Significance
DNA replication poses a unique logistical challenge for cells in that structural features of chromatin and their regulatory functions must be carefully coordinated with the passage of replication machinery so faithful duplication of both the genome and its chromatin structures may be achieved. Nucleosome assembly is fundamental to reestablishment of chromatin in the wake of DNA replication. Here, a mechanism for coordinating nucleosome assembly with DNA replication to maintain silenced chromatin is described.
Keywords: PCNA, nucleosome assembly, heterochromatin, CAF-1, Elg1
Abstract
In Saccharomyces cerevisiae, heterochromatin structures required for transcriptional silencing of the HML and HMR loci are duplicated in coordination with passing DNA replication forks. Despite major reorganization of chromatin structure, the heterochromatic, transcriptionally silent states of HML and HMR are successfully maintained throughout S-phase. Mutations of specific components of the replisome diminish the capacity to maintain silencing of HML and HMR through replication. Similarly, mutations in histone chaperones involved in replication-coupled nucleosome assembly reduce gene silencing. Bridging these observations, we determined that the proliferating cell nuclear antigen (PCNA) unloading activity of Elg1 was important for coordinating DNA replication forks with the process of replication-coupled nucleosome assembly to maintain silencing of HML and HMR through S-phase. Collectively, these data identified a mechanism by which chromatin reassembly is coordinated with DNA replication to maintain silencing through S-phase.
Transcriptional repression within heterochromatin occurs through mechanisms that are typically insensitive to the identity of the genes encoded in the DNA of the repressed domains. The specific composition of proteins and epigenetic signatures that distinguishes heterochromatin from euchromatin varies somewhat among species and indeed even from one chromosome region to another. For example, in humans, regions of constitutive heterochromatin are typically enriched for H3K9 trimethylation, whereas regions of facultative heterochromatin such as those found in inactivated X chromosomes and at loci that regulate cell identity are typically enriched for H3K27 trimethylation (1, 2). Despite the range of mechanisms and proteins involved in heterochromatin formation and maintenance, certain characteristics appear universal and underlie a fundamental relationship between heterochromatin structure and function: Heterochromatin is structurally compact, localizes to distinct areas of the nucleus, and promotes transcriptional repression by limiting access of RNA polymerases to DNA (3–10). The replication and epigenetic inheritance of heterochromatin are enigmatic in at least three ways. First, for heterochromatin that is epigenetically inherited, the unit of memory that allows inheritance of the chromatin structure and where it resides is unclear. Second, the processes necessary to propagate that memory and reestablish the chromatin structure every cell cycle are poorly understood. Third, it is unclear how temporary and at least partial disassembly and reassembly of chromatin during DNA replication are coordinated and balanced with maintaining repression of genes in heterochromatin.
To gain further insight into how heterochromatin disassembly and reassembly during DNA replication occur without loss of gene repression, we examined the well-characterized chromatin domains of the transcriptionally silent HML and HMR loci in Saccharomyces cerevisiae, which share structural features of heterochromatin in other eukaryotes. The chromatin at HML and HMR is composed of highly ordered nucleosomes, each bound by the Sir2–Sir3–Sir4 complex, forming a compact heterochromatin structure necessary for transcriptional silencing (11). This structure is inherited, at least in part, through an epigenetic mechanism (12, 13). Establishment of silencing is initiated through the recruitment of the Sir complex to regulatory sites known as “silencers” that flank HML and HMR (14–16). Silencing is ultimately achieved upon Sir complex binding across HML and HMR where it deacetylates key positions on H3 and H4 N-terminal tails through the enzymatic activity of Sir2 (17–19). The resulting compact chromatin structure constrains access of RNA Pol II or Pol III at HML and HMR sufficiently to block transcription (10, 20, 21).
In dividing yeast cells, silencing is transiently lost in approximately one cell per 1,000 cell divisions (22). This result implies that nearly all cells maintain silencing at HML and HMR through S-phase despite the need to replicate the silent chromatin structure in the face of partial nucleosome disassembly and other chromatin changes that occur during DNA replication. Hints at how the maintenance of silencing is balanced with DNA replication come from studies demonstrating that mutations in replication fork proteins and mutations affecting replication-coupled nucleosome assembly result in decreased silencing of HML and HMR. Replication-coupled nucleosome assembly is a multistep process that incorporates histones into newly replicated DNA within a few hundred bases from the replication fork (23, 24). Initially, Asf1 delivers newly synthesized H3–H4 dimers to replication forks where the H3–H4 dimers are transferred to either the CAF-1 complex or Rtt106, both of which deposit histone H3–H4 tetramers onto newly replicated DNA (25–30). Mutants of asf1Δ, rtt106Δ, and cac1Δ result in decreased silencing of HML and HMR, demonstrating the importance of proper chromatin assembly for heterochromatin function (31–39). CAF-1, a heterotrimeric histone chaperone composed of Cac1, Cac2, and Cac3, is coupled to DNA replication through a physical interaction with proliferating cell nuclear antigen (PCNA) (40–42). PCNA travels with replication forks on both the leading and lagging strands where it increases the processivity of replicative DNA polymerases by physically tethering them to DNA. PCNA also serves as an interaction scaffold to coordinate proteins involved in telomere maintenance, chromatin assembly, DNA damage-response signaling, and DNA repair with the replication fork (43). Certain mutations of POL30, which encodes PCNA, including some mutations that disrupt the physical interaction between PCNA and CAF-1, disrupt silencing at HML and HMR and at telomeres (31, 40, 44). These data suggest that coordination of nucleosome assembly machinery with DNA replication forks is important to maintain silencing. PCNA is regulated, in part, by controlling when and where it resides on chromatin through the combined actions of two five-membered protein complexes that load and unload PCNA onto DNA (45). The RFC complex consisting of Rfc1–5 loads PCNA, and the other complex, Elg1–Rfc2–5, unloads PCNA (45–49). The PCNA-unloading activity of Elg1 is important for multiple chromatin-based processes such as DNA repair, telomere-length maintenance, and telomeric silencing (50–53). elg1Δ mutants do not appear to impair Okazaki fragment processing or the completion of DNA replication (48). Thus, the phenotypes caused by elg1Δ are not solely explained by DNA-replication defects.
To drill into the processes that allow duplication of heterochromatin in particular and all chromatin structures in general, we tested the extent to which proteins that act at the replication fork were necessary for the faithful propagation of heterochromatin’s gene-silencing capability. Elg1 stood out as a major determinant of heterochromatin stability. This study revealed the mechanistic basis of Elg1’s contribution to heterochromatin stability, providing insight into the coordination between DNA replication and chromatin assembly by the factors that promote or limit PCNA.
Results
Elg1 Was Necessary for Full Silencing at HML.
We measured silencing at HML using the previously described CRASH (Cre-reported altered states of heterochromatin) assay (22, 54). Briefly, the Cre recombinase gene resides at the transcriptionally silent HML locus (Fig. 1A). In these strains, even transient expression of Cre due to a loss of silencing at HML leads to site-specific recombination between LoxP sites and a permanent switch from RFP to GFP expression. Loss-of-silencing events, which manifest as GFP-expressing sectors in otherwise RFP-expressing yeast colonies, are detected at low levels in wild-type cells (Fig. 1B) (22). elg1Δ mutations caused a substantial increase in silencing loss compared with wild type, suggesting that control of PCNA unloading by Elg1 was important for silencing (Fig. 1B). It was possible to quantify the frequency of silencing loss captured by the CRASH assay with flow cytometry. In CRASH strains, cells that had recently lost silencing contained both RFP and GFP for approximately three cell cycles. We utilized this property for quantification because the frequency of cells in a population that have recently lost silencing is proportional to the rate of silencing loss in that population (Materials and Methods). For each sample, we calculated the ratio of cells in the population that had recently lost silencing to the number of cells in the population that had the potential to switch from RFP to GFP. We referred to this ratio as the “apparent silencing-loss rate.” The apparent silencing-loss rate in elg1Δ mutants was eightfold greater than in wild type (Fig. 1C).
Fig. 1.
