasma insulin at baseline and 15 min post glucose stimulation in females (E) and males (F). Results are means ± SEM *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t test or two-way ANOVA.
Mild Metabolic Changes in Zbed6−/− Mice.
Respiratory exchange ratios (RERs) were measured during 2 consecutive days and nights in metabolic cages. This revealed an overall increased reliance on carbohydrates, rather than lipids, as a fuel source for male Zbed6−/− mice, with higher RER values than Zbed6+/+ mice during both day and night (Fig. 3D). A corresponding difference was not noted for female mice (Fig. 3D). The assessment of whole-body glucose tolerance showed that deletion of Zbed6 or the IGF2 mutation had no effect on overall glucose tolerance in males or females (Fig. 3 E and F). However, male Zbed6−/− mice achieved the same glucose clearance from the blood, with significantly reduced plasma insulin levels, indicating increased whole-body insulin sensitivity (Fig. 3F).
Up-Regulation of Igf2 Expression Promotes the Growth of Internal Organs.
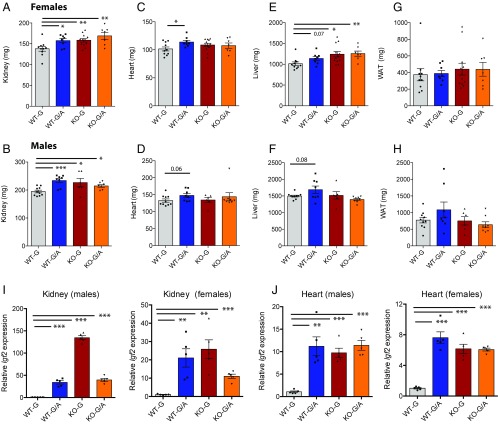
The weights of internal organs (kidney, heart, and liver) were higher in Zbed6−/− and Igf2pA/mG mice compared with wild-type littermates. This effect was most pronounced for kidney, where a 14–20% increased organ weight was observed for both females and males of the three types of transgenic mice (Fig. 4 A and B). Moreover, the heart size tended to be larger for all three classes of mutant mice, but the trend was only statistically significant (females) or close to significant (males) for Zbed6 wild-type–Igf2 knock-in mice (Fig. 4 C and D). The pattern of increased growth was also apparent in liver from females of all three types of transgenic mice (Fig. 4E), whereas only Zbed6 wild-type–Igf2 knock-in males showed a tendency for increased liver weight that approached statistical significance. In contrast to the increased weight of internal organs in Zbed6/Igf2 mutant mice, no significant changes were observed in the weight of gonadal white adipose tissue (Fig. 4 G and H). qPCR analysis revealed a dramatic up-regulation of Igf2 mRNA in kidney of both Igf2pA/mG and Zbed6−/− mice and in both sexes (Fig. 4I). An elevation of Igf2 mRNA expression was also documented in heart (Fig. 4J). In general, the differences in the growth of internal organs were more pronounced in females than in males, whereas the up-regulation of Igf2 expression was similar in males and females.
Fig. 4.
The ZBED6–Igf2 axis affects the growth of internal organs. (A–H) Weight of dissected kidney, heart, liver and gonadal white adipose tissue (WAT) in females and males in milligrams (mg). (I) Expression analysis of Igf2 mRNA using qPCR in kidney tissue from males (Left) and females (Right). (J) Expression analysis of Igf2 mRNA in heart tissue from males (Left) and females (Right). Results are means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t test.
The ZBED6–Igf2 Axis Has a Major Effect on Transcriptional Regulation at the Igf2 Locus.
Igf2 and the H19 noncoding RNA are imprinted genes in which Igf2 is expressed from the paternal allele and H19 from the maternal allele. The transcriptional regulation of these two genes is controlled by several factors. For instance, disruption of the imprinting control region between Igf2 and H19 leads to biallelic expression of both Igf2 and H19 (13, 14). Here, we explored transcriptional changes caused by the disruption of the ZBED6–Igf2 interaction on the whole transcriptome and on the Igf2 locus. We conducted whole-transcriptome analysis of multiple tissues of wild-type, Zbed6−/−, and Igf2pA/mG mice; the double-mutant mice were not included in this analysis. Around 10 million sequenced reads per sample were aligned to the mouse reference genome (mm10), and the uniquely mapped reads were used for differential expression (DE) analysis, using Cufflinks (15). Zbed6 is located in the first intron of another zinc-finger protein gene Zc3h11a, and the ZBED6 protein is expressed after intron retention in transcripts containing the entire coding sequence of this “host” gene (9). However, the RNAseq data indicated that the expression of Zc3h11a was not affected by Zbed6 inactivation (Fig. S1).
