Significance
Insights from natural applications of antibiotics are important to gain a deeper understanding of the evolutionary processes that underlie the maintenance of an antibiotic defense and prevent the rise and spread of antibiotic resistance. Using 25 species and subspecies of beewolf digger wasps that engage in a defensive symbiosis with Streptomyces bacteria, we tracked evolutionary changes in the antibiotic cocktail that protects the wasps’ larval offspring against mold fungi. Our results yield insights into the mechanistic basis as well as the ecological and evolutionary implications of producing a complex cocktail of antimicrobial compounds in a symbiotic setting.
Keywords: defensive symbiosis, protective mutualism, antibiotic resistance, Philanthus, Streptomyces philanthi
Abstract
The increasing resistance of human pathogens severely limits the efficacy of antibiotics in medicine, yet many animals, including solitary beewolf wasps, successfully engage in defensive alliances with antibiotic-producing bacteria for millions of years. Here, we report on the in situ production of 49 derivatives belonging to three antibiotic compound classes (45 piericidin derivatives, 3 streptochlorin derivatives, and nigericin) by the symbionts of 25 beewolf host species and subspecies, spanning 68 million years of evolution. Despite a high degree of qualitative stability in the antibiotic mixture, we found consistent quantitative differences between species and across geographic localities, presumably reflecting adaptations to combat local pathogen communities. Antimicrobial bioassays with the three main components and in silico predictions based on the structure and specificity in polyketide synthase domains of the piericidin biosynthesis gene cluster yield insights into the mechanistic basis and ecoevolutionary implications of producing a complex mixture of antimicrobial compounds in a natural setting.
Since the discovery of penicillin in 1928 by Alexander Fleming, the use of antibiotics for the treatment of infectious diseases has revolutionized human medicine (1). However, the past 60 y have seen a dramatic increase in antibiotic-resistant pathogens, rendering many of the previously potent antibiotics effectively useless (2–5). The rapid evolution of resistance through mutation (6) and the spread of resistance genes by horizontal gene transfer (7–9) raise the question of how antibiotics can maintain their efficiency in a natural context over long evolutionary timescales. Unfortunately, our knowledge about the ecology and evolution of antibiotics remains severely limited. While they can be used as chemical weapons under certain symbiotic or free-living conditions (10–12), in other situations they seem to serve as signaling molecules that affect gene expression in con- or heterospecific microorganisms (13–19).
Two of the main challenges associated with studying the ecology of antibiotics are (i) the difficulty of monitoring complex microbial interactions in nature (20) and (ii) the detection and quantification of antibiotic production in situ (21). A solution for the first issue is to consider associations that involve only a limited number of interacting organisms. In particular, defensive symbioses between insects and antibiotic-producing symbionts represent attractive model systems, since a limited number of partners have interacted over millions of years to ensure protection against coevolving or opportunistic pathogens (11, 12, 20). A prime example is the tripartite symbiosis of leafcutter ants, their nutritional fungus gardens, and Actinobacteria that protect the gardens from coevolved parasitic microfungi (22–26) as well as the ants themselves against entomopathogens (27–29). One major limitation in the study of leafcutter ants and many other defensive symbioses has been the focus on bioactive microbial metabolites produced in vitro, with several antibiotic compounds characterized from Pseudonocardia and Streptomyces isolates that are associated with the leafcutter ants (25, 30–33). However, the presence of these compounds in situ and their relevance for the protective activity of the insects’ nutritional resources have received little attention (32). By contrast, HPLC–high-resolution MS, NMR, and imaging MS were successfully used for in situ characterization of the chemical mediators in another defensive symbiosis involving solitary beewolf wasps and antibiotic-producing Streptomyces bacteria (19, 21, 34).
This specialized protective symbiosis occurs in the clade of Philanthini digger wasps that are associated with host-specific strains of the actinobacterium Streptomyces philanthi (35). Beewolf females harbor the symbiotic bacteria in antennal reservoirs (36) and secrete them into their brood cells, where they are transferred to the larval cocoon and protect the developing larva against opportunistic mold fungi from the environment (37). In the European beewolf, Philanthus triangulum, this defensive activity is achieved through the symbiont-mediated production of a mixture of antimicrobial compounds, including streptochlorin and at least eight different piericidins on the cocoon (34). Despite the opportunity for environmental acquisition of nonspecific bacteria and phylogenetic evidence for occasional horizontal transfer of symbionts between host species, the symbiosis has remained specific since its origin about 68 Mya, involving monophyletic clades of wasps and bacteria, respectively (35). Considering the high selection pressure exerted by antagonistic soil microbes as well as the specificity and long coevolutionary history in the beewolf–Streptomyces symbiosis, it seems surprising that the symbiont-produced antibiotic mixture has provided an efficient defense against pathogens over the past 68 My.
Here, we addressed three key questions for understanding the evolutionary dynamics of the multicomponent antimicrobial defense in beewolf wasps and their ecological relevance. (i) How did the symbiont-produced antibiotic mixture change over evolutionary timescales and to what extent do the patterns reflect phylogenetic constraints and ecological adaptations? (ii) How is the diversity of antimicrobial compounds generated on the molecular level? (iii) Can synergistic or antagonistic interactions of compounds present in the beewolves’ antibiotic mixture affect their activity against potential antagonists?
Results
Composition of Symbiont-Produced Antibiotic Mixtures.
In the European beewolf (P. triangulum), the symbiont-produced antibiotics have been detected on the cocoon surface as well as in the antennal gland reservoirs of adult females (34). To characterize the antibiotic mixture produced by the symbiotic Streptomyces bacteria of 25 beewolf host species and subspecies in three genera, we subjected methanol extracts from antennae and/or cocoons to HPLC coupled to high-resolution electrospray ionization (ESI)-MS/MS (data available from the Dryad Digital Repository: doi:10.5061/dryad.6907h).
Comparisons of the accurate molecular masses of all detected substances with 2,583 Streptomyces-produced bioactive substances in Antibase 2005 (38) revealed actinopyrone, Mer-A2026, and traces of nigericin in addition to the previously reported compounds piericidin and streptochlorin. Automated as well as manual searches yielded additional predicted derivatives of these five core structures. While streptochlorin and eight piericidins exhibiting different modifications had been described previously as symbiont-produced substances from cocoons of the European beewolf (34), pimprinine (=SF2583B) and its derivative, several Mer-A2026 and actinopyrone derivatives, and nigericin had only been known from free-living Streptomyces. The pimprinines represent chlorine-free streptochlorin analogs, and Mer-A2026 and the actinopyrones are structurally closely related to the piericidins, exhibiting a pyridine ring that lacks a methoxy group and a pyranone instead of the pyridine ring, respectively. A scan of the MS/MS fragmentation patterns for the pyridine/pyranone fragment yielded additional derivatives, resulting in a total of 45 piericidin, actinopyrone, and Mer-A2026 derivatives (Fig. S1 and Table S1). A screen for streptochlorin-related compounds based on the characteristic chlorine isotope pattern revealed a previously unknown pentenyl-streptochlorin.
The proposed structures of streptochlorin and piericidin A1 and B1 isolated from the cocoons of European beewolves (P. triangulum) were previously confirmed by NMR (34). Furthermore, these three substances as well as actinopyrone A and nigericin were confirmed by comparison with synthetic standards (Fig. S2). For all other reported compounds, structures were predicted by comparing experimental with expected MS/MS fragmentation patterns (39) (Fig. S1 and Dataset S1). In total, 49 symbiont-produced putative bioactive compounds were identified across beewolf host species and subspecies (Fig. 1), 18 of which had been described previously from other Streptomyces species, including 9 previously found on the cocoons of European beewolves (34), while 31 constitute unique derivatives (Table S1).
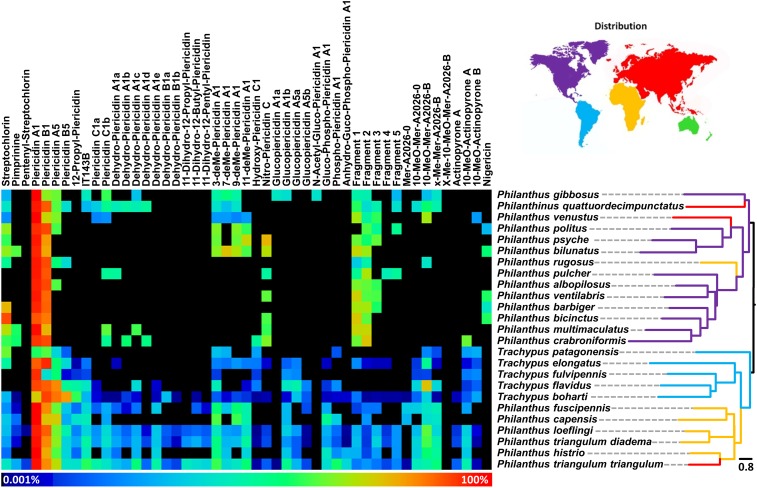
Fig. 1.