The PCNA-unloading activity of Elg1 contributed to silencing HML. (A) Illustration of the CRASH assay to measure silencing of HML. The CRASH assay contains two features: (i) Cre inserted at the HML locus controlled by the α2 promoter and (ii) an RFP-GFP reporter cassette on chromosome V at the URA3 locus. Cells that maintain silencing of HML constitutively express RFP driven by the GPD promoter. Loss of silencing at HML leads to Cre expression and results in Cre-mediated recombination between two LoxP sites, resulting in removal of the RFP and HygMX genes, repositioning the GFP gene so that it becomes constitutively expressed. (B) Representative images of colonies from wild-type (JRY10790) or elg1Δ mutant (JRY10799) strains containing the CRASH assay. (C) The apparent silencing-loss rates of the strains in B were quantified by flow cytometry, as described in Materials and Methods. A P value was calculated using an unpaired, two-sided t test. (D) Representative images of colonies from wild type (JRY10790) and pol30-D150E (JRY10800), elg1Δ (JRY10799), and elg1Δ pol30-D150E (JRY10801) mutant strains containing the CRASH assay. (E) Quantification of apparent silencing-loss rates by flow cytometry of the strains in D. P values were calculated by ANOVA and Tukey’s test post-hoc analysis. In C and E, the red line in the box plots represents the median value calculated from at least 10 cultures. The blue boxes represent the 25th and 75th percentiles. Whiskers represent the range of values within 1.5× the interquartile range. Values extending beyond 1.5× the interquartile range are marked as outliers (red plus sign).
It was formally possible that the increased loss of silencing associated with elg1Δ was due to an Elg1 function beyond its known role in unloading PCNA from DNA. To test whether the silencing defect was due to the retention of PCNA on DNA, we utilized a previously characterized PCNA mutant (pol30-D150E) that disrupts the interaction interface between individual subunits of the PCNA trimer, causing it to spontaneously disassemble from chromatin even in the absence of Elg1 (55). These PCNA trimer-interface mutations rescue multiple other elg1Δ phenotypes (56). On its own, the pol30-D150E single mutant had no effect on silencing as measured by the CRASH assay; however, the pol30-D150E mutant nearly completely rescued the silencing defect of an elg1Δ mutation (Fig. 1 D and E). These results implied that the silencing defects of elg1Δ were specifically due to the increased retention of PCNA on chromatin.
Histone Chaperones Asf1, CAF-1, and Rtt106 Independently Contributed to the Maintenance of Silencing at HML.
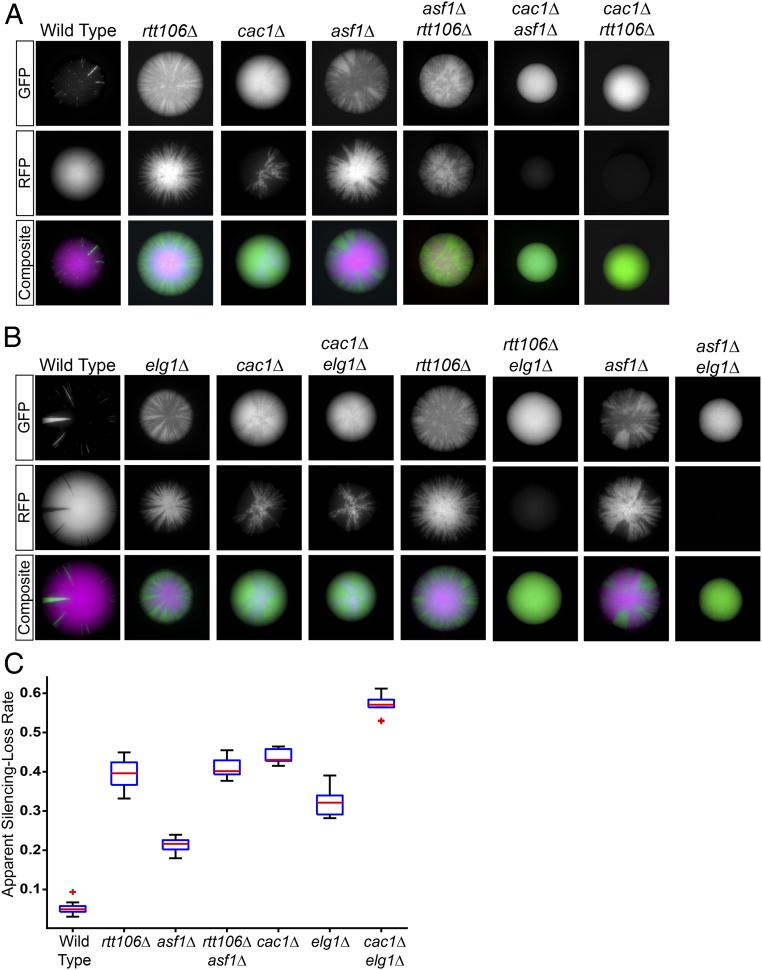
How might increased retention of PCNA on chromatin in an elg1Δ mutant lead to a defect in silencing? Disruption of the interaction between CAF-1 and PCNA causes silencing defects, and PCNA acts as an important scaffold to control the recruitment of histone chaperone activity to the right time and place during S-phase (31, 40, 41). This led us to the hypothesis that removal of PCNA from chromatin following replication might be important for the regulation of histone chaperone function and that failure to remove PCNA might impact nucleosome assembly and also silencing. Conventional genetic assays that measure the steady-state level of silencing at either HML or HMR averaged over a population of cells are capable of detecting silencing defects in cac1Δ mutants but not in asf1Δ and rtt106Δ mutants. Instead, silencing phenotypes of asf1Δ and rtt106Δ mutants are detected only in genetic backgrounds that sensitize cells to silencing defects, such as strains in which multiple histone chaperones are mutated (31, 42, 57, 58). These observations are consistent with two possible interpretations: asf1Δ and rtt106Δ mutants lack silencing phenotypes at HML and HMR because other histone chaperones compensate in those mutants or the lack of phenotype reflects insufficient sensitivity of the assays. To distinguish between these two possibilities, the effect of mutant histone chaperones on silencing at HML was assessed with the CRASH assay, which is tuned to detect transient effects that reveal the dynamics of silenced chromatin (22). Deletion of CAC1, which encodes the largest subunit of the CAF-1 complex, resulted in increased loss of silencing (Fig. 2A). Both the asf1Δ and rtt106Δ mutations also resulted in increased silencing loss but to a lesser degree than did CAF-1 complex mutants (Fig. 2 A and C). Thus, individual histone chaperones independently contributed to silencing in ways that escaped detection by previous silencing assays.
Fig. 2.
Genetic analysis of histone chaperones and Elg1 function in silencing. (A) Representative images of colonies from wild type (JRY10790); the single mutants rtt106Δ (JRY10802), cac1Δ (JRY10803), and asf1Δ (JRY10804); and the double mutants cac1Δ asf1Δ (JRY10805), cac1Δ rtt106Δ (JRY10806), and asf1Δ rtt106Δ (JRY10807), all of which carried the two components of the CRASH assay. Double-mutant strains of cac1Δ rtt106Δ and cac1Δ asf1Δ obtained from crosses of the single mutants uniformly expressed GFP, indicating they were unable to maintain silencing. (B) Representative images of colonies with elg1Δ (JRY10799) in combination with the same histone chaperone single mutants as in A. elg1Δ (JRY10799) was crossed to histone chaperone mutants to obtain the elg1Δ cac1Δ (JRY10808), elg1Δ asf1Δ (JRY10809), and elg1Δ rtt106Δ (JRY10810) double mutants. (C) Quantification of apparent silencing-loss rates by flow cytometry of the single-mutant strains and the elg1Δ cac1Δ and asf1Δ rtt106Δ double mutants from B. The rates of silencing loss of double-mutant strains that uniformly expressed GFP could not be quantified. See the legend of Fig. 1 for a description of box plots.