The visualization of the mapped reads in female skeletal muscle tissue (tibialis anterior) confirmed a complete knock-out of Zbed6 expression in Zbed6−/− mice, whereas Zbed6 expression was not affected in Igf2pA/mG mice as expected (Fig. 5A). In tibialis anterior muscle of female mice, 108 and 82 DE genes were identified in Zbed6−/− and Igf2pA/mG mice, respectively, compared with wild-type mice (P < 0.05 after Benjamini–Hochberg correction for multiple testing; Fig. 5B), with 42 shared DE genes between the two groups (Fig. 5C). Igf2 was the most significant DE gene in skeletal muscle of both Zbed6−/− and Igf2pA/mG mice, with more than 20-fold up-regulation (Fig. 5D), in agreement with the qPCR analysis (Fig. 3C). The Igf2 opposite-strand (Igf2os) transcript was also up-regulated 20-fold, whereas no significant changes were detected for the H19 RNA (Fig. 5D).
Fig. 5.
Transcriptome analysis of the tibialis anterior muscle in Igf2pA/mG and Zbed6−/− female mice. (A) Uniquely mapped reads at the Zbed6 locus in Zbed6−/− (KO-G), Igf2G/A (WT-G/A), and WT (WT-G) mice. (B) Volcano plot showing differentially expressed (DE) genes in skeletal muscle of KO-G (Left) and WT-G/A (Right) vs. WT mice. Red dots represent statistically significant differential expression (P < 0.05). (C) Overlap between DE genes in skeletal muscle of KO-G and WT-G/A mice. (D) The expression of Igf2, Igf2os, and H19 transcripts as measured by FPKM values. (E) Expression analysis of two putative direct ZBED6 targets (Tmem8 and Enho) that were up-regulated in KO-G mice, but not in WT-G/A mice. Results are means ± SEM *P < 0.05; **P < 0.01; ***P < 0.001, after Benjamini–Hochberg correction for multiple testing.
A number of genes showed differential expression in Zbed6−/−, but not in Igf2pA/mG, mice, and vice versa (Fig. 5C). For example, the expression of the transmembrane protein 8 (Tmem8) and energy homeostasis-associated (Enho) genes were significantly up-regulated in skeletal muscle of Zbed6−/− mice, but not Igf2pA/mG mice (Fig. 5E). Both Tmem8 and Enho were previously identified as putative direct targets of ZBED6 based on ChIP-seq analysis (10). ZBED6 is expected to affect transcriptional regulation of many genes other than Igf2. The significant differences in the transcriptome between Zbed6−/− and Igf2pA/mG mice, although they both overexpress Igf2, is consistent with this notion. The gene ontology analysis of DE genes in skeletal muscle in both Zbed6−/− and Igf2pA/mG mice revealed a significant enrichment of genes belonging to the extracellular region category (Dataset S1).
The results of the transcriptome analysis of skeletal muscle tissue from male mice were in excellent agreement with the results obtained for female mice (Fig. S2 and Dataset S2). In agreement with expression analysis of the skeletal muscle tissues of the females, Igf2 was the most up-regulated gene in both Zbed6−/− and Igf2pA/mG muscle tissues, whereas Enho and Tmem8 were only up-regulated in the skeletal muscle of Zbed6−/− mice (Fig. S2).
We also carried out transcriptome analysis of heart and kidney tissue (SI Text and Figs. S3 and S4). The results were in line with those obtained for the muscle transcriptome, and the results for males and females were highly consistent. The numbers of DE genes per genotype (Zbed6−/− and Igf2pA/mG) were in the range of 26–255. We observed a dramatic increase in Igf2 expression in both genotypes; in contrast, the putative ZBED6 targets Enho, Tmem8, and Pianp (paired immunoglobin-like type 2 receptor-associating neural protein) only showed differential expression between wild-type and Zbed6−/− mice. Interestingly, Tmem8 showed highly significant differential expression in heart, but not in kidney, suggesting tissue-specific ZBED6 regulation (Fig. S3 D and K).
