Significance
Regulatory T cells (Tregs) play a critical role in inflammatory, autoimmune, and antitumor immune responses. Increased expression of transcription factor Helios by tumor-infiltrating Tregs can enhance immune-suppressive activity, while deletion of Helios promotes an effector T helper (Th) cell phenotype that can contribute to the host antitumor immune response. We report that chronic inflammatory conditions of tumors induce Helios-deficient Tregs to express increased levels of genes associated with T cell activation and Th cell differentiation. Helios-dependent changes in gene expression are restricted to tumor sites and not observed in peripheral lymphoid tissues. We suggest that Helios-deficient Tregs that recognize tumor-associated self-antigens may become unstable in the tumor microenvironment and undergo reprogramming into effector T cells that can inhibit tumor growth.
Keywords: Treg, transcriptome, tumor microenvironment
Abstract
Regulatory T cells (Tregs) are key modulators of immune tolerance, capable of suppressing inflammatory immune responses and promoting nonlymphoid tissue homeostasis. Helios, a transcription factor (TF) that is selectively expressed by Tregs, has been shown to be essential for the maintenance of Treg lineage stability in the face of inflammatory conditions that include autoimmune disease and cancer. Helios-deficient Tregs within tumors acquire effector T cell function and contribute to immune responses against cancer. However, the underlying genetic basis of this Treg reprogramming is not well understood. Here, we report that Helios-deficient Tregs within the chronic inflammatory tumor microenvironment (TME) derepress genetic programs associated with T helper (Th) cell differentiation by up-regulating Th cell-associated TFs and effector cytokines. These genetic changes of Helios-deficient Tregs are most apparent in a Treg subpopulation with high affinity for self-antigens, as detected by both increased GITR/PD-1 expression and increased responsiveness to self-antigens. Their combined effects may promote a phenotype conversion of Tregs into effector T cells within the TME, where TCR engagement and costimulatory receptor expression by Tregs are increased. These data provide a genetic basis for the unstable phenotype of Helios-deficient Tregs within the inflammatory environment of tumors and suggest that immune milieu-dependent alterations in gene expression are a central feature of Treg conversion.
Regulatory T cells (Tregs) are a specialized subset of CD4 T cells that play a key role in the maintenance of self-tolerance and immune regulation during inflammation. The majority of Tregs differentiate in the thymus as an alternative pathway for CD4 single positive cells that express self-reactive TCR. As a result, the TCR repertoire of FoxP3+ Tregs in peripheral tissues is highly shifted toward self-reactive TCR (1). Due to the self-reactivity of their TCRs, stable expression of a suppressive phenotype is critically important to prevent differentiation into proinflammatory effector T cells that can provoke the development of autoimmune disease. Emerging evidence supports the notion that FoxP3 expression alone is not sufficient to maintain the characteristic suppressive and homeostatic features of Tregs; additional genetic elements are required for expression of Treg function, lineage stability, and adaptation to the tissue environment.
While Tregs are classically viewed as regulators of other immune cells, it is increasingly acknowledged that specialized Tregs in tissues are important for maintenance of organ homeostasis. Recent studies revealed that tissue-resident Tregs acquire and stabilize distinct genetic programs, which confers plasticity to Tregs and allows them to adapt to particular tissues and to express tissue-appropriate cellular and molecular components (2–5). Tumor tissues can also be viewed as a newly organized organ that forms a micromilieu that promotes cancer cell survival. It is therefore likely that intratumoral Tregs adopt a tumor tissue-specific genetic program in response to distinct and separately evolving tumor microenvironments (TMEs) (6).
Recent studies suggest that although the Helios transcription factor (TF) is largely dispensable for Treg activity in the steady state, control of the genetic program of FoxP3+ CD4 Tregs by Helios in the context of inflammation is essential to maintain a stable phenotype and potentiate suppressive function (7, 8). The Helios (Ikzf2) gene, a member of the Ikaros family of TFs, differs from other Ikaros family members according to its selective expression by thymocytes undergoing negative selection, as well as by regulatory lineages of CD4 and CD8 T cells. These observations indicate the critical contribution of Helios to self-reactive T cell selection, differentiation, and function. This view receives direct support from recent findings that selective Helios deficiency within CD4 Tregs leads to enhanced antitumor immunity that reflects induction of an unstable phenotype and conversion of Tregs into T effector cells within the TME (9). In view of the increased self-reactivity of TCR in CD4 Tregs compared with conventional T cells, conversion of Tregs might be expected to generate highly potent effector CD4 T cells accompanied by release from Treg-mediated suppression within the TME. While the antitumor activity of converted Tregs that lack Helios is clear, the basis for genetic reprogramming of these Tregs and associated functional changes is poorly understood.
Here, we analyze the transcriptome of Tregs in the spleen and tumor site with respect to Helios expression. These data delineate Helios-dependent influences on the Treg transcriptome within the TME and reveal that effector cell conversion of Helios-deficient Tregs within the tumor-tissue microenvironment is associated with increased expression of genes that control T effector cell phenotype and may be linked to the increased self-reactivity of Tregs that undergo intratumoral conversion.