Double-mutant analysis was performed to determine whether the histone chaperones function to maintain silencing through common or distinct pathways. In the case of cac1Δ asf1Δ and cac1Δ rtt106Δ double mutants, the silencing defects were sufficiently strong that only GFP-expressing strains were recovered. These double mutants confer a further increase in silencing loss compared with each of the single mutants (Fig. 2A). Silencing defects in asf1Δ rtt106Δ double mutants were equivalent to those in rtt106Δ single mutants. These results were consistent with a model in which Asf1 and Rtt106 contribute to silencing through the same pathway. Therefore, all three histone chaperones contributed to the stability of silencing, with the CAF-1 complex functioning in a distinct genetic pathway from Asf1 and Rtt106.
Elg1 and the CAF-1 Complex Function in the Same Pathway to Maintain Silencing.
The histone chaperone activity of the CAF-1 complex depends on its physical interaction with PCNA at replication forks (41, 44). The genome-wide distribution of PCNA bound to chromatin coincides with regions of active replication, which depends on a cycle of PCNA loading at primer–template junctions, including at the initiation of every Okazaki fragment, and rapid unloading from recently replicated regions (47, 59, 60). This pattern is disrupted in elg1Δ mutants in which PCNA is retained broadly across replicated regions of chromatin (47). We reasoned it was possible that proteins that interact with PCNA, such as CAF-1, might also become abnormally distributed across chromatin in the absence of Elg1. If the silencing phenotype of an elg1Δ mutant stems from inadequate CAF-1 function, the phenotype of a cac1Δ mutation on silencing would be qualitatively similar to that of elg1Δ (i.e., both mutations would cause silencing defects), and a cac1Δ elg1Δ double mutant would appear no more defective in silencing than a cac1Δ single mutant. In contrast, if Elg1 and CAF-1 function in silencing through separate pathways, the silencing phenotype of a cac1Δ elg1Δ double mutant would be predicted to be substantially stronger than either of the single mutants. There was only a slight (1.3-fold) increase in silencing loss in the cac1Δ elg1Δ double mutant compared with a cac1Δ single mutant (Fig. 2 B and C). In contrast, all asf1Δ elg1Δ and rtt106Δ elg1Δ double-mutant spores (derived from a cross of the single mutants) lost silencing within the first few cell divisions. All double mutants, but not the single mutants or wild-type segregants, formed uniformly green colonies. Therefore, defects in silencing caused by asf1Δ and rtt106Δ were at least additive with the silencing defect of elg1Δ. We conclude that CAC1 and ELG1 function together to maintain silencing through a contribution to chromatin assembly that parallels but is distinct from that of ASF1 and RTT106. The slight increase in a cac1Δ elg1Δ double-mutant phenotype over that of the cac1∆ single mutant suggests that Elg1 also contributes to the maintenance of silencing through another process yet to be identified.
CAF-1 and PCNA Retention on Chromatin Is Increased in the Absence of Elg1.
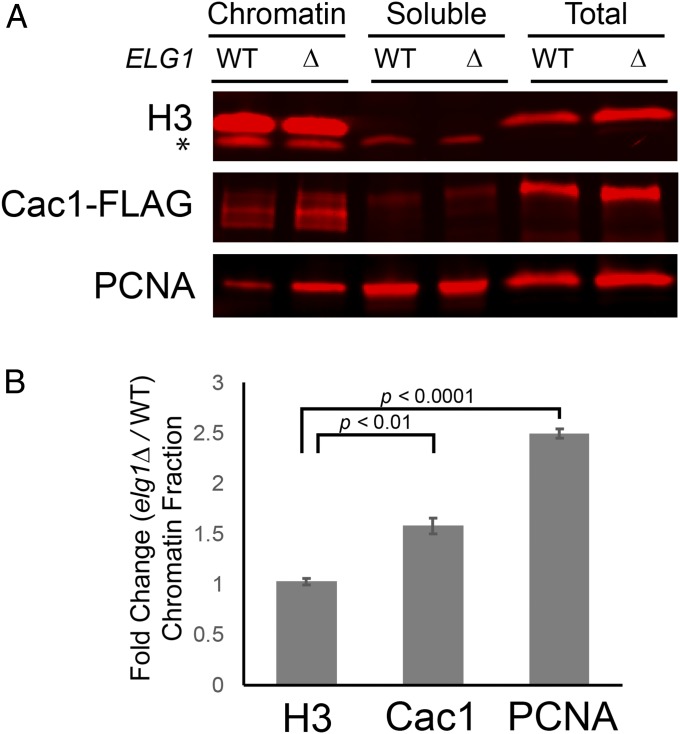
In the absence of Elg1, the amount of chromatin-bound PCNA increases, and PCNA remains on nascent DNA beyond S-phase (46, 56). Because CAF-1 physically interacts with PCNA, we tested whether the absence of Elg1 has a similar effect on CAF-1 association with chromatin. To measure binding of CAF-1 to chromatin, lysates were prepared from cycling cells and separated into chromatin and soluble fractions by centrifugation through a sucrose cushion. As expected, histone H3 was detected by immunoblot almost exclusively in the chromatin fraction (Fig. 3A). Similar to previous observations, the amount of PCNA bound to chromatin increased by more than 50% in elg1Δ mutant cells compared with wild-type cells (Fig. 3). Importantly, the proportion of Cac1 retained in the chromatin fraction also increased significantly in elg1Δ mutant cells compared with wild-type cells (Fig. 3). No changes in the proportion of histone H3 bound to chromatin were observed in elg1Δ mutants relative to wild-type cells, which suggested that the enrichment of CAF-1 (and PCNA) in the chromatin fraction caused by elg1Δ mutants was not attributed to a generalized, nonspecific increase of all chromatin-binding proteins in those samples. Thus, in the absence of PCNA unloading, CAF-1 complex was retained on chromatin in excess of that in wild type.
Fig. 3.
CAF-1 and PCNA association with chromatin increased in an elg1Δ mutant. (A) Immunoblot analysis of histone H3, Cac1-FLAG, and PCNA levels in different subcellular fractions. Lysates prepared from wild-type (JRY10790) and elg1Δ (JRY10799) cells were separated into chromatin-associated material and soluble material. The asterisk indicates nonspecific bands. (B) Quantification of the immunoblot signals of the chromatin fraction from A. The average fold-change values were calculated by dividing chromatin-associated levels measured in elg1Δ mutants by the levels measured in wild-type cells (n = 3 biological replicates). Error bars represent SEM. P values were calculated using unpaired, two-sided t tests.
Overexpression of the CAF-1 Complex Rescues the elg1Δ Silencing Defect.