ZBED6 Primarily Regulates Igf2 Expression Postnatally.
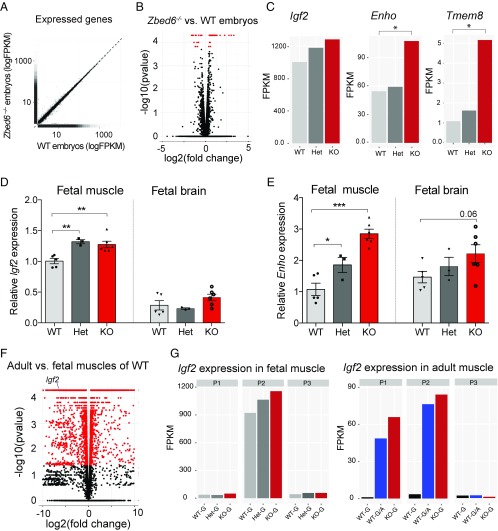
Igf2 is expressed at high levels in fetal tissues and is markedly down-regulated after birth. Our results show that the disruption of ZBED6–Igf2 interaction leads to a profound up-regulation of Igf2 expression in multiple organs in adult mice (see previous section). To investigate the possible role of ZBED6 in regulating Igf2 expression in fetal tissues, we performed transcriptome analysis of fetal muscle tissue using Zbed6−/−, Zbed6+/−, and wild-type embryos at day 12.5. Fifty-seven genes showed significant differential expression between wild-type and Zbed6−/− embryos (Fig. 6 A and B). A closer examination of these differentially expressed genes (Fig. S5) shows that as many as 28 (49%) are associated with a ZBED6 ChIP-seq peak, which is a significant enrichment, as only 23% of all of the 12,228 genes with a detectable expression in this experiment were associated with a ChIP-seq peak (P = 0.002; Fisher’s exact test). The result implies that a large proportion of these 28 genes are direct targets of ZBED6. Furthermore, 25 of the 28 DE genes associated with a ChIP-seq peak showed significant up-regulation of expression in Zbed6−/− mice, consistent with the notion that ZBED6 primarily, or possibly exclusively, acts as a repressor. In fact, the differential expression for the three genes showing the opposite trend was not convincing because of heterogeneity within genotype classes (Fig. S5). Interestingly, for the majority of these 28 genes, showing DE between wild-type and Zbed6−/− mice, as well as for the Zbed6 locus itself, the Zbed6+/− heterozygotes showed intermediate expression indicating that the direct targets of ZBED6 are dose sensitive for the amount of ZBED6 present (Fig. S5).
Fig. 6.
Limited effect of ZBED6 silencing on Igf2 expression in fetal tissues. (A) Transcriptome analysis of fetal muscle tissue of Zbed6−/− and WT embryos at day 12.5 of development. The expression of all genes is plotted as logFPKM. (B) Volcano plot comparing the transcriptome in Zbed6−/− vs. WT embryos; differentially expressed transcripts reaching statistical significance (P < 0.05) are indicated in red. (C) Expression of Igf2, Enho, and Tmem8 measured as FPKM values in wild-type (WT), Zbed6+/− (Het), and Zbed6−/− (KO) mice. (D and E) qPCR validation of the expression of Igf2 (D) and Enho (E) in fetal tissues, results are means ± SEM *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t test. (F) Volcano plot of genes in adult versus fetal muscle of WT mice; differentially expressed transcripts reaching statistical significance (P < 0.05) are indicated in red. (G) Differential expression of Igf2 isoforms generated from different Igf2 promoters (P1–3) in fetal (Left) and adult muscle (Right) of WT and mutant mice.