Results
Helios Expression Regulates the Transcriptome of Intratumoral but Not Splenic Tregs.
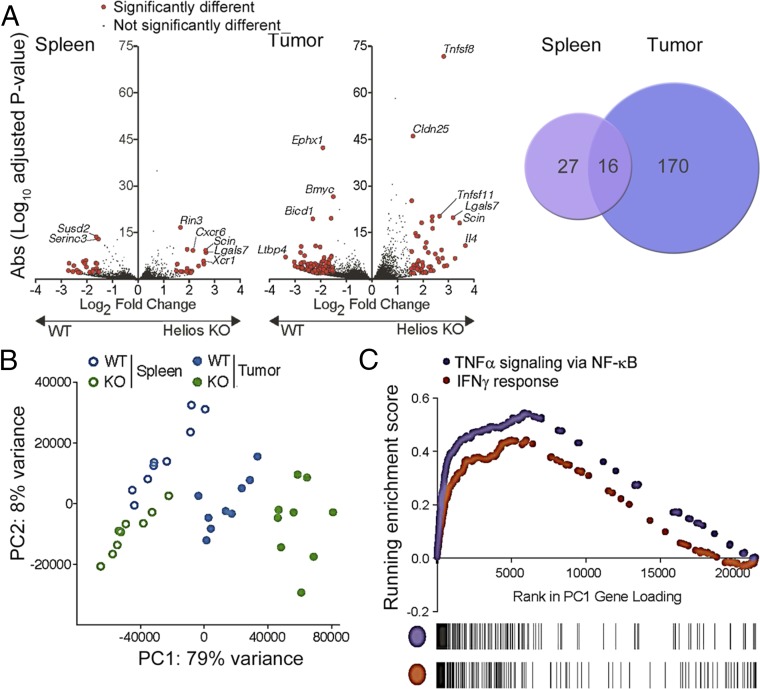
Several studies have linked the Helios TF with maintenance of lineage stability of effector Tregs (eTregs) and expression of suppressive activity under acute or chronic inflammatory conditions (8–10). Earlier analyses showed that mice with Helios-deficient Tregs have smaller tumors in models of B16 melanoma, B16 melanoma with GVAX, and MC38 colon adenocarcinoma (9). The impact of Helios-deficient Tregs on antitumor immunity is cell-intrinsic (i.e., transfer of Helios-deficient CD4 Tregs alone is sufficient to reduce tumor growth). Lineage stability of Helios-deficient Tregs is also impaired and is accompanied by diminished FoxP3 expression; acquisition of an effector phenotype; and production of proinflammatory cytokines, including IFN-γ. Acquisition of an unstable phenotype by Helios-deficient Tregs within the TME, but not in peripheral lymphoid organs (9), suggested that an altered Treg signature might be selectively induced within the chronic inflammatory conditions of growing tumor. To identify dominant genetic pathways involved in this process, we performed RNA-sequencing (RNA-seq) using Tregs isolated from tumors and spleens of FoxP3-Cre (Helios WT) and Heliosfl/fl.FoxP3-Cre (Helios KO) mice that had been inoculated s.c. with B16/F10 melanoma cells. Splenic Tregs from Helios WT and KO mice displayed similar transcriptomes containing ∼43 differentially expressed transcripts (at a ≥1.5 log2-fold change and P value <0.01), while the transcriptomes of intratumoral Tregs from Helios WT and KO mice showed 186 differentially expressed genes (≥1.5-fold change) (Fig. 1A and Table S1). These data show that the magnitude and degree of differential gene expression imposed by Helios deficiency within Tregs depend greatly on the tissue microenvironment.
Fig. 1.
Transcriptome analysis of splenic and tumor Tregs reveals divergent gene expression by intratumoral Helios-deficient Tregs. (A) Helios WT and KO mice were inoculated s.c. with B16/F10. Small numbers (∼400) of splenic and tumor Tregs were sorted for gene expression profiling. Volcano plots show the meta-analysis of differential gene expression of splenic and tumor Tregs from Helios WT or KO mice (P < 0.01). Genes that display differential expression by a log-fold change ≥1.5 are shown in red. (B) Principal component analysis of Treg populations isolated from spleen and tumor from Helios WT and KO mice, showing the top two principal components (PC1 and PC2) and their contribution to intersample variation. (C) Gene set enrichment analysis showing enrichment of signatures for TNF-α signaling via NF-κB and IFN-γ responses, which contribute to PC1 in the principal component analysis.
To understand molecular changes underlying the unstable phenotype of Helios-deficient Tregs within the chronic inflammatory conditions of the TME, we investigated the relatedness of Helios-deficient intratumoral Tregs to Helios-sufficient intratumoral and splenic Tregs using principal component analysis (Fig. 1B). Analysis of the relationship using the first principal component representing 79% of variance revealed segregation of populations based on tissue and genetic origin. Intratumoral Tregs formed groups that were most distant from splenic Tregs independent of Helios genotype. Moreover, intratumoral Tregs from Helios WT and KO mice showed sharp segregation in contrast to splenic Tregs, which did not clearly segregate according to Helios expression along principal component 1, which enriches in gene signatures for inflammation, including TNF-α signaling and IFN-γ response (Fig. 1C). These data suggest that the Helios-deficient Treg phenotype is highly sensitive to the TME, while residence of Helios-deficient Tregs within the relatively noninflammatory splenic environment does not impose a significantly altered phenotype.