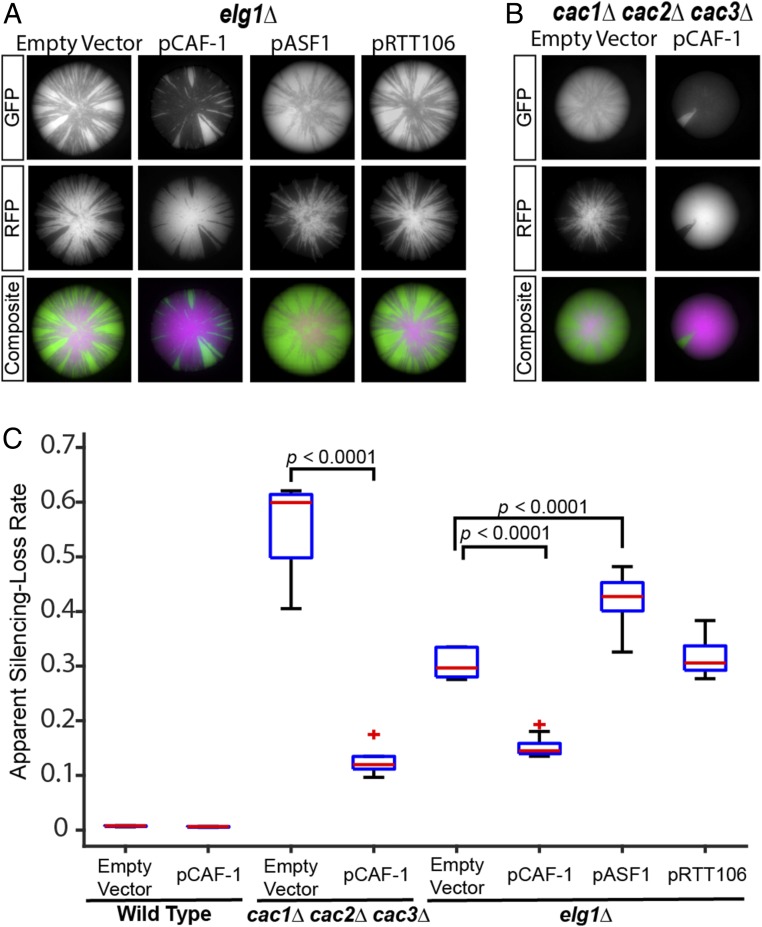
In elg1Δ mutants, co-retention of CAF-1 with PCNA on DNA might cause a significant fraction of the CAF-1 pool to become sequestered from active replication forks where it functions. There are an estimated ∼6,800 homotrimeric PCNA molecules and ∼200–500 molecules of the CAF-1 complex in the nucleus (61). Because of a 13- to 30-fold excess of PCNA compared with CAF-1, CAF-1 may become limiting, leading to the silencing defects observed in elg1Δ, but enough free PCNA may be available at replication forks to support cell division. A simple prediction from this model is that overexpression of the CAF-1 complex should rescue silencing defects caused by elg1Δ mutants. To test this, a high-copy (2-μm) plasmid (pCAF-1) designed to overexpress all three proteins of the CAF-1 complex (Cac1–Cac2–Cac3) in a 1:1:1 stoichiometry was introduced into wild-type cells and an elg1Δ mutant. No phenotype was observed in wild-type cells containing pCAF-1; excess CAF-1 complex did not interfere with normal silencing at HML (Figs. 4C and 5A). As expected, pCAF-1 rescued the silencing defects of the cac1Δ cac2Δ cac3Δ triple mutant as well as cac1Δ, cac2Δ, and cac3Δ single mutants (Figs. 4 B and C and 5B), which demonstrated that pCAF-1 expressed functional CAF-1 complex. Importantly, pCAF-1 rescued the silencing defect of elg1Δ to nearly the same degree as it did the cac1Δ cac2Δ cac3Δ triple mutant (Fig. 4 A and C). In contrast, overexpression of ASF1 or RTT106 was unable to restore silencing in elg1Δ mutants (Fig. 4 A and C), suggesting that the elg1Δ silencing defect was caused specifically by limited CAF-1 and not by limited histone chaperone activity per se. Overexpression of ASF1 further weakened silencing in the elg1Δ mutant (Fig. 4 A and C) and in wild-type cells (Fig. 5 A and F), consistent with previously reported effects of ASF1 overexpression disrupting silencing at telomeres, HML, and HMR (37, 62).
Fig. 4.
Overexpression of the CAF-1 complex rescued elg1Δ silencing defects. The empty-vector control pRS425 and a 2-μm high-copy plasmid containing all three genes of the CAF-1 complex (pCAF-1, alias pJR3418), ASF1 (pASF1, alias pJR3425), or RTT106 (pRTT106, alias pJR3419) were transformed into CRASH assay strains. The cells were grown on a medium that maintained plasmid selection. (A and B) Representative images of colonies containing the indicated plasmid in the elg1Δ (JRY10799) (A) and cac1Δ cac2Δ cac3Δ (JRY10811) (B) strains. (C) Plots of the apparent silencing-loss rates of wild type (JRY10790) transformed with pRS425 or pCAF-1 (pJR3418) and the strains in A and B quantified by flow cytometry. P values were calculated by ANOVA and Tukey’s test post-hoc analysis. See the legend of Fig. 1 for a description of box plots.
Fig. 5.
Impact of histone chaperone overexpression on silencing. (A) Representative images of wild-type (JRY10790) colonies transformed with the empty-vector control pRS425 or a 2-μm high-copy plasmid containing all three genes of the CAF-1 complex (pCAF-1, alias pJR3418), or ASF1 (pASF1, alias pJR3425), or RTT106 (pRTT106, alias pJR3426). (B) Images of cac1Δ (JRY10803) colonies transformed with the same plasmids as in A. (C) Images of cac2Δ (JRY10815) and cac3Δ (JRY10816) colonies transformed with pRS425 or pCAF-1 (pJR3418). (D) Images of asf1Δ (JRY10804) colonies transformed with the same plasmids as in A. (E) Images of rtt106Δ (JRY10802) colonies transformed with the same plasmids as in A. (F) Plots of the apparent silencing-loss rates of the strains with the indicated mutations transformed with the plasmids described in A and quantified by flow cytometry. See the legend of Fig. 1 for a description of box plots.
Quantitative and Qualitative Contributions of Histone Chaperones to Gene Silencing.
The three histone chaperones tested in this study each contributed to the stability of silencing, with the CAF-1 complex functioning in a distinct genetic pathway from Asf1 and Rtt106. It is unknown how each histone chaperone contributes to maintaining silencing. If the silencing defects in individual histone chaperone mutants were caused by a general decrease in the overall capacity to deposit H3–H4 histones onto DNA, restoration of that biochemical activity, rather than the specific histone chaperone, would be expected to restore silencing. Alternatively, if deletion of a histone chaperone resulted in loss of a unique activity required for silencing specific to that protein, overexpression of other histone chaperones should not restore silencing. To distinguish between these models, silencing was measured in wild-type, cac1Δ, asf1Δ, and rtt106Δ strains transformed with high-copy (2-μm) histone chaperone expression plasmids. Overexpression of CAF-1 and RTT106 had no effect on silencing in wild-type cells, while overexpression of ASF1 destabilized silencing, as previously reported (Fig. 5 A and F) (37, 62). Overexpression of CAF-1, but not RTT106 or ASF1, restored silencing in a cac1Δ mutant (Figs. 4B and 5 B and F). Similarly, silencing was restored in asf1Δ mutants by overexpression of ASF1 but not CAF-1 or RTT106 (Fig. 5 D and F). In rtt106Δ mutants, silencing was restored by overexpression of RTT106 as well as CAF-1 and ASF1 (Fig. 5 E and F). Collectively, these data were consistent with a model where Asf1 and CAF-1 contribute unique activities to silencing that cannot be supplanted by an excess of other histone chaperones, while the absence of Rtt106 could be compensated for by an overabundance of either Asf1 or CAF-1.
Silencing at HML Required S-Phase–Specific Expression of Elg1.