In contrast to what we observed in adult tissues, Igf2 showed only marginally increased expression when ZBED6 was knocked out in fetal muscle tissue (Fig. 6C). Two other putative direct targets for ZBED6, Enho and Tmem8, were both significantly up-regulated in Zbed6−/− embryos (Fig. 6C). The differential expression of Enho and Tmem8 and other putative direct targets (Fig. S5) showed that ZBED6 must be functionally active in fetal tissues, despite the lack of a strong up-regulation of Igf2 expression. The observed differential expression patterns from RNA-seq analysis of embryos were validated by qPCR assay, using fetal muscle and brain tissues. The change in Igf2 mRNA expression in Zbed6−/− embryos was limited to a 1.2-fold up-regulation in fetal muscle tissues, whereas no significant changes were detected in brain tissue (Fig. 6D). In agreement with RNA-seq data of embryos, the expression of Enho mRNA was up-regulated threefold in Zbed6−/− and approximately twofold in Zbed6+/− fetal muscle tissues (Fig. 6E).
A comparison of transcriptome data from fetal and adult skeletal muscle tissues of wild-type mice demonstrated dramatic changes in the expression profile of thousands of genes. As expected, Igf2 was one of the most significantly DE genes between fetal and adult tissues, with high expression in fetal muscle and low expression in adult skeletal muscle (Fig. 6F). An isoform quantification analysis using Cufflinks revealed that the majority of Igf2 expression in fetal tissues exists as the NM_010514 isoform, which is mainly initiated from the P2 promoter (Fig. 6G, Left). Surprisingly, the disruption of the ZBED6–Igf2 interaction led to increased transcriptional activities from both the P1 and P2 promoters in adult muscle. The elevated Igf2 expression in Igf2pA/mG and Zbed6−/− muscles was derived 60% from the NM_010514 isoform and 40% from the NM_00112273 isoform (Fig. 6G, Right). The results suggest that ZBED6 acts as a more important repressor at the Igf2 locus in adult tissues than in fetal tissue.
Discussion
The evolution of ZBED6 is an innovation that took place during the development of placental mammals. The high sequence conservation of the Zbed6 gene across placental mammals shows that it is an essential gene in this taxonomic group (9). The ZBED6–Igf2 interaction also appears to be essential. This is based on the fact that the 16 bp, which include the ZBED6 binding site in an intronic region of Igf2, are highly conserved among placental mammals (8). The present study shows that the Zbed6 knock-out mice develop normally and show no obvious pathological conditions. However, we show the ZBED6–Igf2 axis plays an essential role in regulating postnatal growth of muscles and internal organs; findings that provide a reasonable explanation for the high sequence conservation of ZBED6 and its binding site in the Igf2 gene. Thus, the evolution of ZBED6 in placental mammals has added another layer of regulation on the action of the highly potent growth factor IGF2.
The observed effects of Zbed6 knock-out and Igf2 knock-in on Igf2 expression and serum IGF2 concentrations, as well as on muscle and organ growth, were very similar, and there was no clear additive effect of combining the two mutations. This strongly suggests that ZBED6 is regulating muscle and organ growth through its interaction with Igf2. Does this mean that the main functional role of ZBED6 is to regulate Igf2 expression? This is probably not the case, because we also show that Zbed6 inactivation and disruption of the ZBED6 binding site in Igf2 only led to partially overlapping changes in the transcriptome (Fig. 5C). We identified a number of putative direct ZBED6 targets that showed differential expression in the Zbed6 knock-out mice, but not in Igf2 knock-in mice (e.g., Fig. 6E). Because ZBED6 has a very broad tissue distribution and is expressed during different developmental stages, it is plausible that Zbed6 inactivation is causing additional phenotypic effects on traits that were not measured in the current study. The identification of increased muscle and organ growth was guided by our previous discovery that disruption of the ZBED6 binding site in IGF2 is the causal mutation for a major QTL affecting muscle and heart growth in pigs (8).
Zbed6 inactivation and the Igf2 knock-in mutation both resulted in dramatic changes in Igf2 expression (up to 100-fold higher expression) compared with the rather modest phenotypic changes. Therefore, an interesting topic for future research will be to explore how the complex physiological regulation of IGF2 signaling may hamper the consequences of the dramatic up-regulation of IGF2 expression in muscle, kidney, and other tissues. IGF2 signaling is regulated by the IGF binding proteins that sequester IGF2 in circulation, the IGF2 receptor (IGF2R) that reduces the bioactivity of IGF2 (16), and regulation of the insulin receptor and IGF1 receptor that can both be activated by IGF2 (17). The Igf2 locus also has a complex transcriptional regulation involving several regulatory factors such as CTCF that maintains the imprinted expression pattern of Igf2 and H19 (18, 19). This study adds to the characterization of transcriptional regulation at the Igf2 locus and shows that the expression of several forms of the Igf2 transcript, including the opposite-strand (Igf2os) transcript, are affected by the interaction with ZBED6. In contrast, the ZBED6–Igf2 interaction does not appear to have a significant effect on transcriptional regulation of the neighboring H19 gene.