Genes Associated with Th Cell Differentiation Are Up-Regulated by Intratumoral Helios-Deficient Tregs.
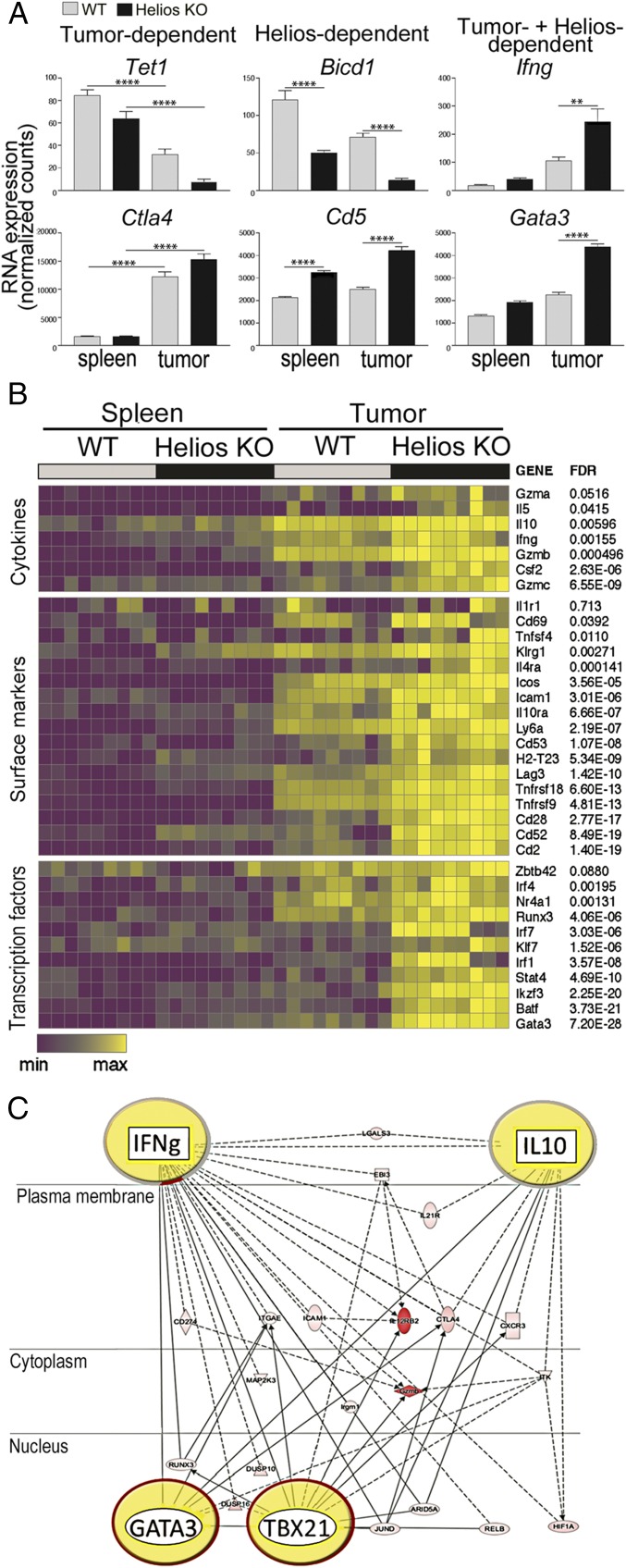
We used transcriptional profiling to identify major gene networks that might contribute to the conversion of Helios-deficient Tregs within the TME but not elsewhere in spleen. We identified genes that were differentially expressed according to the location of Tregs [spleen vs. tumor (e.g., Tet1, CTLA4)], according to Helios expression [Helios WT vs. KO (e.g., Bicd1, CD5)], or according to both location and Helios expression (e.g., IFN-γ, GATA3) (Fig. 2A). To identify genes that are selectively up-regulated by Helios-deficient intratumoral Tregs, we compared the profiles of Helios WT and KO intratumoral Tregs. Genes up-regulated by Helios-deficient intratumoral Tregs were then compared with genes expressed by Helios-deficient intrasplenic Tregs (Fig. 2B). This analysis revealed that Helios-deficient intratumoral Tregs displayed increased levels of Treg surface receptors, including ICOS, Tnfrsf18, Tnfrsf9, Klrg1, and Il1r1, that are associated with final Treg differentiation (Fig. 2B). TFs involved in Th lineage determination, such as Ikzf3, Irf1, Gata3, Batf, and Stat4, were also up-regulated. Of note, expression of Nr4a1, a TF induced upon TCR engagement that reflects TCR signaling strength (11), was up-regulated by intratumoral Tregs, suggesting that recognition of tumor-associated antigens by tumor-resident Tregs influenced the phenotype of these cells. Expression of cytokines involved in T cell effector functions were also increased by Helios-deficient intratumoral Tregs. Gene network analysis showed that these genes collectively formed a network that represents the Th1 and Th2 differentiation programs and contains major gene modules that regulate expression of canonical Th1/Th2 cytokines and TFs, including IFN-γ, IL-10, GATA3, and TBX21 (Fig. 2C).