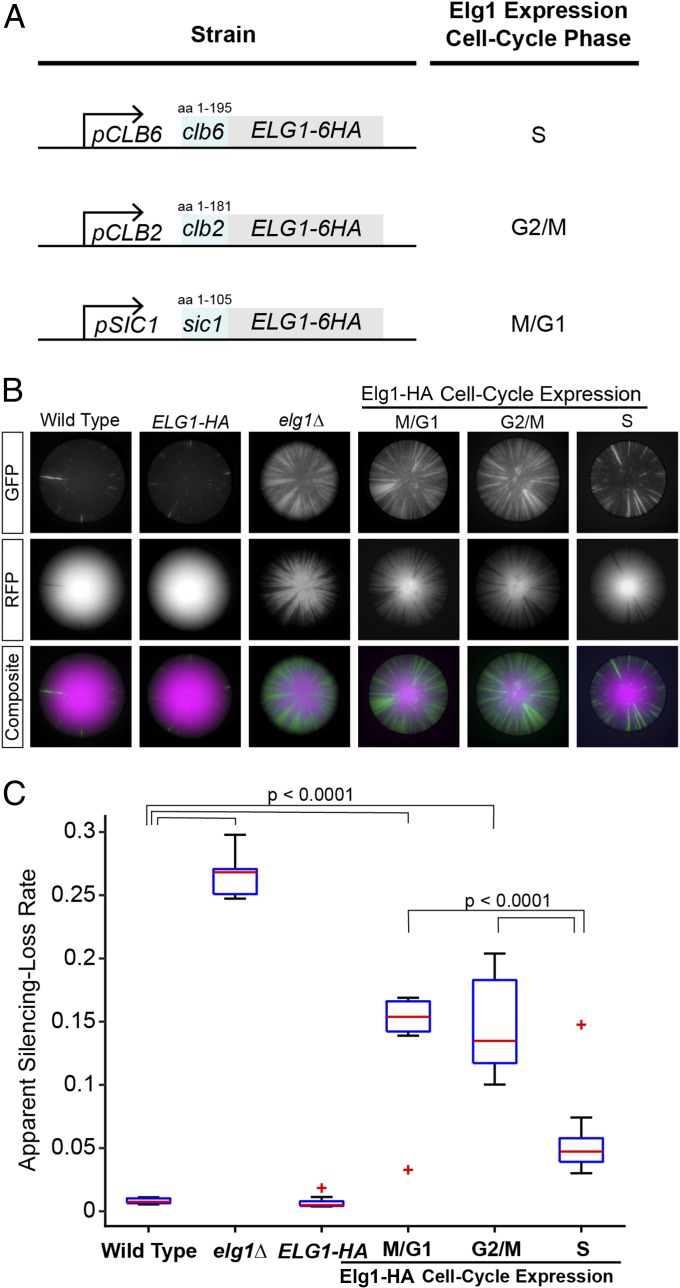
To determine if there was a specific cell-cycle stage in which Elg1-dependent unloading of PCNA was critical for silencing, the CRASH assay was introduced into strains that precisely control the expression and stability of Elg1 at specific cell-cycle stages (Fig. 6A) (56). Expression of Elg1 during S-phase results in PCNA unloading activity while DNA replication takes place. In strains expressing Elg1 during G2/M- or M/G1-phases, PCNA is retained on DNA from the last round of DNA replication until cells reach the cell-cycle stage when Elg1 is expressed. Silencing at HML was maintained to a much higher degree in cells when Elg1 was expressed in S-phase compared with cells in which Elg1 was expressed in M/G1-phase or G2/M-phase (Fig. 6B). Although PCNA is unloaded when Elg1 is expressed during M/G1 or G2/M (56), this activity was insufficient to rescue the elg1Δ silencing defect. These results demonstrated that the PCNA unloading activity of Elg1 was necessary specifically during S-phase to ensure maximum maintenance of silencing at HML.
Fig. 6.
Cell-cycle–specific expression of Elg1 controlled the stability of gene silencing. (A) Diagram illustrating replacement of the native ELG1 promoter sequence and fusion of N-terminal degron sequences for cell-cycle–specific expression of Elg1. A Clb6-degron-Elg1 fusion protein expressed from a CLB6 promoter is limited to S-phase (JRY10814), a Clb2-degron-Elg1 fusion protein expressed from a CLB2 promoter is limited to G2/M-phase (JRY10813), and a Sic1-degron-Elg1 fusion expressed from a SIC1 promoter is limited to M/G1-phase (JRY10812) (56). (B) Representative colony images from wild type (JRY10790), an ELG1-6HA–tagged strain (JRY10827), elg1Δ (JRY10799), or strains with cell-cycle–specific Elg1 expression as described in A. The cell-cycle expression stage of Elg1 is indicated. (C) The apparent silencing-loss rates of the strains in B quantified by flow cytometry. P values were calculated by ANOVA and Tukey’s test post-hoc analysis. See the legend of Fig. 1 for a description of box plots.
Discussion
This study revealed a previously uncharacterized role of Elg1 in S-phase for maintaining silenced chromatin and supported a model in which the PCNA-unloading activity of Elg1 coordinates CAF-1–dependent nucleosome assembly with replication forks. PCNA serves as the scaffold that coordinates CAF-1 nucleosome assembly activity with DNA replication forks, and loss of the interaction between PCNA and CAF-1 disrupts CAF-1 nucleosome assembly function (31, 41, 44). Our data built on this model and demonstrated that the function of CAF-1 also depends on maintaining PCNA’s ability to cycle on and off DNA during replication. Several key observations supported such a model: First, double-mutant analysis demonstrated that silencing defects caused by the loss of either elg1Δ or cac1Δ resulted from defects in a common process (CAF-1–dependent nucleosome assembly). Second, the silencing phenotype of an elg1Δ mutation could be suppressed in two ways: (i) through alleles that allow Elg1-independent unloading of PCNA by destabilizing the PCNA trimer ring (pol30-D150E) and (ii) through overexpression of the CAF-1 complex. These results genetically pinpointed persistence of PCNA on DNA as the cause for the silencing defect and showed that this defect could be compensated for by more CAF-1 complex.
Molecular and cell-cycle experiments provided mechanistic support for this model. In the absence of Elg1, PCNA accumulates nonspecifically on recently replicated chromatin in the wake of replication forks (47, 53). We observed an increase in the fraction of PCNA bound to chromatin in elg1Δ mutants similar to the previously reported values. The fraction of CAF-1 associated with chromatin also increased significantly in the absence of Elg1. Controlled expression of Elg1 limited to S-phase was sufficient to maintain normal silencing, but Elg1 expression limited to the G2/M- or M/G1-phase resulted in defects in silencing that were similar to those of an elg1Δ mutant. A previous study that first reported the Elg1 promoter/degron cell cycle-expression tools used here demonstrated that PCNA is efficiently unloaded from chromatin specifically during the intervals of Elg1 expression but is retained on chromatin to excess when Elg1 is absent (56). Therefore, because silencing was maintained when Elg1 expression was limited to S-phase, PCNA unloading during S-phase was critical for silencing, presumably to ensure proper CAF-1 function, which acts specifically during replication-coupled chromatin assembly.
Why does PCNA need to cycle on and off DNA in S-phase? A combination of PCNA loading by Rfc1 and PCNA unloading by Elg1 ensures that PCNA is rapidly cycled on and off chromatin during replication. PCNA is essential for viability because it is required to complete DNA replication, as is the activity of loading PCNA onto chromatin by Rfc1–5. In contrast, cells lacking Elg1 complete DNA replication with only a slight delay (10–15 min) compared with wild type (48, 63). Thus, Elg1 is not critical for the completion of DNA replication, per se. Instead, Elg1 may be critical for the cycling of PCNA on chromatin to provide interaction sites for proteins that function specifically at replication forks and newly replicated DNA (41, 64). Unloading of PCNA in a timely manner by Elg1 may ensure that PCNA is present only at active sites of replication or recently replicated DNA where it coordinates the activity of proteins involved in DNA metabolism. In the absence of Elg1, widespread accumulation of PCNA on chromatin would preclude PCNA’s ability to serve as a marker that is specific for replicating DNA, resulting in miscoordination of PCNA-interacting proteins due to their recruitment to chromatin at the wrong time and place behind the replication fork.
We demonstrated that maintenance of the PCNA cycle by Elg1 is important for CAF-1 function. In the absence of Elg1, a larger fraction of CAF-1 became bound to chromatin. Because CAF-1 depends on interaction with PCNA for its recruitment to chromatin, it is possible that in elg1 mutants a significant fraction of CAF-1 becomes bound to PCNA that is not associated with active replication. There are estimated to be 200–500 CAF-1 molecules per cell, and PCNA is estimated to be 13- to 30-fold more abundant than CAF-1 (61). However, considering there are ∼350–400 annotated origins of replication in budding yeast, if only a fraction of these fire during S-phase, a substantial portion of the CAF-1 pool (and potentially other PCNA-interacting proteins) may become sequestered with PCNA to chromatin, to the point that the free pool of the CAF-1 complex becomes limiting at active replication forks, where it normally functions. Components of the CAF-1 complex are nonessential, and in this model other histone chaperones would be adequate to support bulk chromatin assembly, but in the absence of CAF-1, the quality of that chromatin would be imperfect for heterochromatin formation and insufficient to maximally maintain silencing at HML and HMR. Consistent with this model, the silencing defects in an elg1 mutant were suppressed by overexpression of the CAF-1 complex. Overall, our model suggests that one primary function of Elg1 is to maintain PCNA in a dynamic state during DNA replication so that PCNA and PCNA-bound proteins are able to cycle from nascent replicated DNA back to active sites of DNA replication.