In sharp contrast to the dramatic up-regulation in adult tissues, only a modest effect of Zbed6 silencing on Igf2 expression was noted in fetal tissues. This result is in perfect agreement with our previous characterization of IGF2 mRNA expression in fetal and adult tissues in pigs carrying the point mutation disrupting the ZBED6 site in IGF2 (8). The minor effect in fetal tissues is unlikely to be a result of a lack of ZBED6 expression, as Zbed6 mRNA levels were similar in adult and fetal tissue. Furthermore, RNA-seq data from fetal tissue indicated that ZBED6 represses the expression of other putative direct ZBED6 targets such as Enho and Tmem8 previously identified by ChIP-seq analysis (9, 10). A more plausible explanation for the modest effect of Zbed6 silencing on Igf2 expression is epigenetic regulation, as gel shift assays indicated that ZBED6 does not bind its target sequence (GGCTCpG) when it is methylated (8).
The phenotypic consequences of Zbed6 inactivation and the Igf2 regulatory mutation in transgenic mice showed both striking similarities and differences compared with domestic pigs carrying the IGF2 mutation (8). There was an excellent agreement between transgenic mice and mutant pigs regarding the minor role for ZBED6 in the regulation of Igf2 expression in fetal tissues in contrast to the major role in postnatal tissue. In addition, the increased muscle growth is consistent with the pig phenotype, but the increased muscle growth in female transgenic mice (+15–20% increase) was more pronounced than in pigs (+4%). A possible explanation for this difference is that the relative effect of the IGF2 mutation in pigs is reduced because of the long history of selection for increased muscle growth in pigs. This selective breeding will have affected many genes in the genome, although IGF2 was by far the most important locus explaining differences in muscle growth in an intercross between the European wild boar and Large White domestic pigs (20). However, the marked sex difference in mice, including a more pronounced effect on muscle growth in females than in males, is not observed in pigs. A possible explanation for this species difference is that most pigs used for meat production are castrated. Thus, the phenotypic consequences of the up-regulated expression of Igf2 in male transgenic mice may have been masked by the action of testosterone. This interpretation is supported by the fact that the up-regulation of both Igf2 expression and serum IGF2 levels were as dramatic in male as in female mice. The up-regulation of Igf2 expression in mice (about 30-fold) was much larger than in mutant pigs (about threefold). Both mutant pigs and transgenic mice had a larger heart than wild-type animals. In contrast, the weights of both liver and kidney were larger in transgenic mice than in wild-type littermates, whereas no significant effect on the weight of kidney or liver was observed in pigs, despite the fact that the number of individuals in the pig experiment was an order of magnitude higher than in the current mouse study (20). Another striking difference was that the pig mutation did not affect serum IGF2 concentrations (8) whereas transgenic mice had about eightfold higher IGF2 concentration. A plausible explanation for this is the corresponding striking difference in the effects on IGF2 mRNA expression in liver in pigs and transgenic mice, as liver is the major source of circulating IGF2. This could also explain why transgenic mice showed a significant general effect on growth, whereas mutant pigs showed normal body weight but altered body composition. The elevated serum IGF2 concentrations in our transgenic mice imply that disturbed ZBED6–IGF2 interaction could be an explanation for elevated IGF2 serum concentrations in some human patients.
Taken together, this study demonstrates the important role of the ZBED6–Igf2 axis in regulating Igf2 expression, muscle growth, and the growth of internal organs in placental mammals. The study has also established two unique mouse models for further characterization of the functional importance of ZBED6 and the phenotypic consequences of a very specific up-regulation of Igf2 expression. For instance, up-regulated expression of IGF2 occurs in many cancers and is associated with poor prognosis (17). The Igf2 knock-in mouse with its dramatic up-regulation of Igf2 expression can now be exploited to study how IGF2 affects origin, growth and cancer progression.