Composition of the symbiont-produced antimicrobial mixture in antennal extracts of 25 different beewolf species and subspecies. Colors indicate relative abundance of individual compounds in the antibiotic cocktail (columns). Host species (rows) are sorted based on the dendrogram displaying the chemical distances (Right). Branches in the dendrogram are colored according to the geographic origin of the host species (purple, North America; blue, South America; yellow, Africa; red, Eurasia).
As beewolf cocoons are exceptionally difficult to collect for a large number of species, most species were represented by antennal extracts only. To find out whether the composition of the antibiotic mixture in the antennae is representative of the ecologically more relevant composition on the cocoon, we compared the amount (based on ion counts rather than absolute quantification due to the lack of commercial standards for most compounds, which prevented the assessment of ionization efficiencies) and composition of the antibiotic mixture between antennae and cocoons for three geographically and phylogenetically disparate species (P. triangulum, Philanthus gibbosus, and Trachypus elongatus). Cocoons exhibited significantly higher total amounts of putative antibiotics than antennae in T. elongatus (Mann–Whitney U of sum of peak areas; P < 0.001, U = 7.0) and P. triangulum (Mann–Whitney U of sum of peak areas; P < 0.001 species, U = 20.0). In P. gibbosus, the concentrations of single antennal extracts were too low for a thorough chemical analysis, and the pooled antennal extract still contained lower amounts of antibiotics than single cocoon extracts. The composition of the mixture differed significantly among species (Wilk’s Λ = 7.6 × 10−5, df = 270, P < 0.001) as well as between antennae and cocoons (Wilk’s Λ = 0.178, df = 10, P < 0.001) based on a discriminant analysis of the first 10 principal components. However, antennae and cocoons of the same species clustered together in a discriminant analysis based on all antennal and cocoon extracts (Fig. 2 and Fig. S3A), indicating that chemical differences across species are larger than between antennal and cocoon extracts of the same species. Thus, the composition of the antibiotic mixture in the antennae can be used as a proxy for the cocoon.
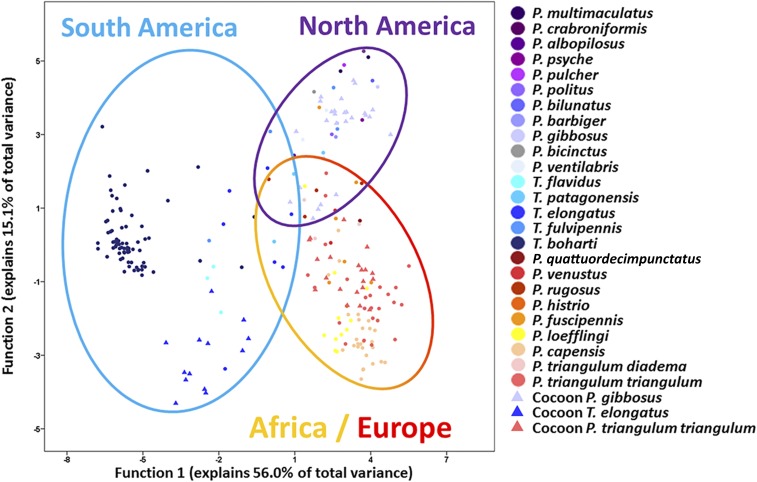
Fig. 2.
Discriminant analysis of the antibiotic mixtures produced by the symbionts of 25 different beewolf species and subspecies. The analysis is based on the 10 principal components extracted from the chemical composition of antennal (circles) and cocoon (triangles) extracts (n = 242 extracts, Wilk’s Λ = 7.6 × 10−5, df = 270, P < 0.001).
Phylogenetic and Geographic Influence on Antibiotic Mixtures.
A comparison of the chemical profiles of antennal extracts across 25 beewolf species and subspecies revealed a high degree of conservation in the composition of the antibiotic mixture, with piericidin A1 and B1 as the major components (based on ion counts) followed by streptochlorin and piericidin A5, while nigericin was detected only in a few North American beewolves at low relative amounts (Fig. 1). Piericidin A1 was the dominant compound across most host taxa, but some South American Trachypus species displayed higher amounts of piericidin B1 and/or A5 (specifically Trachypus patagonensis, T. elongatus, and Trachypus boharti). Most of the North American beewolves formed a distinct group in the chemical clustering with a considerably lower number of minor compounds in the antibiotic profile (Fig. 1). This was likely caused by the comparatively low absolute amounts of antibiotics in the antennae of North American beewolves, as the number of detected compounds correlated strongly with the total amount of antibiotics in the antennal extracts across species (Spearman rank correlation: rho = 0.855, P < 0.001, n = 29).
To assess interspecific differences in the antibiotic mixture, we performed a principal component analysis and a subsequent discriminant analysis based on the first 10 principal components, with species and tissue (antenna or cocoon) as grouping variables (i.e., without a priori grouping according to geography). The analysis significantly separated the symbiont-produced antibiotic profiles between the different host species, with 81.4% correct classifications based on the 10 discriminant functions (Wilk’s Λ < 7.6 × 10−5, df = 270, P < 0.001) (Fig. 2). When mapping the geographic origin of the host species onto the discriminant analysis, a clear pattern emerged, with the first discriminant function (representing 56.0% of the variance) separating the antibiotic profiles of the South American Trachypus species from the Philanthus and Philanthinus samples and the second function (representing 15.1% of the variance) distinguishing the North American from the European and African species. Concordantly, a discriminant analysis with a priori grouping according to the continental distribution of the host species (South America, North America, Europe/Africa) provided highly significant support for a geographic differentiation in the antibiotic mixtures (Wilk’s Λ = 0.049, df = 20, P < 0.001) (Fig. S3B).
To identify the underlying factors of the significant geographic pattern in antibiotic profiles, we investigated the effect of three different but mutually correlated variables on the composition of the antibiotic mixture: (i) the symbionts’ evolutionary history, (ii) the hosts’ evolutionary history, and (iii) the geographic distance itself, which we hypothesize to correlate with ecological differences. As an approximation for these factors, we used genetic distance matrices calculated from host and symbiont phylogenetic trees (38) as well as a geographic distance matrix based on the beewolf sampling locations. All three matrices correlated significantly with the antibiotic profiles in both Procrustes analyses and Mantel tests (Table 1). Partial Mantel tests showed a significant correlation between the antibiotic profiles and host phylogeny when controlling for symbiont phylogeny but not vice versa. In the partial Mantel tests controlling for the geographic distance, the correlation of antibiotic profiles with host phylogeny was no longer significant, and the correlation with symbiont phylogeny was only marginally significant and comparatively weak. However, the geographic distance remained strongly correlated with the antibiotic profiles when considering either host or symbiont phylogenies as covariate matrices (Table 1).
Table 1.
Mantel test, Procrustes analysis, and partial Mantel test results for correlations of the antibiotic profiles of 25 different beewolf species with the host phylogeny, symbiont phylogeny, and geographic distance of sampling localities
| Main effect | Controlled for | Correlation with antibiotic profile | |||
| Procrustes analysis | Mantel tests | ||||
| r | P | ρ | P | ||
| Host phylogeny | 0.741 | <0.0001 | 0.253 | 0.0036 | |
| Symbiont phylogeny | 0.519 | <0.0001 | 0.196 | 0.0136 | |
| Geography | 0.753 | <0.0001 | 0.381 | <0.0001 | |
| Host phylogeny | Symbiont phylogeny | 0.195 | 0.0162 | ||
| Host phylogeny | Geography | 0.139 | 0.0863 | ||
| Symbiont phylogeny | Host phylogeny | 0.107 | 0.1026 | ||
| Symbiont phylogeny | Geography | 0.161 | 0.0375 | ||
| Geography | Host phylogeny | 0.323 | 0.0004 | ||
| Geography | Symbiont phylogeny | 0.366 | 0.0002 | ||
Mantel test and Procrustes analysis only correlate two matrices, while partial Mantel tests control for an additional factor, a third matrix. Spearman rank coefficients (Mantel tests; ρ) or correlation coefficients of symmetric Procrustes rotation (r) and P values are given. Significant correlations are highlighted in bold.