Fig. 2.
Identification of genes that are selectively up-regulated by Helios-deficient intratumoral Tregs. (A) Example of genes that are differentially expressed by Tregs based on tissue (Left, spleen vs. tumor) and genotype (Center, Helios WT vs. KO). An example of genes that are up-regulated selectively by intratumoral Helios-deficient Tregs is also shown (Right). (B) Expression of genes for cell surface proteins, TFs, and cytokines that are selectively up-regulated by Helios-deficient intratumoral Tregs is presented as a heat map. (C) Pathway analysis of genes that are up-regulated by Helios-deficient intratumoral Tregs. The four most prominent nodules are highlighted. Filled symbols indicate genes up-regulated by intratumoral Helios-deficient Tregs, and white symbols indicate neighboring genes that are functionally associated with, but not included in, these genes. **P ≤ 0.01; ****P ≤ 0.0001.
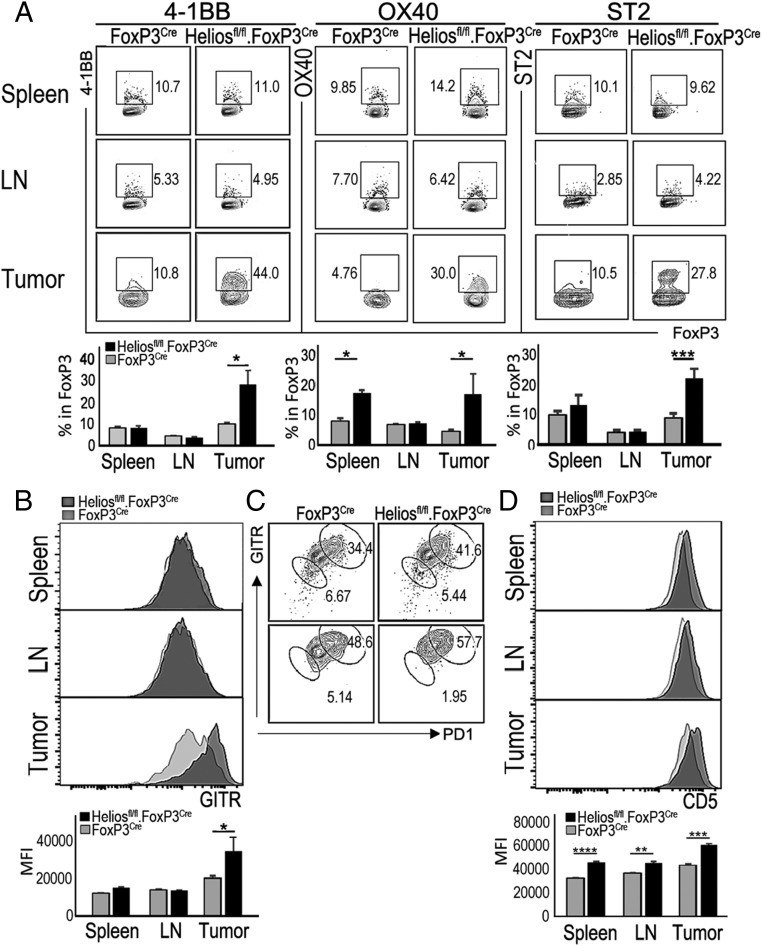
Recent analysis of Tregs has indicated that expression of costimulatory molecules 4-1BB (Tnfrsf9) and OX-40 (Tnfrsf4) may regulate the generation and activity of Tregs (12). Thus, OX-40 and 4-1BB signals may promote proliferation and/or survival of Tregs in vitro (13–15), while ligation of OX-40 or 4-1BB in vivo can alter suppressive activity in multiple disease settings, including inflammatory bowel disease and allogeneic bone marrow or skin transplantation (13, 16, 17). Although the underlying mechanisms have not been delineated, we hypothesized that increased expression and ligation of OX-40 and 4-1BB may potentiate induction of an unstable Treg phenotype within the TME. Fluorescence-activated cell sorting (FACS) analysis of Tregs from B16 tumor-bearing mice showed that Helios-deficient intratumoral Tregs expressed significantly higher levels of OX-40 and 4-1BB than Tregs isolated from lymphoid tissues (spleen and lymph node) (Fig. 3A). Engagement of these costimulatory receptors within the TME may potentiate the induction of an unstable Treg phenotype and Treg conversion through amplification of TCR-mediated signaling.
Fig. 3.