Materials and Methods
Animal Models.
The Zbed6−/− and Igf2pA/mG models were both generated by homologous recombination in mouse C57BL/6 ES cells. The targeted Zbed6 locus included a Neomycin (Neo) selection cassette flanked by FRT sites and two loxP sites flanking the single Zbed6 exon. Neo was removed by mating the mice with a strain expressing FlpE in germ line, leaving the flanking loxP sites and a single FRT site within the region flanked by loxP sites (Fig. 1A). These mice were in turn mated with PGK-Cre mice (21) expressing the Cre recombinase in the germ line, resulting in the removal of the entire Zbed6 coding sequence, leaving only a single loxP site in a nonconserved region in this intronic region (Fig. 1A).
Gene targeting at the Igf2 locus introduced the single G-A base change at the ZBED6 binding site in the CpG island in an intronic region of Igf2 together with two loxP sites flanking the CpG island and a Neo cassette flanked by FRT sites. Neo was removed by mating the mice with a line expressing the FlpE recombinase in the germ line, resulting in the allele used in the current study (Fig. 1B). The loxP sites flanking the CpG island were introduced to make it possible to delete the entire CpG island to explore whether it has any functions other than harboring a ZBED6 binding site.
The two transgenic mouse models were both maintained on a C57BL/6 background. Phenotypic characterization was carried out using littermates segregating for the four genotype classes of major interest. The growth curve was generated by measuring body weight starting from 6 wk until 20 wk of age. Littermates of both males and females were used in all presented experiments, and the wild-type animals were used as control. Total lean and fat mass were assessed in conscious mice (23 wk of age), using the EchoMRI-100 system (EchoMRI LLC). Tissue samples collected from 4-h fasted Avertin-anesthetized mice; 16 μL/g body weight 2.5% avertin (2,2,2-tribromo ethanol and tertiary amyl alcohol) were used for this purpose.
All animals were group-housed with free access to food and water in the pathogen-free facilities of Uppsala University and Karolinska Institutet. All procedures described in this study were approved by the Uppsala Ethical Committee on Animal Research (#C63/15 and #C143/15) and the Stockholm Ethical Committee (#N38/15), following the rules and regulations of the Swedish Animal Welfare Agency, and were in compliance with the European Communities Council Directive of 22 September 2010 (2010/63/EU). All efforts were made to minimize animal suffering and to reduce the number of animals used.
Serum Concentration of IGF2.
Concentrations of IGF2 was determined in mouse serum, using a time-resolved immunofluorometric assay (TR-IFMA). Before assay, 50 μL mouse serum was added to 450 μL acetic acid (200 mM) with 0.05% (vol/vol) HSA, 0.5% (vol/vol) Tween20, 0.9% (wt/vol) NaCl, reducing pH to 2.66 and hereby dissociating IGF2 from the IGF binding proteins (IGFBPs). The IGFBPs were saturated with molar excess of IGF1 (100 ng/mL; GF-050–8; Austral Biologicals) and incubated for 2 h at 5 °C. Samples were neutralized by the addition of 500 μL Tris buffer [1.8% (wt/vol) Tris⋅HCL, 3.5% (wt/vol) Tris base] with 1% (vol/vol) EDTA, 0.5% HSA, 0.2% Tween20, 0.9% NaCl. Thus, in the final solution, mouse serum was diluted 1:20. Recombinant mouse IGF2 (792-MG-050; RnD Systems) served as calibrator and was subjected to the same procedures.
IGF2 concentrations were determined in triplicates using antibodies from a commercial assay (DY792; RnD Systems). Biotinylated detection antibody was diluted in assay buffer [PBS at pH 8.0, 0.05% (wt/vol) NaN3, 0.5% HSA, 0.5% Tween20, 0.9% NaCl], mixed 1:1,000 with streptavidin-europium (PerkinElmer) and determined as a TR-IFMA. Calibrators were prepared at final concentrations ranging from 0 to 1 ng/mL, and the calibration curve behaved linearly within this range. There was no cross-reactivity with mouse IGF1 (tested at 100 ng/mL). Intra-assay and interassay coefficients of variation (CVs) were <10% and limit of detection was 0.05 ng/mL Because of limited sample material, measurements were not obtained from one female Zbed6−/− mouse. All other sample measurements were above limit of detection.