Measurement of transient silencing loss revealed individual contributions of histone chaperones to silencing. The three histone chaperones that function during replication-coupled chromatin assembly, Asf1, Rtt106, and CAF-1, have known roles in silencing of HML and HMR. However, the individual contributions of each factor to silencing have been difficult to study, in part because of an overlap in functions among the histone chaperones. Previous genetic studies of the contribution of histone chaperones toward silencing have mostly relied on transgene reporters that reflect the steady-state expression of either HML or HMR from a population of cells. Those reporters generally lacked sufficient sensitivity to detect the effects of asf1Δ and rtt106Δ single mutants. The ability to measure transient losses of silencing with the CRASH assay uncovered the contribution of individual histone chaperones to silencing and enabled double-mutant analysis that revealed which operated together and which operated at different steps. The inability to rescue the silencing phenotypes associated with loss of Asf1 and CAF-1 by overexpression of other histone chaperones established that each played a unique role in nucleosome assembly in heterochromatin that was not bypassed by increasing the abundance of other histone chaperones. Asf1 is required for H3K56 acetylation by Rtt109, a mark that stimulates the histone deposition activity of both CAF-1 and Rtt106 (27, 65). Both CAF-1 and Rtt106 deposit histone H3–H4 onto DNA. The silencing defect in an rtt106Δ mutant was rescued by overexpression of either ASF1 or CAF-1; however, the silencing defect in a cac1Δ mutant could not be rescued by RTT106 overexpression. Thus, CAF-1 fulfills a specialized role during nucleosome assembly, while the function of Rtt106 could be met with the same biochemical activity of a different chaperone (in this case, CAF-1). An intriguing possibility is that CAF-1 has a specialized function in the assembly of nucleosomes on the lagging strand, where PCNA is more abundant.
DNA replication poses a unique logistical challenge for the cell in that the structural features of chromatin and their regulatory functions must be carefully coordinated with the passage of replication machinery so faithful duplication of both the genome and its chromatin structures may be achieved. Nucleosome assembly is fundamental to the reestablishment of chromatin in the wake of DNA replication, and here we demonstrated that control of PCNA by Elg1 was necessary to coordinate nucleosome assembly to maintain transcriptional silencing through S-phase.
Materials and Methods
Strains and Plasmids.
All strains used in this study were derived from W303 (Table 1). CRASH assay strains, which use the switch from RFP to GFP expression upon derepression of HML::cre, were generated as described previously (22). Gene deletions were generated by integration of PCR-amplified disruption cassettes and confirmed by PCR using primers to amplify across the junctions at the site of integration (66, 67). The pol30-D150E point mutation was generated using Cas9 technology with the guide RNA sequence 5′-aaacAATTCTCTAAAATTGTTCGTaa-3′, as described previously (68). The repair template was generated by annealing the oligo 5′GTACGACTCCACCCTGTCATTGCCATCTTCTGAATTCTCTAAAATTGTTCGTG-3′ to oligo 5′-GATATTAATAGAATCACTCAATTGGGACAATTCACGAACAATTTTAGAGAATT-3′ and subsequently extending the 3′ ends using Phusion Polymerase (New England Biolabs).
Table 1.
Strains used in this study
| JRY no. | Genotype |
| 10790 | ADE2 lys2 TRP1 hmlα2∆::CRE ura3∆::pGPD:loxP:yEmRFP;tCYC1:hygMX:loxP:yEGFP:tADH1 |
| 10799 | ADE2 lys2 TRP1 hmlα2∆::CRE ura3∆::pGPD:loxP:yEmRFP:tCYC1:hygMX:loxP:yEGFP:tADH1 elg1∆::HISMX |
| 10800 | ADE2 lys2 TRP1 hmlα2∆::CRE ura3∆::pGPD:loxP:yEmRFP;tCYC1:hygMX:loxP:yEGFP:tADH1 pol30-D150E |
| 10801 | ADE2 lys2 TRP1 hmlα2∆::CRE ura3∆::pGPD:loxP:yEmRFP:tCYC1:hygMX:loxP:yEGFP:tADH1 elg1∆::HISMX pol30-D150E |
| 10802 | ADE2 lys2 TRP1 hmlα2∆::CRE ura3∆::pGPD:loxP:yEmRFP;tCYC1:hygMX:loxP:yEGFP:tADH1 rtt106Δ::NATMX |
| 10803 | ADE2 lys2 TRP1 hmlα2∆::CRE ura3∆::pGPD:loxP:yEmRFP:tCYC1:hygMX:loxP:yEGFP:tADH1 cac1∆::URA3K. lactis |
| 10804 | ADE2 lys2 TRP1 hmlα2∆::CRE ura3∆::pGPD:loxP:yEmRFP:tCYC1:hygMX:loxP:yEGFP:tADH1 asf1∆::URA3K. lactis |
| 10805 | ADE2 lys2 TRP1 hmlα2∆::CRE ura3∆::pGPD:yEGFP:tADH1 cac1∆::URA3K. lactis asf1∆::HISMX |
| 10806 | ADE2 lys2 TRP1 hmlα2∆::CRE ura3∆::pGPD:yEGFP:tADH1 cac1∆::URA3K. lactis rtt106∆::HISMX |
| 10807 | ADE2 lys2 TRP1 hmlα2∆::CRE ura3∆::pGPD:yEGFP:tADH1 asf1∆::HISMX rtt106∆::HISMX |
| 10808 | ADE2 lys2 TRP1 hmlα2∆::CRE ura3∆::pGPD:loxP:yEmRFP:tCYC1:hygMX:loxP:yEGFP:tADH1 cac1∆::ura3K. lactis elg1∆::HISMX |
| 10809 | ADE2 lys2 TRP1 hmlα2∆::CRE ura3∆::pGPD:yEGFP:tADH1 asf1∆::ura3K. lactis elg1∆::HISMX |
| 10810 | ADE2 lys2 TRP1 hmlα2∆::CRE ura3∆::pGPD:loxP:yEGFP:tADH1 rtt106Δ::NATMX elg1Δ::HISMX |
| 10811 | ADE2 lys2 TRP1 hmlα2∆::CRE ura3∆::pGPD:loxP:yEmRFP;tCYC1:hygMX:loxP:yEGFP:tADH1 cac1∆::hisMX cac2∆::natMX cac3Δ::hisMX |
| 10812 | bar1Δ::hisG ADE2 can1-100 his3-11,15 leu2-3,112 trp1-1 natNT2-M/G1-pSIC1:sic11-315-ELG1-6HA::hphNT1 hmlα2∆::CRE ura3∆::pGPD:loxP:yEmRFP;tCYC1:hygMX:loxP:yEGFP:tADH1 |
| 10813 | bar1Δ::hisG ADE2 can1-100 his3-11,15 leu2-3,112 trp1-1 natNT2-G2/M-pCLB2:clb21-543-ELG1-6HA::hphNT1 hmlα2∆::CRE ura3∆::pGPD:loxP:yEmRFP;tCYC1:hygMX:loxP:yEGFP:tADH1 |
| 10814 | bar1Δ::hisG ADE2 can1-100 his3-11,15 leu2-3,112 trp1-1 natNT2-S-pCLB6:clb61-585-ELG1-6HA::hphNT1 hmlα2∆::CRE ura3∆::pGPD:loxP:yEmRFP;tCYC1:hygMX:loxP:yEGFP:tADH1 |
| 10815 | ADE2 lys2 TRP1 hmlα2∆::CRE ura3∆::pGPD:loxP:yEmRFP;tCYC1:hygMX:loxP:yEGFP:tADH1 cac2Δ::natMX |
| 10816 | ADE2 lys2 TRP1 hmlα2∆::CRE ura3∆::pGPD:loxP:yEmRFP;tCYC1:hygMX:loxP:yEGFP:tADH1 cac3Δ::hisMX |
| 10827 | ADE2, lys2, TRP1, hmlα2∆::CRE, ura3∆::pGPD:loxP:yEmRFP;tCYC1:kanMX:loxP:yEGFP:tADH1 ELG1-6HA::hphNT1 |
Unless noted otherwise, all strains were derived from W303 with the following genotype: can1-100 his3-11 leu2-3, 11 lys2 TRP1 ADE2 ura3-1 MATa.