Helios-deficient Tregs display an activated phenotype that is associated with antigen recognition. (A) Flow cytometric analysis of expression of 4-1BB, OX-40, and ST2, which were shown to be up-regulated by intratumoral Helios-deficient Tregs in RNA-seq analysis. Tregs (TCR+CD4+FoxP3+) in spleen, lymph node (LN), and tumor from B16/F10 tumor-bearing mice were analyzed for the expression of indicated molecules at day 21 after tumor inoculation. (B) Expression levels of GITR by Tregs in spleen, LN, and tumor from B16/F10 tumor-bearing Helios WT or KO mice. MFI, mean fluorescence intensity. (C) Percentage of PD-1+GITR+ Tregs in spleen and tumor from B16/F10 tumor-bearing Helios WT or KO mice. (D) Levels of CD5 expression by Tregs from spleen, LN, and tumor from B16/F10 tumor-bearing Helios WT or KO mice. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Interestingly, the set of genes up-regulated by Helios-deficient intratumoral Tregs is reminiscent of the recently characterized Treg population called “tissue Tregs” that typically express ST2 (Il1r1), KLRG1, GATA3, and BATF (5) and are involved in maintenance of tissue integrity. ST2 expression was also selectively increased by intratumoral Helios-deficient Tregs in contrast to both Helios-deficient and Helios WT splenic and lymph node Tregs (Fig. 3A).
These data suggest that Helios-deficient Tregs within the chronic inflammatory TME derepress genetic programs associated with Th differentiation and effector T cell activation.
Helios-Deficient Tregs Display Increased Self-Reactivity.
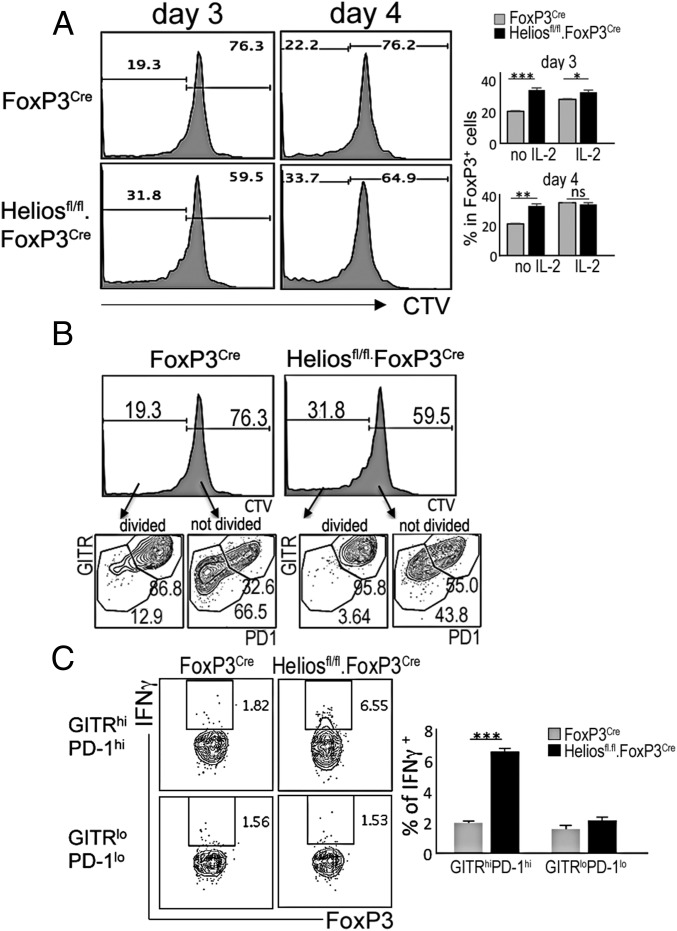
Expression levels of TNF receptor superfamily members, including GITR (Tnfrsf18) and OX-40 (Tnfrsf4), have been positively correlated with strength of TCR signaling during Treg development and in mature Tregs (18). Earlier studies indicating that increased expression of GITR and OX-40 by self-reactive Tregs confers a selective advantage that promotes Treg maturation (18) suggest that GITR/OX-40 costimulation may shape Treg repertoire selection. Our RNA-seq analysis showed that Helios-deficient intratumoral Tregs express significantly higher levels of GITR compared with Helios WT intratumoral Tregs (Fig. 3B). Recent studies have demonstrated that Tregs are divisible into GITRhiPD1hi and GITRloPD1lo cells that are independent of central Treg and eTreg status, and reflect their TCR affinity for self-antigens (19). Our analysis of Helios-deficient Tregs revealed increased proportions of GITRhiPD1hi Tregs in both spleen and tumor (Fig. 3C). This may indicate that the TCR repertoire expressed by Helios-deficient Tregs is skewed toward increased self-reactivity. To directly test whether Helios WT and KO Tregs differ in their degree of self-reactivity, we first compared CD5 protein levels, since expression of this receptor indicates T cell activation and correlates with TCR affinity for its peptide-MHC ligand (20). We observed higher CD5 expression by Helios-deficient CD4 Tregs compared with Helios WT Tregs in both spleen and tumor (Fig. 3D), suggesting that Helios-deficient Tregs may be more self-reactive than Helios WT Tregs. Expression of CD5 is more prominent in intratumoral Tregs, which may reflect increased TCR signaling intensity (20, 21) upon encounter with tumor-associated antigens that represent self-antigens. To further test this hypothesis, we incubated Tregs isolated from Helios WT and KO mice with syngeneic WT activated B cells that can efficiently present MHC class II-associated self-antigens to Tregs. We observed increased proliferation of Helios-deficient Tregs compared with Helios WT Tregs in these cocultures (Fig. 4A). Responsiveness of Tregs to self-antigens was associated with an early response of Tregs that express high-affinity TCR, according to highly increased GITR and PD1 levels on proliferating Tregs (Fig. 4B). This differential responsiveness to self-antigen was abolished when IL-2 was provided to cultures. Increased recognition of tumor-associated antigens by Helios-deficient Tregs harboring high-affinity TCR may also be associated with increased levels of their activation and subsequent conversion associated with production of IFN-γ, since only GITRhiPD-1hi Tregs from Helios-deficient Tregs expressed IFN-γ upon stimulation with antigens presented by syngeneic antigen-presenting cells (Fig. 4C). These data suggest that Helios-deficient Tregs may display a TCR repertoire skewed toward high-affinity anti–self-MHC/peptides, which can promote robust activation in appropriate microenvironments, such as the TME.