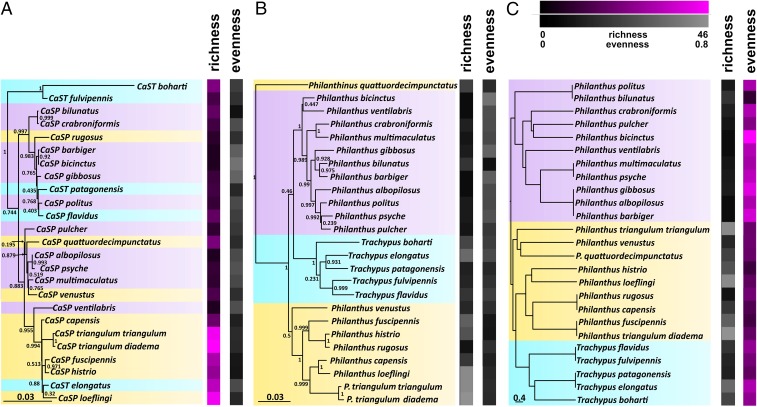
To refine the Procrustes analyses and Mantel tests, we used Blomberg’s K statistic and phylogenetic generalized least square (PGLS) models to test for an influence of phylogeny and geography, respectively. Because antibiotic profiles varied both in the number of compounds and their relative abundance, we calculated classical diversity measures (richness, evenness, Shannon’s H, and Simpson’s λ) to characterize the antibiotic mixtures. While the beewolf host phylogeny had no influence on any of the diversity measures, the symbiont phylogeny had a significant influence on the number of produced compounds (richness; K = 1.030, P = 0.001, indicating a deviation from randomly evolving traits) but not on the diversity or evenness (Fig. 3 and Table 2). However, compound evenness was significantly influenced by the geographic location even after correcting for symbiont (PGLS, P < 0.05) (Fig. 3 and Table 3) or host phylogenetic influence (PGLS, P < 0.05) (Fig. 3 and Table 3), assuming the better fitting Ornstein–Uhlenbeck model of trait evolution (in comparison with a Brownian motion model based on Akaike’s Information Criterion values) (Table S2). A closer look at the symbiont phylogeny and the chemical profiles revealed that the African/European beewolf symbionts produce an especially high number of compounds, whereas extracts from North American beewolves were generally characterized by a higher evenness than those from South American and African/European species (Fig. 3). The patterns observed for presumed horizontal symbiont transfer events between host species confirmed the influence of symbiont phylogeny on compound richness and of geographic distance on evenness. The symbionts of the Eurasian/African Philanthinus quattuordecimpunctatus, Philanthus rugosus, and Philanthus venustus as well as the South American T. patagonensis and Trachypus flavidus exhibited the low to medium compound richness that was characteristic of the North American symbiont clade that they were genetically assigned to. However, they retained low to medium evenness values, like their geographic neighbors. Similarly, the antibiotic profile of T. elongatus is characterized by high compound richness, such as that of its African symbiont relatives, but also comparatively high evenness that is typical for its South American distribution.
Fig. 3.
Phylogenetic and geographic influence on richness (number of compounds) and evenness (Shannon’s E) of the beewolf symbiont-produced antibiotic mixture. (A) Symbiont phylogeny, (B) host phylogeny, and (C) dendrogram based on a logarithmic distance matrix of sampling locations, all in comparison with heat maps displaying compound richness and evenness. Species are color-coded by sampling location (purple, North America; blue, South America; orange, Europe/Africa). Richness and evenness heat maps are presented in color if a significant phylogenetic (Blomberg’s K) or geographic (PGLS) influence was detected; otherwise, they are presented in gray. Branch numbers in the phylogenies are Bayesian posterior probability values.
Table 2.
Influence of symbiont and host phylogeny on the richness, evenness, and diversity of antibiotic substances in extracts of beewolf antennae
| Tested variable | Richness | Evenness | Shannon’s H | Simpson’s λ | ||||||||
| K | PR | PBM | K | PR | PBM | K | PR | PBM | K | PR | PBM | |
| Symbiont phylogeny | 1.030 | 0.001 | 0.954 | 0.165 | 0.508 | 0.001 | 0.135 | 0.702 | 0.001 | 0.110 | 0.845 | 0.001 |
| Host phylogeny | 0.516 | 0.103 | 0.156 | 0.353 | 0.338 | 0.015 | 0.351 | 0.338 | 0.013 | 0.269 | 0.065 | 0.023 |
The influence of symbiont and host phylogeny was measured as Blomberg’s K and tested for deviations from randomly evolving traits (K = 0; PR) and traits following a Brownian motion model of evolution (K = 1; PBM). Significant P values after correction for multiple testing are in bold.
Table 3.
Geographic signal in the richness, evenness, and diversity of antibiotic substances in extracts of different beewolf antennae
| Tested variable | Richness | Evenness | Shannon’s H | Simpson’s λ | ||||||||
| Coefficient ± SE | t | P | Coefficient ± SE | t | P | Coefficient ± SE | t | P | Coefficient ± SE | t | P | |
| Corrected for symbiont phylogeny | ||||||||||||
| Log(easting) | −668.9 ± 546.5 | −1.224 | 0.235 | 32.7 ± 10.4 | 3.156 | 0.0048 | Model | −23.9 ± 11.5 | −2.081 | 0.050 | ||
| Log(northing) | −517.5 ± 455.8 | −1.135 | 0.269 | 26.3 ± 8.5 | 3.071 | 0.0058 | did not | −19.7 ± 9.5 | −2.073 | 0.051 | ||
| Interaction | 96.9 ± 80.9s | 1.197 | 0.245 | −4.8 ± 1.5 | −3.124 | 0.0051 | converge | 3.5 ± 1.7 | 2.064 | 0.052 | ||
| Corrected for host phylogeny | ||||||||||||
| Log(easting) | −222.3 ± 831.4 | −0.267 | 0.792 | 31.9 ± 10.5 | 3.024 | 0.0077 | 51.7 ± 26.6 | 1.948 | 0.065 | −23.7 ± 11.4 | −2.077 | 0.050 |
| Log(northing) | −122.3 ± 687.4 | −0.178 | 0.861 | 25.5 ± 8.7 | 2.924 | 0.0081 | 42.7 ± 21.9 | 1.947 | 0.065 | −19.6 ± 9.4 | −2.071 | 0.051 |
| Interaction | 29.2 ± 122.8 | 0.238 | 0.814 | −4.7 ± 1.6 | −2.997 | 0.0069 | −7.6 ± 3.9 | 1.944 | 0.066 | 3.5 ± 1.7 | 2.060 | 0.052 |
The influence of geography was assessed by PGLS analyses using an Ornstein–Uhlenbeck model of trait evolution. Estimated regression coefficients, including SE, test statistic, and P value, for geographic coordinates and their interaction are given below. Easting and northing denote eastward and northward measured coordinates in the Universal Transverse Mercator coordinate system, respectively. Test results indicating a significant influence of geographic coordinates on antibiotic profiles after correction for multiple testing are printed in bold.
Molecular Basis of Generating Diversity in Antimicrobial Compounds.
The large diversity of piericidin and actinopyrone derivatives detected across beewolf host species compelled us to take a closer look at the underlying molecular basis of their production. The genetic background and biosynthesis of piericidin A1, including post-polyketide synthase (PKS) modification steps, have already been described in related Streptomyces species (40, 41). We sequenced the piericidin biosynthesis gene cluster of “S. philanthi biovar triangulum” strain tri23Af2 and compared it with the previously reported ones to pinpoint the steps in the biosynthesis that lead to the diversity in the antibiotic mixture (Fig. S4). The tri23Af2 cluster is highly similar to the piericidin A1 gene clusters from Streptomyces piomogenus (40) (Fig. S5), Streptomyces sp. SCSIO 03032 (41), and Streptomyces mobaraensis (National Center for Biotechnology Information accession numbers AB431381.1–AB31384.1), comprising the same genes in identical order, with average sequence similarities to S. piomogenus of 73.3 and 66.4% on the nucleotide and amino acid levels, respectively (Fig. S5). The PKS cluster itself is responsible for the elongation of the underlying linear polyketide chain of the piericidins (40), while several post-PKS tailoring steps are required to convert the intermediate into the final product by amidation, cyclization, hydration, and methylation (41). Piericidin A1 is the primary product, but the predicted biosynthetic pathway and the structures of the beewolf symbiont-produced piericidin and actinopyrone derivatives suggest six major ways to deviate from the main product.
The incorporation of different precursors in the (i) initiation as well as (ii) elongation steps explains variation in the length of the polyketide backbone and the presence of methyl branches. An analysis of the eight extending acyltransferase (AT) domains of the S. philanthi biovar triangulum piericidin gene cluster revealed that five AT domains are very similar on the amino acid level, whereas the other three AT domains are divergent. Based on a computational analysis (42–45), these five AT domains are most similar to AT domains that specifically incorporate methylmalonyl-CoA (Table S3). However, the 13 amino acid residues that control substrate specificity of these five AT domains in the S. philanthi piericidin gene cluster show a hybrid pattern of residues typical for methylmalonyl-CoA and malonyl-CoA incorporation. Interestingly, this pattern apparently results in a low degree of promiscuity of these domains, with 99.1–99.9% methylmalonyl-CoA and 0.1–0.9% malonyl-CoA incorporation into the nascent polyketide chain, based on the abundance of the corresponding piericidin derivatives detected by liquid chromatography tandem-MS (LC-MS/MS) (Table S3). By contrast, the corresponding piericidin AT domains of the free-living bacterium S. piomogenus var hangzhouwanensis contain the typical amino acid sequence motifs of methylmalonyl-CoA–specific incorporation (42, 43) (Table S3). One of the remaining three AT domains in the S. philanthi piericidin gene cluster is too dissimilar from all other AT domains in the SBSPKS (Structure Based Sequence Analysis of Polyketide Synthases) database to allow for a prediction of its specificity, while the final two show substrate binding motifs indicative of malonyl-CoA incorporation.