To generate plasmid pJR3418, the ORFs and 5′ and 3′ UTRs of CAC1, CAC2, and CAC3 were PCR amplified from genomic DNA using Gibson Assembly-compatible primers. The PCR fragments were assembled with plasmid pRS425 (69) linearized by SmaI digestion (New England Biolabs) using Gibson Assembly (New England Biolabs). To generate ASF1 and RTT106 overexpression plasmids, the ORF and 5′ and 3′ UTR of RTT106 and ASF1 were PCR amplified from genomic DNA using Gibson Assembly-compatible primers. The PCR fragments were assembled with pRS425 (69) linearized by SmaI digestion using Gibson Assembly to generate pJR3419 (RTT106) and pJR3425 (ASF1).
Colony Growth and Imaging.
Colonies were plated onto 1.5% agar plates containing yeast nitrogen base without amino acids (Difco-Becton-Dickinson), 2% dextrose, and supplemented with complete supplement mixture (CSM)-Trp or CSM-Trp-Leu (Sunrise Science Products), as indicated. Colonies were incubated for 5–7 d at 30 °C. Colonies were imaged as described previously (70).
Quantification of Silencing Loss by Flow Cytometry.
For each CRASH strain, 10 single colonies were inoculated separately into 2 mL of liquid yeast extract-peptone-2% dextrose or CSM-Trp-Leu medium in 96-well deep-well plates (VWR) and were grown overnight to saturation at 30 °C on a microplate orbital shaker (VWR). Overnight cultures were diluted in 1 mL of fresh medium at a density of 105 cells/mL in 96-well deep-well plates and were grown at 30 °C on a microplate orbital shaker until midlog phase. For each culture, a minimum of 50,000 events were collected using an Attune NxT Flow Cytometer (Life Technologies). Scatterplots of forward-scatter (height) and forward-scatter (width) measurements were generated, and gating was established to include only unbudded and budded cells and to exclude debris and clumped cells for further analysis. Gating was used to measure separately the number of GFP+ cells and the number of RFP+ cells. Finally, a Boolean logic gate, RFP+ AND GFP+, was used to determine the number of cells that were both GFP- and RFP-fluorescent. Such cells were inferred to have very recently undergone the Cre-mediated recombination event leading to GFP expression but retained RFP expressed in the recent past. The number of cells in a population that had very recently lost silencing (cells that were both GFP- and RFP-fluorescent) was divided by the number of cells in the population that had the potential to lose silencing (cells that were only RFP-fluorescent plus cells that were both GFP-and RFP-fluorescent) to obtain an apparent rate of silencing loss. Perdurance of RFP molecules in cells for two or three generations after switching from expression of RFP to GFP precluded direct calculation of true rates of silencing loss using this method. However, because the apparent rates of silencing loss directly reflected true silencing-loss rates, these values could be used to quantitatively compare silencing-loss rates across different genetic backgrounds.
Chromatin Enrichment from Whole-Cell Extracts.
An equivalent of 10 OD600 units (1 OD = ∼107 cells/mL) of cells were resuspended in 100 mM Pipes buffer (pH = 9.4) with 10 mM DTT and were incubated for 10 min at room temperature. The cells were pelleted by centrifugation and resuspended in 1 mL of spheroplast buffer containing 0.6 M sorbitol, 20 mM potassium phosphate (pH = 7.4), and 5 mM DTT. To generate spheroplasts, Zymolyase 100T (MP Biomedicals) was added at a final concentration of 80 μg/mL, and the cells were incubated at 37 °C for 10 min. The cells were repeatedly pelleted and were washed once with 1 mL spheroplast buffer and twice with 1 mL of a wash solution of 50 mM Hepes, 100 mM potassium chloride, 2.5 mM magnesium chloride, and 0.4 M sorbitol. The spheroplasts were resuspended in 200 μL of lysis buffer containing 50 mM Hepes (pH = 7.5), 100 mM potassium chloride, 2.5 mM magnesium chloride, and complete protease inhibitor cocktail (Roche). To lyse spheroplasts, Triton X-100 was added to a final concentration of 0.25% and incubated for 5 min on ice with vortexing every minute. Fifty microliters of the whole-cell extract was gently pipetted on top of a 33% sucrose-lysate buffer solution, and the chromatin fraction was separated by centrifugation at 13,525 × g. The supernatant and chromatin pellet were collected separately, and the chromatin was washed with lysis buffer. The fractions were mixed with Laemmli buffer and subjected to separation by SDS/PAGE.
Immunoblotting.
Samples were run on 4–20% gradient SDS/PAGE gels (Bio-Rad) and were transferred to nitrocellulose membranes (EMD-Millipore). The membranes were blocked in LI-COR Odyssey Blocking Buffer (LI-COR Biosciences), and the following antibodies were used for immunodetection: anti-histone H3 (Ab1791; Abcam), anti-PCNA (Ab221196; Abcam), and anti-FLAG (F3165; Sigma). Membranes were incubated with infrared dye-conjugated antibodies IRDye800CW goat anti-mouse antibody and IRDye680RD goat anti-rabbit antibody (LI-COR Biosciences). Membranes were imaged using a LI-COR Odyssey scanner in the 700 nm and 800 nm channels.
Acknowledgments
We thank Takashi Kubota and Richard Kolodner for providing yeast strains and Paul Kaufman for sharing plasmids. This work was supported by NIH F32 Fellowship GM115074 (R.J.), grants from the Berkeley-TAU fund (to M.K. and J.R.) and from the Israel Science Foundation and the Volkswagen Foundation (to M.K.), and NIH Grants R01GM31105 and R01GM120374 (to J.R.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Nishibuchi G, Déjardin J. The molecular basis of the organization of repetitive DNA-containing constitutive heterochromatin in mammals. Chromosome Res. 2017;25:77–87. doi: 10.1007/s10577-016-9547-3. [DOI] [PubMed] [Google Scholar]
- 2.Trojer P, Reinberg D. Facultative heterochromatin: Is there a distinctive molecular signature? Mol Cell. 2007;28:1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Wallrath LL, Elgin SC. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 1995;9:1263–1277. doi: 10.1101/gad.9.10.1263. [DOI] [PubMed] [Google Scholar]
- 4.Loo S, Rine J. Silencers and domains of generalized repression. Science. 1994;264:1768–1771. doi: 10.1126/science.8209257. [DOI] [PubMed] [Google Scholar]
- 5.Singh J, Klar AJ. Active genes in budding yeast display enhanced in vivo accessibility to foreign DNA methylases: A novel in vivo probe for chromatin structure of yeast. Genes Dev. 1992;6:186–196. doi: 10.1101/gad.6.2.186. [DOI] [PubMed] [Google Scholar]
- 6.Gottschling DE. Telomere-proximal DNA in Saccharomyces cerevisiae is refractory to methyltransferase activity in vivo. Proc Natl Acad Sci USA. 1992;89:4062–4065. doi: 10.1073/pnas.89.9.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabianca DS, Gasser SM. Spatial segregation of heterochromatin: Uncovering functionality in a multicellular organism. Nucleus. 2016;7:301–307. doi: 10.1080/19491034.2016.1187354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strom AR, et al. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241–245. doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larson AG, et al. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature. 2017;547:236–240. doi: 10.1038/nature22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steakley DL, Rine J. On the mechanism of gene silencing in Saccharomyces cerevisiae. G3 (Bethesda) 2015;5:1751–1763. doi: 10.1534/g3.115.018515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thurtle DM, Rine J. The molecular topography of silenced chromatin in Saccharomyces cerevisiae. Genes Dev. 2014;28:245–258. doi: 10.1101/gad.230532.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pillus L, Rine J. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell. 1989;59:637–647. doi: 10.1016/0092-8674(89)90009-3. [DOI] [PubMed] [Google Scholar]