Fig. 4.
Increased responsiveness of Helios-deficient Tregs to self-antigens. (A and B) Analysis of the responsiveness of Tregs to self-antigens. Sorted Tregs from Helios WT or KO mice were labeled with CTV and cocultured with activated syngeneic B cells in the presence or absence of IL-2 (19). The percentage of cells that underwent division during the indicated culture time periods and the percentage of Tregs with high-affinity TCR (PD1+GITR+) (19) within dividing or nondividing cells are depicted. (C) Percentage of IFN-γ–expressing cells in high-affinity (GITRhiPD-1hi) and low-affinity (GITRloPD-1lo) FoxP3+ cells from Helios WT or KO mice that were cocultured with activated syngeneic B cells for 3 d. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Discussion
We report that Helios deficiency in Tregs under chronic inflammatory conditions of the TME activates genetic modules that lead to their differentiation into effector Th cells as judged from comparative transcriptome analysis. Divergent expression of core gene sets associated with T cell activation were observed by intratumoral Helios-deficient Tregs compared with Helios-sufficient Tregs, and this genetic difference was not observed in intrasplenic Tregs.
Selective acquisition of effector T cell program(s) by intratumoral Helios-deficient Tregs suggests that recognition of self-antigen under chronic inflammatory conditions may trigger signaling cascades that collectively lead to breakdown of core Treg genetic programs. The TCR repertoire of Tregs is skewed to a large extent to recognizing self-antigens and potentially quasi–self-tumor antigens. The self-reactive nature of Tregs may equip this population of cells to mediate potent antitumor immunity when they acquire the phenotype of effector T cells. Indeed, earlier studies have indicated that isolated Helios-deficient Tregs alone can exert antitumor immune responses in the absence of CD8 T cells (9).
While the core function of Tregs as key regulators of immune tolerance, capable of suppressing multiple immune cell types and dampening excessive inflammatory responses, is well recognized, it is becoming increasingly clear that Tregs also play an important role in promoting nonlymphoid tissue homeostasis (22). This additional role of tissue Tregs, apparent from their regulation of adipose tissue metabolism, facilitation of muscle repair, and enforcement of colonic homeostasis, has recently been extended to maintenance of hematopoietic stem cells in bone marrow and hair follicle stem cells in the skin (4, 23). These tissue Tregs not only express genetic programs that coopt their respective tissue microenvironments but also display clonal expansion of distinctive TCR (2, 24), suggesting a history of migration and expansion of Tregs reactive to tissue specific self-antigens.
Solid tumors develop through complex interactions between different cellular and molecular components that favor tumor cell survival and expansion that may be analogous to developing organs. Tumor tissue may also recruit Tregs reactive to tumor-associated antigens to the TME, which generally represents a site of chronic inflammation. Analysis of epigenetic modifications defining molecular characteristics of tissue Tregs has revealed that all tissue Tregs are demethylated at the Ikzf2 locus (5), which may suggest both the thymic Treg (tTreg) origin of tissue Tregs as well as a universal contribution of Helios expression to tissue Tregs. In addition, tissue Tregs characterized by the expression of markers, including ST2, KLRG1, TIGIT, and GATA3, almost exclusively occupy the CD44hi effector-memory compartment, suggesting that tissue Tregs have previously been activated via TCR signaling. Our comparative transcriptome analysis showed that tumor Tregs (Helios WT) up-regulated ST2, GATA3, KLRG1, and BATF, reminiscent of extensively characterized fat and skin tissue Tregs (5, 25). Helios-deficient Tregs within the TME displayed increased expression of these tissue Treg markers compared with Helios WT intratumoral Tregs, as well as expression of markers, including Nr4a1 and CD69, that reflect antigen recognition by TCR and relatively strong signaling strength. Although earlier studies did not detect the impact of Helios expression on thymic Treg development (7, 8, 26), enhanced expression of Nr4a1 and CD5 by Helios-deficient Tregs indicates their skewed TCR repertoire toward high-affinity TCR. The reductionist in vitro approach taken here showing that Helios-deficient Tregs display enhanced responsiveness to self-antigens supports the notion that the increased activation of Tregs within TME may reflect their somewhat increased TCR affinity to self-antigens.