Additional diversity in piericidin derivatives is generated by (iii) incomplete dehydration or dehydrogenation steps during PKS chain elongation or later reduction, which lead to unsaturated or hydroxylated derivatives. (iv) The formation of the pyridine ring occurs after the side chain is released from the PKS cluster. If cyclization occurs without the preceding transamination of the terminal hydroxyl group, a pyranone is formed instead of a pyridine, resulting in the formation of actinopyrones as end products. (v) The methyl and methoxy groups of the pyridine ring and the hydroxyl group of the aliphatic side chain can be substituted with methoxy or glycosyl groups, presumably by unspecific enzymes encoded in genomic regions outside of the piericidin gene cluster. (vi) Finally, one of the double bonds in the piericidin A1 backbone can undergo epoxidation, resulting in piericidin C and its derivatives. A combination of these six modifications can explain 38 of 45 piericidin, Mer-A2026, and actinopyrone derivatives. The remaining compounds show the entire pyridine ring but a truncated side chain. We exclude a sampling artifact, as the North American specimens were collected in two separate sampling trips, which show similar products that were also found in individual species sampled in Europe and South Africa. The derivatives with truncated side chain could arise either from degradation of complete precursors or by an incomplete synthesis. As the pyridine/pyranone ring is formed last in the piericidin and actinopyrone biosynthesis and is always present in the truncated derivatives, the nascent polyketide cannot have been prematurely released from the PKS. However, it is possible that several PKS modules were skipped during elongation. While not common, this process has been documented for several Streptomyces species, leading to an increase in chemical diversity in single bacterial strains (46).
In Vitro Activity of Antibiotic Combinations.
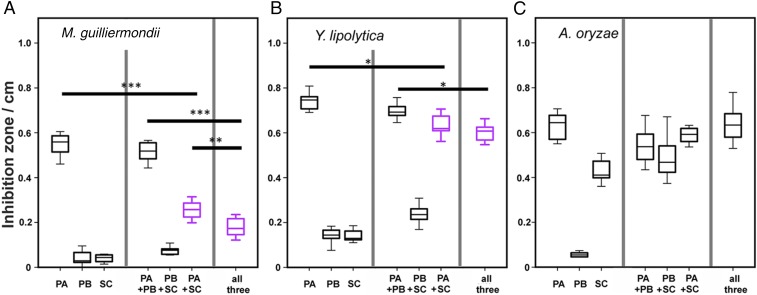
To investigate whether the observed differences in antibiotic mixtures between beewolf species may be ecologically relevant by affecting antimicrobial activity, we performed agar diffusion assays with different combinations of the three most abundant compounds, piericidin A1 and B1 and streptochlorin, against three different fungal model species [Aspergillus oryzae, Yarrowia lipolytica, and Meyerozyma guilliermondii (formerly Candida guilliermondii)]. Piericidin A1 was generally most active, while streptochlorin was similarly active against A. oryzae but less so against both yeasts, and piericidin B1 had a very low inhibitory effect against all three tested organisms (Fig. 4). Piericidin B1 neither increased nor reduced the activity of piericidin A1, but it enhanced the inhibition of streptochlorin against Y. lipolytica (one-way ANOVA, P < 0.001, df = 6, F = 385.95, Tukey’s honest significant difference (HSD) post hoc test P < 0.001) and decreased the combined effectivity of piericidin A1 and streptochlorin against M. guilliermondii (one-way ANOVA, P < 0.001, df = 6, F = 129.45, Tukey HSD post hoc test P = 0.0095). However, streptochlorin generally increased the activity of piericidin B1 (M. guilliermondii: one-way ANOVA, P < 0.001, df = 6, F = 129.45, Tukey HSD post hoc test P = 0.016; Y. lipolytica: one-way ANOVA, P < 0.001, df = 6, F = 385.95, Tukey HSD post hoc test P = 0.042; A. oryzae: one-way ANOVA, P < 0.001, df = 6, F = 114.87, Tukey HSD post hoc test P < 0.001) but reduced the inhibition of yeasts in mixtures containing piericidin A1 (piericidin A1 alone: M. guilliermondii: one-way ANOVA, P < 0.001, df = 6, F = 129.45, Tukey HSD post hoc test P < 0.001; Y. lipolytica: one-way ANOVA, P < 0.001, df = 6, F = 385.95, Tukey HSD post hoc test P = 0.023; piericidin A1 + B1: M. guilliermondii: one-way ANOVA, P < 0.001, df = 6, F = 129.45, Tukey HSD post hoc test P < 0.001; Y. lipolytica: one-way ANOVA, P < 0.001, df = 6, F = 385.95, Tukey HSD post hoc test P = 0.042). Hence, changes in the antimicrobial mixture differentially affect potential antagonists and may allow for fine-tuning the antimicrobial defense to local pathogen communities.
Fig. 4.
Bioactivity of different combinations of piericidin A1 (PA) and B1 (PB) and streptochlorin (SC) in agar diffusion assays against (A) M. guilliermondii, (B) Y. lipolytica, and (C) A. oryzae. Antagonistic effects (i.e., significantly lower inhibition zones of mixtures than one of the single substances) are highlighted by magenta boxes (according to ANOVA and Tukey HSD post hoc tests; *P < 0.05, **P < 0.01, ***P < 0.001).
Discussion
In addition to the nine previously described symbiont-produced antibiotic compounds on the surface of European beewolf (P. triangulum) cocoons (34), we identified another 40 putative antimicrobial compounds across different beewolf host species that were present on the cocoons and in the antennae of these solitary wasps. With the exception of nigericin, which was only present in few species in low amounts, all of these compounds represent modifications of only two core structures (streptochlorin and piericidin). Although the antibiotic mixture was qualitatively similar across beewolf species, with piericidin A1 and B1 as the major components across all 25 examined species and subspecies, the abundance and relative composition varied between species. While the number of compounds was influenced by the phylogenetic background of the Streptomyces symbiont strains, the evenness of the mixture was significantly influenced by geographic location, suggesting an adaptation to the local environment. Antimicrobial bioassays with all possible combinations of the three main antibiotics against three fungal species revealed positive and negative interaction effects, indicating that changes in the mixture may indeed affect its antimicrobial efficacy as well as the target spectrum. Molecular analyses of the gene cluster underlying piericidin biosynthesis allow for predicting the mechanistic basis of generating the high diversity of derivatives observed in the beewolf symbiosis.
Composition of the Mixture and Activity of the Individual Components.
Previous studies have shown the broad-spectrum activity of the European beewolf’s antibiotic mixture against fungal and bacterial antagonists (34) as well as the activity of single compounds in the beewolves’ mixture that were also discovered in other Streptomyces species. Pimprinine (47), streptochlorin (34), piericidin A (34, 48, 49) and B (34, 50), 7-demethylpiericidin A1 (51), glucopiericidin A and B (34, 52), IT143B (53), and Mer-2026-A and B (54) have been shown to exhibit antibacterial and antifungal activity. 11-Demethylpiericidin A1 and glucopiericidin A5 showed bioactivity against bacteria, and the actinopyrones have been described to exhibit specific activity against Helicobacter pylori (55), whereas piericidin B5 did not show activity against any of the tested microorganisms (56). Nigericin is known as a K+ ionophore (57), displaying strong antibacterial (58) but only weak antifungal activity on its own, but a high potential for synergistic interactions (59). For the remaining compounds, antimicrobial activity remains speculative and is hypothesized exclusively based on the structural similarity to the active piericidins, actinopyrones, and streptochlorin. However, various studies on the cytotoxicity in cancer cell lines and the inhibition of the electron transport complexes of the respiratory chain by synthetic piericidin derivatives showed that most modifications of the side chain had only a minor influence on their activity as long as the pyridine core remained intact (60), suggesting antimicrobial activity of many of the reported compounds. Furthermore, it needs to be emphasized that even subinhibitory concentrations of individual compounds may be ecologically relevant in a complex mixture with synergistic and antagonistic effects (61). Concordantly, our bioassays with different combinations of the three major components (ions) of the beewolf antibiotic mixture (piericidin A1 and B1 and streptochlorin) highlight the importance of compound interactions, including antagonistic and synergistic effects between active compounds as well as potentially modulatory activities of compounds that are not active on their own.