RNA Sequencing.
Tissues were placed in RNAlater (Sigma) directly after collection. RNA extraction was performed using the RNeasy Mini kit (QIAGEN). The RNA quality and integrity were measured with a RNA ScreenTape assay (TapeStation; Agilent Technologies). Strand-specific mRNA sequencing libraries from five individuals per genotype were generated using the SENSE RNA-Seq Library Prep Kit (Lexogen) following the manufacturer’s instructions. The libraries were sequenced as 125-bp single reads, using an Illumina HiSeq instrument. Sequenced reads were mapped to the reference mouse genome (mm10) using STAR with default parameters (22). Cufflinks and Cuffdiff were used as described by Trapnell et al. (23) to identify differentially expressed genes and isoforms based on gene models for mm10 downloaded from the UCSC Genome Browser (https://genome.ucsc.edu/), and the abundance of gene expression was calculated as fragment per kilobase of transcript per million mapped reads (FPKM). The cummerbund R package was used for gene expression data visualization (23). The mapped reads were visualized using the integrative genomics viewer (24).
Quantitative PCR.
Total RNA was extracted using the RNeasy Mini kit (Qiagen), and the samples were treated with DNase I to eliminate genomic DNA. The High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) was used to generate cDNA from RNA. qPCR analysis was performed in ABI MicroAmp Optical 384-well Reaction plates on an ABI 7900 real-time PCR instrument, using TaqMan gene expression reagents (Applied Biosystems). The amplification and detection of each gene was performed using TaqMan Gene Expression Assays that consisted of forward and reverse primers with TaqMan minor groove binder probe for each gene (Zbed6: Mm04178798_s1; Igf2: Mm00439564_m1; Tbp: Mm01277042_m1; Enho: Mm01223541_m1; 18S: Mm03928990_g1; Applied Biosystems); 18S and Tbp were used as housekeeping genes.
Glucose Tolerance Test.
Mice (24 wk of age) were fasted for 4 h and injected intraperitoneally with 2 g/kg glucose. Blood glucose was measured 0, 15, 30, 60, and 120 min after injection (OneTouch Ultra 2 glucose meter; LifeScan). Plasma insulin concentration was measured at 0 and 15 min, using an ELISA with mouse insulin as standard (Crystal Chem).
Metabolic Cages.
Mice (24–26 wk of age) were acclimated for 24 h in single cages. Subsequently, mice were monitored for 2 d in the TSE PhenoMaster home cage system (TSE Systems) in a controlled environment (12 h light/12 h dark, 25 °C, 45% humidity) with ad libitum access to standard rodent chow (R34; Lantmännen) and water. The RER was calculated from the continuously measured O2 consumption (V̇o2) and CO2 production (V̇co2).
Statistical Analysis.
The sample sizes for phenotypic characterization were 5–10 per male genotype and 5–11 per female genotype, and five per group for gene expression analysis. Data are presented as mean ± SEM. Values were analyzed by two-tailed unpaired Student’s t tests, one-way analysis of variance followed by Tukey’s post hoc test, or two-way analysis of variance followed by Sidak’s test for multiple comparisons, using GraphPad Prism version 6. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
The work was funded by grants from the Knut and Alice Wallenberg Foundation (L.A.), the Strategic Research Programme in Diabetes at Karolinska Institutet, Swedish Research Council (2012-1760 to A.K., 2015-00165 to J.R.Z.), and the Aarhus University Research Foundation (to R.H. and J.F.). Sequencing was performed by the SNP&SEQ Technology Platform, supported by Uppsala University and Hospital, SciLifeLab and Swedish Research Council (80576801 and 70374401). Computer resources were provided by Uppsala Multidisciplinary Center for Advanced Computational Science, Uppsala University.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. SRP128271).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719278115/-/DCSupplemental.