In view of the antigen-primed status of Tregs, it is likely that Tregs recognizing particular self- or tumor antigens within this tissue microenvironment are more easily activated than conventional T cells. Helios expression may safeguard these tissue Tregs and promote Treg lineage stability in the face of high levels of activation within the chronic inflammatory conditions of the TME. It is therefore feasible that extensive inflammatory signaling via costimulatory molecules in addition to TCR engagement may lead to the induction of differentiation of conventional T cells with antitumor activity in the absence of the stabilizing influence of the Helios TF.
Materials and Methods
Mice.
C57BL/6J (B6) and FoxP3YFP-Cre mice were obtained from the Jackson Laboratory. Heliosfl/fl mice were provided by Ethan Shevach, NIH, Bethesda, MD, and Heliosfl/fl.FoxP3YFP-Cre mice were generated by crossing them to FoxP3YFP-Cre mice. CD45.1+ C57BL/6 mice were obtained from Taconic Farms. All mice were housed in specific pathogen-free conditions in the animal facilities at the Dana–Farber Cancer Institute. All experiments were performed in compliance with federal laws and institutional guidelines as approved by the Dana–Farber Cancer Institute’s Animal Care and Use Committee.
Antibodies and Flow Cytometry.
Fluorescence dye-labeled Abs specific for CD4 (L3T4), CD8 (53-6.7), TCR (H57-597), CD25 (PC61), CD45.2 (104), ST2 (DIH9), CD5 (53-7-3), GITR (DTA-1), OX-40 (OX-86), 4-1BB (17-B5), CD44 (IM7), CD62L (MEL-14), PD-1 (J43), Helios (22F6), FoxP3 (NRRF-30), and IFN-γ (XMG1.2) were purchased from Becton Dickinson, eBioscience, and Biolegend. Intracellular staining for Helios, FoxP3, and IFN-γ was performed using a FoxP3 staining buffer set (eBioscience).
Tumor Induction and Cell Preparation.
B16/F10 melanoma cells were s.c. inoculated into Helios WT or KO mice, and the tumor growth was monitored. Twenty days after tumor induction, cells were prepared from spleen and tumor. Single-cell suspensions of total splenocytes and lymphocyte-enriched tumor cells were stained with CD45, TCR, and CD4 for FACS analysis. FoxP3+ CD4 cells (CD4+YFP+) were sorted in pools of 100 cells into a microplate containing 5 μL of RLT buffer (Qiagen) + 1% (vol/vol) β-mercaptoethanol in each well. Immediately after sorting, plates were sealed, vortexed briefly, spun down at 400 × g for 1 min, and flash-frozen on dry ice for storage at −80 °C until library preparation.
RNA-Seq Analysis of Tregs.
Three wells, each containing four pools from each sample (equivalent to 400 cells), were combined to generate three technical replicates from three biological replicates (a total of nine samples per group) to generate next-generation sequencing libraries. Single-cell lysates were converted to cDNA following capture with Agencourt RNA Clean beads using the SmartSeq2 protocol as previously described (27). The cDNA was amplified using 18 PCR enrichment cycles before quantification and dual-index barcoding with an Illumina Nextera XT kit. The libraries were enriched with 12 cycles of PCR and then combined in equal volumes before final bead clean-up and sequencing on a NextSeq500 instrument by 37-bp paired-end reads. After demultiplexing, low-quality base reads were trimmed with Trimmomatic (28) using the following parameters: LEADING: 15, TRAILING: 15, SLIDINGWINDOW: 4:15, and MINLEN: 16. Trimmed reads were then aligned to the mm10 mouse genome using Bowtie2. HTSeq was used to map aligned reads to genes and to generate a gene count matrix. Normalized counts and differential expression analysis were performed using the DESeq2 R package. We performed gene set enrichment analysis as previously described (29). Gene network analysis was performed using Ingenuity Pathway Analysis (Qiagen).
In Vitro Stimulation and Proliferation.
B cells were isolated from B6 CD45.1 mice and stimulated with LPS (50 ng/mL) overnight. FoxP3+ CD4 Tregs (YFP+CD4+) were isolated from Helio WT or KO mice by sorting for YFP+ cells with FACS after labeling with CD45, TCR, CD4, and CD25 (purity > 97%). Purified CD4 Tregs were labeled with CellTrace Violet (CTV; ThermoFisher Scientific) according to the manufacturer’s instructions. CTV-labeled CD4 Tregs were cocultured with activated B cells with or without IL-2 (20 ng/mL). Proliferation of CD4 Tregs was measured by FACS based on levels of CTV signal.
Statistical Analysis.
Statistical analysis was performed according to unpaired, two-tailed Student’s t tests using Prism software (GraphPad). A P value of ≤0.05 was considered statistically significant (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).
Supplementary Material
Acknowledgments
We thank A. Thornton and E. Shevach (NIH) for provision of Heliosfl/fl mice and A. Angel for manuscript/figure preparation. These studies were supported, in part, by NIH Grant R01AI37562 and by a grant from the LeRoy Schecter Research Foundation (to H.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720447115/-/DCSupplemental.