Biosynthetic Basis of Diversity in the Antibiotic Mixture.
The majority of the variation in the beewolf antibiotic mixture is based on the high number of piericidin derivatives. Sequencing of the S. philanthi biovar triangulum piericidin gene cluster and comparing it with that of S. piomogenus var hangzhouwanensis with its known biosynthesis (40, 41) allowed us to attribute the majority of the diversity in predicted piericidin and actinopyrone derivatives found in the beewolf symbionts (Fig. S4) to the following six mechanisms: (i) different starting or (ii) extender units during polyketide synthesis; (iii) skipping the reduction of a double bond during extension; (iv) ring formation with or without prior aminotransferase reaction (leading to piericidins and actinopyrones, respectively); and (v) methylation, glycosylation, or phosphorylation of hydroxyl groups as well as (vi) oxidation or epoxidation of double bonds.
Initiating PKS biosynthesis with different starting units is a well-known feature of Streptomyces PKSs (62). In contrast, variation in the incorporation of fatty acid-CoA units by extender modules during the elongation of the polyketide is usually lower due to the high substrate specificity of the ATs (62, 63). Surprisingly, four of the eight AT domains exhibit hybrid sequence motifs of malonyl-CoA– and methylmalonyl-CoA–specific AT domains, and these domains indeed incorporate both methylmalonyl-CoA and malonyl-CoA based on the characterized products. Thus, substrate specificity of PKS-AT domains cannot only be experimentally manipulated to broaden the spectrum of available PKS (63) but also naturally evolves to accept different substrates (64). In contrast to the symbiotic S. philanthi bacteria, the AT domains of the free-living S. piomogenus var hangzhouwanensis piericidin PKS cluster do not show this promiscuity pattern but retain methylmalonyl-CoA–specific amino acid residues, suggesting that the S. philanthi piericidin AT domains may have been subject to selection for generating diversity in bioactive metabolites.
After the release from the PKS protein, the polyketide is usually subjected to several post-PKS tailoring steps, some of which are encoded within the PKS gene cluster, like two methyltransferases, one aromatase, a monooxygenase, and an asparagin-synthase–like enzyme in the case of the piericidin cluster (40, 41). Other enzymes involved in post-PKS modifications are often encoded by separate genes, yielding additional diversity in final compounds (65–67).
Although previous genomic screens for secondary metabolite gene clusters across microbial taxa revealed the potential of individual strains to produce a diversity of compounds, the large number of putative antibiotics in the beewolf symbionts seems particularly extensive. However, recent studies on the chemical ecology of multipartite interactions reported similarly complex chemistries, especially in bacteria living in a symbiotic context (reviews in refs. 11 and 12). Specifically, symbiotic γ-Proteobacteria in entomopathogenic nematodes provide an arsenal of compounds that are responsible for overcoming the immune system of the attacked insect as well as for monopolizing the carcass by warding off scavengers and microbial competitors (68, 69). Likewise, several symbionts associated with marine animals provide a number of defense compounds derived from multiple (70) or individual PKS clusters (70–73). Remarkably, Lin et al. (74) characterized 13 nocapyrone derivatives from a Nocardiopsis alba strain that is a putative symbiont of the marine cone snail Conus rolani. Containing a stable pyranone ring and a side chain with different modifications, these nocapyrone derivatives exhibit a similar pattern to the structural variation observed in the piericidins and actinopyrones that are produced by the beewolf symbionts. Unfortunately, the ecological function of the nocapyrones and many other symbiotically produced secondary metabolites remains elusive. However, the diversity of compounds produced in the beewolf mutualism as well as in other defensive alliances indicates that (i) a combination of antibiotics allows for an evolutionary stable defense against a broad spectrum of potential antagonists and/or that (ii) the different compounds may serve other functions in addition to defense (e.g., signaling to modulate the composition or activity of the microbial community in their particular microenvironment) (10, 13–17, 75).
Evolutionary Stability of the Antibiotic Mixture.
Many defensive symbioses are of a dynamic nature, with intermediate symbiont infection frequencies, multiple coinfecting symbionts, and/or frequent replacements of symbionts by environmentally acquired microbes (12, 31, 76, 77). In a Red Queen-like scenario, such a dynamic association is to be expected to keep pace with coevolving pathogens. While we cannot completely exclude the possibility that we missed additional antibiotic compounds produced by the beewolves’ symbionts, we found that all beewolf species seem to rely on a very similar antimicrobial mixture for defense. The conserved nature of the antibiotic mixture strongly indicates that it represents the antibiotic profile of the Streptomyces bacterium that was acquired by the common ancestor of all Philanthini digger wasps in the late Cretaceous (35). How, then, could the beewolves’ antibiotic defense remain efficient over the past 68 My and prevent the evolution of resistance in antagonistic microbes? Two alternative hypotheses may account for the long-term maintenance and efficacy of the beewolf symbionts’ antibiotic mixture: (i) the complexity of the antibiotic mixture has prevented the evolution of resistance in antagonistic bacteria, or (ii) the selective pressures on antagonistic microorganisms were not sufficiently high to favor the evolution of multidrug resistance.
A combination of antibiotics can be beneficial and slow down the evolution of resistance for two main reasons. First, if at least two compounds act antagonistically, a microorganism that evolves resistance to only one of the compounds experiences a higher effective inhibition by the remaining compound and thus, has a decreased fitness compared with nonresistant cells (78, 79). Synergism, however, despite the obvious expectation of being initially more effective, favors a faster evolution of resistance, as an arising mutation providing resistance against one compound also removes the synergistic inhibition (80, 81). Second, intrinsically nontoxic compounds can restore the activity of toxins by neutralizing resistance mechanisms (such as β-lactamase inhibitors with β-lactam antibiotics) (82, 83). Inhibition bioassays with different combinations of three of the beewolf antibiotics revealed antagonistic interactions between some of the major compounds, suggesting that the composition of the beewolf antibiotic mixture may disfavor the evolution of resistant antagonists while maintaining broad-spectrum efficacy, which could be especially important to ward off partially resistant mold strains during the long development and hibernation period.
Given the 68 My of beewolf–Streptomyces symbiosis, however, a specialized pathogen may still be expected to evolve resistance, even against a complex mixture of antibiotics. However, are beewolves and their symbionts likely to exert selection pressures on soil microbes that are sufficiently strong to favor the evolution of a specialized multiresistant pathogen? In contrast to other insects with defensive microbial symbionts (e.g., leafcutter ants, aphids), beewolves occur in comparatively small aggregations (usually dozens to several hundred individuals), and their nesting sites are often located in disturbed and ephemeral habitats (84, 85). Thus, they represent a scarce and unpredictable resource for microbes, and indeed, only common mold fungi that are abundant in the beewolves’ nesting environments have been observed to infect the beewolf offspring, while specialized microbial pathogens are unknown (86). Thus, the key for the long-term success of the symbiont-provided antibiotic mixture in beewolves was probably the lack of coevolving pathogens. Presumably, the complexity of the mixture mainly served to ensure its broad-spectrum efficacy against opportunistic microbial antagonists (34), with the ability to suppress the evolution of resistance being a secondary effect.
Despite the high degree of conservation in the qualitative composition of the antibiotic mixture, we detected consistent quantitative differences across beewolf species. These differences were of a dual nature. The number of detected compounds (richness) was influenced by the symbiont phylogenetic background, while the relative abundance of the single compounds (evenness) was affected by geographic distance. The first finding is consistent with evolutionary changes in the genetic basis of secondary metabolite biosynthesis that limit or expand the number of produced compounds. The second observation suggests an environmental influence on the quantitative expression of compound biosynthesis. These expression changes presumably allow for rapid adaptations to the local (micro-)environment (temperature, humidity, soil microbial community) by quantitatively adjusting the composition of the antibiotic mixture. Concordantly, beewolves that acquired a symbiont strain horizontally from another host species (35, 87) exhibited antibiotic mixtures that quantitatively resemble those of co-occurring species rather than those of hosts with closely related symbiont strains (Fig. 3).
Evidence for a geographic signal in biosynthetic capabilities of secondary metabolites is not restricted to the beewolf symbionts but has also recently been provided for free-living bacteria. Charlop-Powers et al. (88) sequenced the NRPS adenylation and PKS ketosynthase domains of bacterial communities from multiple locations on different continents and found a higher relatedness of produced compounds in bacteria from similar environments in close proximity than from more distant habitats. Similarly, Lemetre et al. (89) found the composition of bacterial biosynthetic domains in soil samples to be influenced by the geographic location, most notably latitude, but not by altitude, temperature, or annual precipitation, which are common predictors of microbial diversity. Thus, environmental factors correlating with geographic distance seem to be important selective forces driving the composition of natural product biosynthesis in microbes.
A Unique Strategy for Protection?