References
- 1.Daughaday WH, et al. Somatomedin: Proposed designation for sulphation factor. Nature. 1972;235:107. doi: 10.1038/235107a0. [DOI] [PubMed] [Google Scholar]
- 2.Dulak NC, Temin HM. Multiplication-stimulating activity for chicken embryo fibroblasts from rat liver cell conditioned medium: A family of small polypeptides. J Cell Physiol. 1973;81:161–170. doi: 10.1002/jcp.1040810205. [DOI] [PubMed] [Google Scholar]
- 3.Rinderknecht E, Humbel RE. Amino-terminal sequences of two polypeptides from human serum with nonsuppressible insulin-like and cell-growth-promoting activities: Evidence for structural homology with insulin B chain. Proc Natl Acad Sci USA. 1976;73:4379–4381. doi: 10.1073/pnas.73.12.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Constância M, et al. Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature. 2002;417:945–948. doi: 10.1038/nature00819. [DOI] [PubMed] [Google Scholar]
- 5.Kaneda M. Genomic imprinting in mammals-epigenetic parental memories. Differentiation. 2011;82:51–56. doi: 10.1016/j.diff.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Sullivan MJ, Taniguchi T, Jhee A, Kerr N, Reeve AE. Relaxation of IGF2 imprinting in Wilms tumours associated with specific changes in IGF2 methylation. Oncogene. 1999;18:7527–7534. doi: 10.1038/sj.onc.1203096. [DOI] [PubMed] [Google Scholar]
- 7.de Pagter-Holthuizen P, et al. The human insulin-like growth factor II gene contains two development-specific promoters. FEBS Lett. 1987;214:259–264. doi: 10.1016/0014-5793(87)80066-2. [DOI] [PubMed] [Google Scholar]
- 8.Van Laere A-S, et al. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature. 2003;425:832–836. doi: 10.1038/nature02064. [DOI] [PubMed] [Google Scholar]
- 9.Markljung E, et al. ZBED6, a novel transcription factor derived from a domesticated DNA transposon regulates IGF2 expression and muscle growth. PLoS Biol. 2009;7:e1000256. doi: 10.1371/journal.pbio.1000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang L, et al. ZBED6 modulates the transcription of myogenic genes in mouse myoblast cells. PLoS One. 2014;9:e94187. doi: 10.1371/journal.pone.0094187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akhtar Ali M, et al. Transcriptional modulator ZBED6 affects cell cycle and growth of human colorectal cancer cells. Proc Natl Acad Sci USA. 2015;112:7743–7748. doi: 10.1073/pnas.1509193112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, et al. Transcription factor ZBED6 affects gene expression, proliferation, and cell death in pancreatic beta cells. Proc Natl Acad Sci USA. 2013;110:15997–16002. doi: 10.1073/pnas.1303625110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaffer CR, Grinberg A, Pfeifer K. Regulatory mechanisms at the mouse Igf2/H19 locus. Mol Cell Biol. 2001;21:8189–8196. doi: 10.1128/MCB.21.23.8189-8196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trapnell C, et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown J, Jones EY, Forbes BE. Keeping IGF-II under control: Lessons from the IGF-II-IGF2R crystal structure. Trends Biochem Sci. 2009;34:612–619. doi: 10.1016/j.tibs.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Livingstone C. IGF2 and cancer. Endocr Relat Cancer. 2013;20:R321–R339. doi: 10.1530/ERC-13-0231. [DOI] [PubMed] [Google Scholar]
- 18.Schoenherr CJ, Levorse JM, Tilghman SM. CTCF maintains differential methylation at the Igf2/H19 locus. Nat Genet. 2003;33:66–69. doi: 10.1038/ng1057. [DOI] [PubMed] [Google Scholar]
- 19.Engel N, Thorvaldsen JL, Bartolomei MS. CTCF binding sites promote transcription initiation and prevent DNA methylation on the maternal allele at the imprinted H19/Igf2 locus. Hum Mol Genet. 2006;15:2945–2954. doi: 10.1093/hmg/ddl237. [DOI] [PubMed] [Google Scholar]
- 20.Jeon JT, et al. A paternally expressed QTL affecting skeletal and cardiac muscle mass in pigs maps to the IGF2 locus. Nat Genet. 1999;21:157–158. doi: 10.1038/5938. [DOI] [PubMed] [Google Scholar]
- 21.Lallemand Y, Luria V, Haffner-Krausz R, Lonai P. Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site-specific recombinase. Transgenic Res. 1998;7:105–112. doi: 10.1023/a:1008868325009. [DOI] [PubMed] [Google Scholar]
- 22.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.