References
- 1.Hogquist KA, Baldwin TA, Jameson SC. Central tolerance: Learning self-control in the thymus. Nat Rev Immunol. 2005;5:772–782. doi: 10.1038/nri1707. [DOI] [PubMed] [Google Scholar]
- 2.Burzyn D, et al. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155:1282–1295. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cipolletta D, et al. PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ali N, et al. Regulatory T cells in skin facilitate epithelial stem cell differentiation. Cell. 2017;169:1119–1129.e11. doi: 10.1016/j.cell.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delacher M, et al. Genome-wide DNA-methylation landscape defines specialization of regulatory T cells in tissues. Nat Immunol. 2017;18:1160–1172. doi: 10.1038/ni.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plitas G, et al. Regulatory T cells exhibit distinct features in human breast cancer. Immunity. 2016;45:1122–1134. doi: 10.1016/j.immuni.2016.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thornton AM, et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol. 2010;184:3433–3441. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim HJ, et al. Stable inhibitory activity of regulatory T cells requires the transcription factor Helios. Science. 2015;350:334–339. doi: 10.1126/science.aad0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakagawa H, et al. Instability of Helios-deficient Tregs is associated with conversion to a T-effector phenotype and enhanced antitumor immunity. Proc Natl Acad Sci USA. 2016;113:6248–6253. doi: 10.1073/pnas.1604765113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sebastian M, et al. Helios controls a limited subset of regulatory T cell functions. J Immunol. 2016;196:144–155. doi: 10.4049/jimmunol.1501704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moran AE, et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.So T, Lee SW, Croft M. Immune regulation and control of regulatory T cells by OX40 and 4-1BB. Cytokine Growth Factor Rev. 2008;19:253–262. doi: 10.1016/j.cytogfr.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeda I, et al. Distinct roles for the OX40-OX40 ligand interaction in regulatory and nonregulatory T cells. J Immunol. 2004;172:3580–3589. doi: 10.4049/jimmunol.172.6.3580. [DOI] [PubMed] [Google Scholar]
- 14.Elpek KG, et al. Ex vivo expansion of CD4+CD25+FoxP3+ T regulatory cells based on synergy between IL-2 and 4-1BB signaling. J Immunol. 2007;179:7295–7304. doi: 10.4049/jimmunol.179.11.7295. [DOI] [PubMed] [Google Scholar]
- 15.Zheng G, Wang B, Chen A. The 4-1BB costimulation augments the proliferation of CD4+CD25+ regulatory T cells. J Immunol. 2004;173:2428–2434. doi: 10.4049/jimmunol.173.4.2428. [DOI] [PubMed] [Google Scholar]
- 16.Vu MD, et al. OX40 costimulation turns off Foxp3+ Tregs. Blood. 2007;110:2501–2510. doi: 10.1182/blood-2007-01-070748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valzasina B, et al. Triggering of OX40 (CD134) on CD4(+)CD25+ T cells blocks their inhibitory activity: A novel regulatory role for OX40 and its comparison with GITR. Blood. 2005;105:2845–2851. doi: 10.1182/blood-2004-07-2959. [DOI] [PubMed] [Google Scholar]
- 18.Mahmud SA, et al. Costimulation via the tumor-necrosis factor receptor superfamily couples TCR signal strength to the thymic differentiation of regulatory T cells. Nat Immunol. 2014;15:473–481. doi: 10.1038/ni.2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wyss L, et al. Affinity for self antigen selects Treg cells with distinct functional properties. Nat Immunol. 2016;17:1093–1101. doi: 10.1038/ni.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandl JN, Monteiro JP, Vrisekoop N, Germain RN. T cell-positive selection uses self-ligand binding strength to optimize repertoire recognition of foreign antigens. Immunity. 2013;38:263–274. doi: 10.1016/j.immuni.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith K, et al. Sensory adaptation in naive peripheral CD4 T cells. J Exp Med. 2001;194:1253–1261. doi: 10.1084/jem.194.9.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panduro M, Benoist C, Mathis D. Tissue Tregs. Annu Rev Immunol. 2016;34:609–633. doi: 10.1146/annurev-immunol-032712-095948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujisaki J, et al. In vivo imaging of Treg cells providing immune privilege to the haematopoietic stem-cell niche. Nature. 2011;474:216–219. doi: 10.1038/nature10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolodin D, et al. Antigen- and cytokine-driven accumulation of regulatory T cells in visceral adipose tissue of lean mice. Cell Metab. 2015;21:543–557. doi: 10.1016/j.cmet.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayatsu N, et al. Analyses of a mutant Foxp3 allele reveal BATF as a critical transcription factor in the differentiation and accumulation of tissue regulatory T cells. Immunity. 2017;47:268–283.e9. doi: 10.1016/j.immuni.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 26.Cai Q, Dierich A, Oulad-Abdelghani M, Chan S, Kastner P. Helios deficiency has minimal impact on T cell development and function. J Immunol. 2009;183:2303–2311. doi: 10.4049/jimmunol.0901407. [DOI] [PubMed] [Google Scholar]
- 27.Trombetta JJ, et al. Preparation of single-cell RNA-seq libraries for next generation sequencing. Curr Protoc Mol Biol. 2014;107:4.22.1–4.22.17. doi: 10.1002/0471142727.mb0422s107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.