Analogous to the beewolf symbiosis, the simultaneous use of multiple symbiont-derived compounds for defense has been described across various terrestrial and marine systems (reviewed in ref. 12), rendering combination prophylaxes a common phenomenon in defensive symbioses. However, the long-term evolutionary stability of the beewolves’ antibiotic mixture is unusual in light of many systems that exhibit dramatic changes in defensive chemistry, often through the dynamic acquisition of novel protective symbionts (27, 90–96). As discussed above, the lack of a specialized coevolving antagonist likely relaxed selection for fundamental changes in defensive chemistry in the beewolf symbiosis. At the same time, the dynamic community of opportunistic soil fungi constituted a high selective pressure to maintain or expand the broad-spectrum antifungal activity and to quantitatively adjust the antibiotic mixture to the local environment. We predict that other symbioses that are threatened by a community of opportunistic antagonists rather than specialized pathogens or predators use a similar strategy (e.g., the Penicillium symbionts that defend a leaf-rolling weevil’s offspring from mold fungi and bacteria) (97). Unfortunately, however, the lack of defensive symbioses for which a thorough description of all involved players, their ecology, and their evolution as well as the in vivo relevant chemistry is available, currently severely hampers our understanding of how the chemistry of an antibiotic defense changes over evolutionary timescales in other symbiotic associations.
Experimental Procedures
Antennae and/or cocoons of 25 species and subspecies of Philanthini digger wasps were collected from the United States, Brazil, Germany, Turkey, and South Africa. Samples were extracted with methanol and analyzed by ultrahigh-performance LC-ESI-MS/MS.
Mass spectra were screened for exact masses of known compounds produced by Streptomyces bacteria published in Antibase 2005 (41) and derivatives of piericidin, streptochlorin, actinopyrone, Mer derivatives, and nigericin by automated and manual prediction of their structure and according mass. Peak areas of all identified compounds were integrated as a proxy for relative compound amounts. Principal component and discriminant analyses were performed to detect grouping patterns between species and geographic patterns with SPSS 23 (IBM). Mantel tests and Procrustes analyses were applied to test for correlations between the chemical composition of the specimen extracts and environmental influence (represented by their geographic origin) as well as host and symbiont phylogeny. We used Blomberg’s K statistic to test for a phylogenetic influence and PGLS models to assess the geographic influence under a phylogenetic background on diversity measures of the chemical substance composition.
The S. philanthi piericidin gene cluster was extracted from a de novo whole-genome assembly based on 454 shotgun and 8-kb paired end libraries of in vitro cultures of S. philanthi biovar triangulum strain tri23Af2 (98). The gene cluster, its substrates, and product were predicted with NP.searcher and SEARCHPKS and by comparison with the S. piomogenus piericidin gene cluster (43).
Fungal inhibition assays were conducted in standard agar diffusion assays using piericidin A1 and B1, streptochlorin, and additive mixtures of all combinations of two and all three compounds against A. oryzae and the yeasts M. guilliermondii, and Y. lipolytica (details are in SI Experimental Procedures).
Supplementary Material
Acknowledgments
We thank Christine Michel, Fred and Sarah Gess, Sabrina Koehler, Gudrun Herzner, Dirk Koedam, and Erol Yildirim for help with field work or generous gifts of specimens. We also thank Riya Christina Menezes for help with the verification of nigericin and Jörn Piel for his constructive comments on the manuscript. Permits were issued by the nature conservation boards of KwaZulu Natal (Permit 4362/2004), Eastern Cape Province (WRO44/04WR, WRO9/04WR,WRO74/06WR, WRO75/06WR, CRO135/11CR, CRO136/11CR, CRO179/10CR, and CRO180/10CR), and Western Cape Province (001-202-00026, 001-506-00001, AAA004-00053-0035, AAA004-00089-0011, AAA004-00683-0035, and 0046-AAA004-00008) of South Africa and the Brazilian Ministry of the Environment (MMA/SISBIO/22861-1). We acknowledge financial support from the Max Planck Society and German Science Foundation Grant DFG KA2846/2-1 (to M. Kaltenpoth).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in GenBank, https://www.ncbi.nlm.nih.gov/genbank (accession nos. KX098584, KU759552–KU759556, and KU759557–KU759562). The MS data reported in this paper have been deposited in the Dryad Digital Repository (doi:10.5061/dryad.6907h).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719797115/-/DCSupplemental.
References
- 1.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Payne DJ. Microbiology. Desperately seeking new antibiotics. Science. 2008;321:1644–1645. doi: 10.1126/science.1164586. [DOI] [PubMed] [Google Scholar]
- 3.Taubes G. The bacteria fight back. Science. 2008;321:356–361. doi: 10.1126/science.321.5887.356. [DOI] [PubMed] [Google Scholar]
- 4.Laxminarayan R, et al. Antibiotic resistance-the need for global solutions. Lancet Infect Dis. 2013;13:1057–1098. doi: 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization . Antimicrobial Resistance: Global Report on Surveillance. WHO; Geneva: 2014. [Google Scholar]
- 6.Woodford N, Ellington MJ. The emergence of antibiotic resistance by mutation. Clin Microbiol Infect. 2007;13:5–18. doi: 10.1111/j.1469-0691.2006.01492.x. [DOI] [PubMed] [Google Scholar]
- 7.Barlow M. 2009. What antimicrobial resistance has taught us about horizontal gene transfer. Horizontal Gene Transfer, Methods in Molecular Biology, eds Gogarten M, Gogarten J, Olendzenski L (Humana, New York), Vol 532, pp 397–411.
- 8.Allen HK, et al. Call of the wild: Antibiotic resistance genes in natural environments. Nat Rev Microbiol. 2010;8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 9.D’Costa VM, et al. Antibiotic resistance is ancient. Nature. 2011;477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 10.Czárán TL, Hoekstra RF, Pagie L. Chemical warfare between microbes promotes biodiversity. Proc Natl Acad Sci USA. 2002;99:786–790. doi: 10.1073/pnas.012399899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Arnam EB, Currie CR, Clardy J. Defense contracts: Molecular protection in insect-microbe symbioses. Chem Soc Rev. July 26, 2017 doi: 10.1039/c7cs00340d. [DOI] [PubMed] [Google Scholar]
- 12.Flórez LV, Biedermann PHW, Engl T, Kaltenpoth M. Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat Prod Rep. 2015;32:904–936. doi: 10.1039/c5np00010f. [DOI] [PubMed] [Google Scholar]
- 13.Yim G, Wang HHM, Davies J. The truth about antibiotics. Int J Med Microbiol. 2006;296:163–170. doi: 10.1016/j.ijmm.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 14.Yim G, Wang HH, Davies J. Antibiotics as signalling molecules. Philos Trans R Soc Lond B Biol Sci. 2007;362:1195–1200. doi: 10.1098/rstb.2007.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fajardo A, Martínez JL. Antibiotics as signals that trigger specific bacterial responses. Curr Opin Microbiol. 2008;11:161–167. doi: 10.1016/j.mib.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Romero D, Traxler MF, López D, Kolter R. Antibiotics as signal molecules. Chem Rev. 2011;111:5492–5505. doi: 10.1021/cr2000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumbhar C, Watve M. Why antibiotics: A comparative evaluation of different hypotheses for the natural role of antibiotics and an evolutionary synthesis. Nat Sci. 2013;5:26–40. [Google Scholar]
- 18.Schlatter DC, Kinkel LL. Antibiotics: Conflict and communication in microbial communities. Microbe. 2014;9:282–288. [Google Scholar]
- 19.van der Meij A, Worsley SF, Hutchings MI, van Wezel GP. Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol Rev. 2017;41:392–416. doi: 10.1093/femsre/fux005. [DOI] [PubMed] [Google Scholar]
- 20.Clardy J, Fischbach MA, Currie CR. The natural history of antibiotics. Curr Biol. 2009;19:R437–R441. doi: 10.1016/j.cub.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaltenpoth M, Strupat K, Svatoš A. Linking metabolite production to taxonomic identity in environmental samples by (MA)LDI-FISH. ISME J. 2016;10:527–531. doi: 10.1038/ismej.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Currie CR, Scott JA, Summerbell RC, Malloch D. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature. 1999;398:701–704. [Google Scholar]
- 23.Currie CR, Bot ANM, Boomsma JJ. Experimental evidence of a tripartite mutualism: Bacteria protect ant fungus gardens from specialized parasites. Oikos. 2003;101:91–102. [Google Scholar]
- 24.Little AEF, Murakami T, Mueller UG, Currie CR. Defending against parasites: Fungus-growing ants combine specialized behaviours and microbial symbionts to protect their fungus gardens. Biol Lett. 2006;2:12–16. doi: 10.1098/rsbl.2005.0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haeder S, Wirth R, Herz H, Spiteller D. Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proc Natl Acad Sci USA. 2009;106:4742–4746. doi: 10.1073/pnas.0812082106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cafaro MJ, et al. Specificity in the symbiotic association between fungus-growing ants and protective Pseudonocardia bacteria. Proc R Soc B. 2011;278:1814–1822. doi: 10.1098/rspb.2010.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sen R, et al. Generalized antifungal activity and 454-screening of Pseudonocardia and Amycolatopsis bacteria in nests of fungus-growing ants. Proc Natl Acad Sci USA. 2009;106:17805–17810. doi: 10.1073/pnas.0904827106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mattoso TC, Moreira DDO, Samuels RI. Symbiotic bacteria on the cuticle of the leaf-cutting ant Acromyrmex subterraneus subterraneus protect workers from attack by entomopathogenic fungi. Biol Lett. 2012;8:461–464. doi: 10.1098/rsbl.2011.0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Souza DJ, et al. Ectosymbionts and immunity in the leaf-cutting ant Acromyrmex subterraneus subterraneus. Brain Behav Immun. 2013;28:182–187. doi: 10.1016/j.bbi.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 30.Oh D-C, Poulsen M, Currie CR, Clardy J. Dentigerumycin: A bacterial mediator of an ant-fungus symbiosis. Nat Chem Biol. 2009;5:391–393. doi: 10.1038/nchembio.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seipke RF, et al. A single Streptomyces symbiont makes multiple antifungals to support the fungus farming ant Acromyrmex octospinosus. PLoS One. 2011;6:e22028. doi: 10.1371/journal.pone.0022028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoenian I, et al. Chemical basis of the synergism and antagonism in microbial communities in the nests of leaf-cutting ants. Proc Natl Acad Sci USA. 2011;108:1955–1960. doi: 10.1073/pnas.1008441108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carr G, Derbyshire ER, Caldera E, Currie CR, Clardy J. Antibiotic and antimalarial quinones from fungus-growing ant-associated Pseudonocardia sp. J Nat Prod. 2012;75:1806–1809. doi: 10.1021/np300380t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kroiss J, et al. Symbiotic streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat Chem Biol. 2010;6:261–263. doi: 10.1038/nchembio.331. [DOI] [PubMed] [Google Scholar]
- 35.Kaltenpoth M, et al. Partner choice and fidelity stabilize coevolution in a Cretaceous-age defensive symbiosis. Proc Natl Acad Sci USA. 2014;111:6359–6364. doi: 10.1073/pnas.1400457111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goettler W, Kaltenpoth M, Herzner G, Strohm E. Morphology and ultrastructure of a bacteria cultivation organ: The antennal glands of female European beewolves, Philanthus triangulum (Hymenoptera, Crabronidae) Arthropod Struct Dev. 2007;36:1–9. doi: 10.1016/j.asd.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Kaltenpoth M, Göttler W, Herzner G, Strohm E. Symbiotic bacteria protect wasp larvae from fungal infestation. Curr Biol. 2005;15:475–479. doi: 10.1016/j.cub.2004.12.084. [DOI] [PubMed] [Google Scholar]
- 38.Laatsch H. Antibase. Wiley VCH; Weinheim, Germany: 2005. [Google Scholar]
- 39.Yoshida S, Yoneyama K, Shiraishi S, Watanabe A, Takahashi N. Chemical structures of new piericidins produced by Streptomyces pactum. Agric Biol Chem. 1977;41:855–862. [Google Scholar]
- 40.Liu Q, et al. Elucidation of piericidin A1 biosynthetic locus revealed a thioesterase-dependent mechanism of α-pyridone ring formation. Chem Biol. 2012;19:243–253. doi: 10.1016/j.chembiol.2011.12.018. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y, et al. Elucidating hydroxylation and methylation steps tailoring piericidin A1 biosynthesis. Org Lett. 2014;16:736–739. doi: 10.1021/ol4034176. [DOI] [PubMed] [Google Scholar]
- 42.Yadav G, Gokhale RS, Mohanty D. Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J Mol Biol. 2003;328:335–363. doi: 10.1016/s0022-2836(03)00232-8. [DOI] [PubMed] [Google Scholar]
- 43.Yadav G, Gokhale RS, Mohanty D. SEARCHPKS: A program for detection and analysis of polyketide synthase domains. Nucleic Acids Res. 2003;31:3654–3658. doi: 10.1093/nar/gkg607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ansari MZ, Yadav G, Gokhale RS, Mohanty D. NRPS-PKS: A knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res. 2004;32:W405–W413. doi: 10.1093/nar/gkh359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anand S, et al. SBSPKS: Structure based sequence analysis of polyketide synthases. Nucleic Acids Res. 2010;38:W487–W496. doi: 10.1093/nar/gkq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moss SJ, Martin CJ, Wilkinson B. Loss of co-linearity by modular polyketide synthases: A mechanism for the evolution of chemical diversity. Nat Prod Rep. 2004;21:575–593. doi: 10.1039/b315020h. [DOI] [PubMed] [Google Scholar]
- 47.Naik SR, Harindran J, Varde AB. Pimprinine, an extracellular alkaloid produced by Streptomyces CDRIL-312: Fermentation, isolation and pharmacological activity. J Biotechnol. 2001;88:1–10. doi: 10.1016/s0168-1656(01)00244-9. [DOI] [PubMed] [Google Scholar]
- 48.Tamura S, et al. Isolation and physiological activities of piericidin A, a natural insecticide produced by Streptomyces. Agric Biol Chem. 1963;27:576–582. [Google Scholar]
- 49.Takahashi N, Suzuki A, Tamura S. Structure of piericidin A. J Am Chem Soc. 1965;87:2066–2068. doi: 10.1021/ja01087a050. [DOI] [PubMed] [Google Scholar]
- 50.Takahashi N, et al. Isolation structure and physiological activities of piericidin B, natural insecticide produced by a Streptomyces. Agric Biol Chem. 1968;32:1115–1122. [Google Scholar]
- 51.Kimura K, Takahashi H, Miyata N, Yoshihama M, Uramoto M. New piericidin antibiotics, 7-demethylpiericidin A1 and 7-demethyl-3′-rhamnopiericidin A1. J Antibiot (Tokyo) 1996;49:697–699. doi: 10.7164/antibiotics.49.697. [DOI] [PubMed] [Google Scholar]
- 52.Matsumoto M, et al. New piericidin glucosides, glucopiericidin-A and glucopiericidin-B. J Antibiot (Tokyo) 1987;40:149–156. doi: 10.7164/antibiotics.40.149. [DOI] [PubMed] [Google Scholar]
- 53.Urakawa A, et al. IT-143-A and B, novel piericidin-group antibiotics produced by Streptomyces sp. J Antibiot (Tokyo) 1996;49:1052–1055. doi: 10.7164/antibiotics.49.1052. [DOI] [PubMed] [Google Scholar]
- 54.Kominato K, et al. Mer-A2026A and B, novel piericidins with vasodilating effect. I. Producing organism, fermentation, isolation and biological properties. J Antibiot (Tokyo) 1995;48:99–102. doi: 10.7164/antibiotics.48.99. [DOI] [PubMed] [Google Scholar]
- 55.Yano K, et al. Actinopyrone-A, actinopyrone-B and actinopyrone-C: New physiologically active substances. 1. Producing organism, fermentation, isolation and biological properties. J Antibiot (Tokyo) 1986;39:32–37. doi: 10.7164/antibiotics.39.32. [DOI] [PubMed] [Google Scholar]
- 56.Nishioka H, et al. Isolation and structure determination of novel phosphatidylinositol turnover inhibitors, piericidin B5 and B5 N-oxide, from Streptomyces sp. J Antibiot (Tokyo) 1993;46:564–568. doi: 10.7164/antibiotics.46.564. [DOI] [PubMed] [Google Scholar]
- 57.Hamidinia SA, et al. The ionophore nigericin transports Pb2+ with high activity and selectivity: A comparison to monensin and ionomycin. Biochemistry. 2004;43:15956–15965. doi: 10.1021/bi048175z. [DOI] [PubMed] [Google Scholar]
- 58.Wu ZX, Bai LQ, Wang MZ, Shen YM. Structure-antibacterial relationship of nigericin derivatives. Chem Nat Compd. 2009;45:333–337. [Google Scholar]
- 59.Fang A, Wong GK, Demain AL. Enhancement of the antifungal activity of rapamycin by the coproduced elaiophylin and nigericin. J Antibiot (Tokyo) 2000;53:158–162. doi: 10.7164/antibiotics.53.158. [DOI] [PubMed] [Google Scholar]
- 60.Zhou X, Fenical W. The unique chemistry and biology of the piericidins. J Antibiot (Tokyo) 2016;69:582–593. doi: 10.1038/ja.2016.71. [DOI] [PubMed] [Google Scholar]
- 61.Sengupta S, Chattopadhyay MK, Grossart HP. The multifaceted roles of antibiotics and antibiotic resistance in nature. Front Microbiol. 2013;4:47. doi: 10.3389/fmicb.2013.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khosla C, Gokhale RS, Jacobsen JR, Cane DE. Tolerance and specificity of polyketide synthases. Annu Rev Biochem. 1999;68:219–253. doi: 10.1146/annurev.biochem.68.1.219. [DOI] [PubMed] [Google Scholar]
- 63.Dunn BJ, Khosla C. Engineering the acyltransferase substrate specificity of assembly line polyketide synthases. J R Soc Interface. 2013;10:20130297. doi: 10.1098/rsif.2013.0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ridley CP, Lee HY, Khosla C. Evolution of polyketide synthases in bacteria. Proc Natl Acad Sci USA. 2008;105:4595–4600. doi: 10.1073/pnas.0710107105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kong R, et al. Elucidation of the biosynthetic gene cluster and the post-PKS modification mechanism for fostriecin in Streptomyces pulveraceus. Chem Biol. 2013;20:45–54. doi: 10.1016/j.chembiol.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 66.Liu XJ, Kong RX, Niu MS, Qiu RG, Tang L. Identification of the post-polyketide synthase modification enzymes for fostriecin biosynthesis in Streptomyces pulveraceus. J Nat Prod. 2013;76:524–529. doi: 10.1021/np300667r. [DOI] [PubMed] [Google Scholar]
- 67.Nguyen H-C, Darbon E, Thai R, Pernodet J-L, Lautru S. Post-PKS tailoring steps of the spiramycin macrolactone ring in Streptomyces ambofaciens. Antimicrob Agents Chemother. 2013;57:3836–3842. doi: 10.1128/AAC.00512-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bode HB. Entomopathogenic bacteria as a source of secondary metabolites. Curr Opin Chem Biol. 2009;13:224–230. doi: 10.1016/j.cbpa.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 69.Koppenhoefer HS, Gaugler R. 2009. Entomopathogenic nematode and bacteria mutualism. Defensive Mutualism in Microbial Symbiosis, Mycology Series, eds White JF, Torres MS (CRC, Boca Raton, FL), Vol 27, pp 99–116.
- 70.Wilson MC, et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature. 2014;506:58–62. doi: 10.1038/nature12959. [DOI] [PubMed] [Google Scholar]
- 71.Hale KJ, Hummersone MG, Manaviazar S, Frigerio M. The chemistry and biology of the bryostatin antitumour macrolides. Nat Prod Rep. 2002;19:413–453. doi: 10.1039/b009211h. [DOI] [PubMed] [Google Scholar]
- 72.Lopanik N, Lindquist N, Targett N. Potent cytotoxins produced by a microbial symbiont protect host larvae from predation. Oecologia. 2004;139:131–139. doi: 10.1007/s00442-004-1487-5. [DOI] [PubMed] [Google Scholar]
- 73.Elshahawi SI, et al. Boronated tartrolon antibiotic produced by symbiotic cellulose-degrading bacteria in shipworm gills. Proc Natl Acad Sci USA. 2013;110:E295–E304. doi: 10.1073/pnas.1213892110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin Z, et al. A bacterial source for mollusk pyrone polyketides. Chem Biol. 2013;20:73–81. doi: 10.1016/j.chembiol.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Davies J. Are antibiotics naturally antibiotics? J Ind Microbiol Biotechnol. 2006;33:496–499. doi: 10.1007/s10295-006-0112-5. [DOI] [PubMed] [Google Scholar]
- 76.Barke J, et al. A mixed community of actinomycetes produce multiple antibiotics for the fungus farming ant Acromyrmex octospinosus. BMC Biol. 2010;8:109. doi: 10.1186/1741-7007-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barke J, Seipke RF, Yu DW, Hutchings MI. A mutualistic microbiome: How do fungus-growing ants select their antibiotic-producing bacteria? Commun Integr Biol. 2011;4:41–43. doi: 10.4161/cib.4.1.13552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chait R, Craney A, Kishony R. Antibiotic interactions that select against resistance. Nature. 2007;446:668–671. doi: 10.1038/nature05685. [DOI] [PubMed] [Google Scholar]
- 79.Yeh PJ, Hegreness MJ, Aiden AP, Kishony R. Drug interactions and the evolution of antibiotic resistance. Nat Rev Microbiol. 2009;7:460–466. doi: 10.1038/nrmicro2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Burnside CE, Library CCMMA. Saprophytic Fungi Associated with the Honey Bee. Univ of Wisconsin Press; Madison, WI: 1927. [Google Scholar]
- 81.Hegreness M, Shoresh N, Damian D, Hartl D, Kishony R. Accelerated evolution of resistance in multidrug environments. Proc Natl Acad Sci USA. 2008;105:13977–13981. doi: 10.1073/pnas.0805965105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chait R, Vetsigian K, Kishony R. What counters antibiotic resistance in nature? Nat Chem Biol. 2011;8:2–5. doi: 10.1038/nchembio.745. [DOI] [PubMed] [Google Scholar]
- 83.Reading C, Cole M. Clavulanic acid: A beta-lactamase-inhiting beta-lactam from Streptomyces clavuligerus. Antimicrob Agents Chemother. 1977;11:852–857. doi: 10.1128/aac.11.5.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Olberg G. Der Bienenfeind Philanthus. Akademische Verlagsgemeinschaft Geest & Portig K.-G.; Leipzig, Germany: 1953. [Google Scholar]
- 85.Simon-Thomas RT, Simon-Thomas AMJ. Some observations on the behaviour of females of Philanthus triangulum, Hymenoptera, Sphecidae. Tijdschr Entomol. 1972;115:123–139. [Google Scholar]
- 86.Engl T, Bodenstein B, Strohm E. Mycobiota in the brood cells of the European beewolf, Philanthus triangulum (Hymenoptera: Crabronidae) Eur J Entomol. 2016;213:271–277. [Google Scholar]
- 87.Kaltenpoth M, Roeser-Mueller K, Stubblefield JW, Seger J, Strohm E. Biogeography of a defensive symbiosis. Commun Integr Biol. 2014;7:e993265. doi: 10.4161/19420889.2014.993265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Charlop-Powers Z, et al. Global biogeographic sampling of bacterial secondary metabolism. eLife. 2015;4:e05048. doi: 10.7554/eLife.05048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lemetre C, et al. Bacterial natural product biosynthetic domain composition in soil correlates with changes in latitude on a continent-wide scale. Proc Natl Acad Sci USA. 2017;114:11615–11620. doi: 10.1073/pnas.1710262114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kost C, et al. Non-specific association between filamentous bacteria and fungus-growing ants. Naturwissenschaften. 2007;94:821–828. doi: 10.1007/s00114-007-0262-y. [DOI] [PubMed] [Google Scholar]
- 91.Thacker R, et al. Phylogenetic relationships among the filamentous cyanobacterial symbionts of Caribbean sponges and a comparison of photosynthetic production between sponges hosting filamentous and unicellular cyanobacteria. In: Custódio M, Lôbo-Hajdu G, Hajdu E, Muricy G, editors. Porifera Research: Biodiversity, Innovation and Sustainability. Vol 28. Série Licros; Rio de Janeiro: 2007. pp. 621–626. [Google Scholar]
- 92.Lim-Fong GE, Regali LA, Haygood MG. Evolutionary relationships of “Candidatus Endobugula” bacterial symbionts and their Bugula bryozoan hosts. Appl Environ Microbiol. 2008;74:3605–3609. doi: 10.1128/AEM.02798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Erpenbeck D, et al. Evolution, radiation and chemotaxonomy of Lamellodysidea, a demosponge genus with anti-plasmodial metabolites. Mar Biol. 2012;159:1119–1127. [Google Scholar]
- 94.Henry LM, et al. Horizontally transmitted symbionts and host colonization of ecological niches. Curr Biol. 2013;23:1713–1717. doi: 10.1016/j.cub.2013.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Flórez LV, Kaltenpoth M. Symbiont dynamics and strain diversity in the defensive mutualism between Lagria beetles and Burkholderia. Environ Microbiol. 2017;19:3674–3688. doi: 10.1111/1462-2920.13868. [DOI] [PubMed] [Google Scholar]
- 96.Flórez LV, et al. Antibiotic-producing symbionts dynamically transition between plant pathogenicity and insect-defensive mutualism. Nat Commun. 2017;8:15172. doi: 10.1038/ncomms15172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang L, et al. Farming of a defensive fungal mutualist by an attelabid weevil. ISME J. 2015;9:1793–1801. doi: 10.1038/ismej.2014.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nechitaylo TY, Westermann M, Kaltenpoth M. Cultivation reveals physiological diversity among defensive ‘Streptomyces philanthi’ symbionts of beewolf digger wasps (Hymenoptera, Crabronidae) BMC Microbiol. 2014;14:202. doi: 10.1186/s12866-014-0202-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.