- 13.Mahoney DJ, Marquardt R, Shei GJ, Rose AB, Broach JR. Mutations in the HML E silencer of Saccharomyces cerevisiae yield metastable inheritance of transcriptional repression. Genes Dev. 1991;5:605–615. doi: 10.1101/gad.5.4.605. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Z, Hayashi MK, Merkel O, Stillman B, Xu R. Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. EMBO J. 2002;21:4600–4611. doi: 10.1093/emboj/cdf468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rusché LN, Kirchmaier AL, Rine J. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2207–2222. doi: 10.1091/mbc.E02-03-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoppe GJ, et al. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol Cell Biol. 2002;22:4167–4180. doi: 10.1128/MCB.22.12.4167-4180.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 18.Smith JS, et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landry J, et al. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schnell R, Rine J. A position effect on the expression of a tRNA gene mediated by the SIR genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:494–501. doi: 10.1128/mcb.6.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Bryant G, Zhao A, Ptashne M. Nucleosome avidities and transcriptional silencing in yeast. Curr Biol. 2015;25:1215–1220. doi: 10.1016/j.cub.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 22.Dodson AE, Rine J. Heritable capture of heterochromatin dynamics in Saccharomyces cerevisiae. eLife. 2015;4:e05007. doi: 10.7554/eLife.05007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasser R, Koller T, Sogo JM. The stability of nucleosomes at the replication fork. J Mol Biol. 1996;258:224–239. doi: 10.1006/jmbi.1996.0245. [DOI] [PubMed] [Google Scholar]
- 24.Dahlin JL, Chen X, Walters MA, Zhang Z. Histone-modifying enzymes, histone modifications and histone chaperones in nucleosome assembly: Lessons learned from Rtt109 histone acetyltransferases. Crit Rev Biochem Mol Biol. 2015;50:31–53. doi: 10.3109/10409238.2014.978975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgess RJ, Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nat Struct Mol Biol. 2013;20:14–22. doi: 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han J, et al. A Cul4 E3 ubiquitin ligase regulates histone hand-off during nucleosome assembly. Cell. 2013;155:817–829. doi: 10.1016/j.cell.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Q, et al. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mattiroli F, et al. DNA-mediated association of two histone-bound complexes of yeast chromatin assembly factor-1 (CAF-1) drives tetrasome assembly in the wake of DNA replication. eLife. 2017;6:e22799. doi: 10.7554/eLife.22799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sauer PV, et al. Insights into the molecular architecture and histone H3-H4 deposition mechanism of yeast chromatin assembly factor 1. eLife. 2017;6:e23474. doi: 10.7554/eLife.23474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fazly A, et al. Histone chaperone Rtt106 promotes nucleosome formation using (H3-H4)2 tetramers. J Biol Chem. 2012;287:10753–10760. doi: 10.1074/jbc.M112.347450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang K, et al. A DNA binding winged helix domain in CAF-1 functions with PCNA to stabilize CAF-1 at replication forks. Nucleic Acids Res. 2016;44:5083–5094. doi: 10.1093/nar/gkw106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zunder RM, Antczak AJ, Berger JM, Rine J. Two surfaces on the histone chaperone Rtt106 mediate histone binding, replication, and silencing. Proc Natl Acad Sci USA. 2012;109:E144–E153. doi: 10.1073/pnas.1119095109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song Y, et al. CAF-1 is essential for Drosophila development and involved in the maintenance of epigenetic memory. Dev Biol. 2007;311:213–222. doi: 10.1016/j.ydbio.2007.08.039. [DOI] [PubMed] [Google Scholar]
- 34.Huang H, et al. Drosophila CAF-1 regulates HP1-mediated epigenetic silencing and pericentric heterochromatin stability. J Cell Sci. 2010;123:2853–2861. doi: 10.1242/jcs.063610. [DOI] [PubMed] [Google Scholar]
- 35.Monson EK, de Bruin D, Zakian VA. The yeast Cac1 protein is required for the stable inheritance of transcriptionally repressed chromatin at telomeres. Proc Natl Acad Sci USA. 1997;94:13081–13086. doi: 10.1073/pnas.94.24.13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Houlard M, et al. CAF-1 is essential for heterochromatin organization in pluripotent embryonic cells. PLoS Genet. 2006;2:e181. doi: 10.1371/journal.pgen.0020181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singer MS, et al. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyler JK, et al. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 39.Huang S, Zhou H, Tarara J, Zhang Z. A novel role for histone chaperones CAF-1 and Rtt106p in heterochromatin silencing. EMBO J. 2007;26:2274–2283. doi: 10.1038/sj.emboj.7601670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Z, Shibahara K, Stillman B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature. 2000;408:221–225. doi: 10.1038/35041601. [DOI] [PubMed] [Google Scholar]
- 41.Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 42.Kaufman PD, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 43.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 44.Krawitz DC, Kama T, Kaufman PD. Chromatin assembly factor I mutants defective for PCNA binding require Asf1/Hir proteins for silencing. Mol Cell Biol. 2002;22:614–625. doi: 10.1128/MCB.22.2.614-625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parnas O, et al. Elg1, an alternative subunit of the RFC clamp loader, preferentially interacts with SUMOylated PCNA. EMBO J. 2010;29:2611–2622. doi: 10.1038/emboj.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shemesh K, et al. A structure-function analysis of the yeast Elg1 protein reveals the importance of PCNA unloading in genome stability maintenance. Nucleic Acids Res. 2017;45:3189–3203. doi: 10.1093/nar/gkw1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kubota T, Katou Y, Nakato R, Shirahige K, Donaldson AD. Replication-coupled PCNA unloading by the Elg1 complex occurs genome-wide and requires Okazaki fragment ligation. Cell Rep. 2015;12:774–787. doi: 10.1016/j.celrep.2015.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kubota T, Nishimura K, Kanemaki MT, Donaldson AD. The Elg1 replication factor C-like complex functions in PCNA unloading during DNA replication. Mol Cell. 2013;50:273–280. doi: 10.1016/j.molcel.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 49.Shiomi Y, Nishitani H. Control of genome integrity by RFC complexes; conductors of PCNA loading onto and unloading from chromatin during DNA replication. Genes (Basel) 2017;8:E52. doi: 10.3390/genes8020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gazy I, Liefshitz B, Parnas O, Kupiec M. Elg1, a central player in genome stability. Mutat Res Rev Mutat Res. 2015;763:267–279. doi: 10.1016/j.mrrev.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 51.Smolikov S, Mazor Y, Krauskopf A. ELG1, a regulator of genome stability, has a role in telomere length regulation and in silencing. Proc Natl Acad Sci USA. 2004;101:1656–1661. doi: 10.1073/pnas.0307796100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mazor Y, Kupiec M. Developmentally regulated MAPK pathways modulate heterochromatin in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:4839–4849. doi: 10.1093/nar/gkp512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu C, et al. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol Cell. 2014;56:551–563. doi: 10.1016/j.molcel.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu T-Y, Dodson AE, Terhorst J, Song YS, Rine J. Riches of phenotype computationally extracted from microbial colonies. Proc Natl Acad Sci USA. 2016;113:E2822–E2831. doi: 10.1073/pnas.1523295113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goellner EM, et al. PCNA and Msh2-Msh6 activate an Mlh1-Pms1 endonuclease pathway required for Exo1-independent mismatch repair. Mol Cell. 2014;55:291–304. doi: 10.1016/j.molcel.2014.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson C, Gali VK, Takahashi TS, Kubota T. PCNA retention on DNA into G2/M phase causes genome instability in cells lacking Elg1. Cell Rep. 2016;16:684–695. doi: 10.1016/j.celrep.2016.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Enomoto S, Berman J. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 1998;12:219–232. doi: 10.1101/gad.12.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharp JA, Fouts ET, Krawitz DC, Kaufman PD. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr Biol. 2001;11:463–473. doi: 10.1016/s0960-9822(01)00140-3. [DOI] [PubMed] [Google Scholar]
- 59.Kelch BA, Makino DL, O’Donnell M, Kuriyan J. How a DNA polymerase clamp loader opens a sliding clamp. Science. 2011;334:1675–1680. doi: 10.1126/science.1211884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gomes XV, Burgers PMJ. ATP utilization by yeast replication factor C. I. ATP-mediated interaction with DNA and with proliferating cell nuclear antigen. J Biol Chem. 2001;276:34768–34775. doi: 10.1074/jbc.M011631200. [DOI] [PubMed] [Google Scholar]
- 61.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 62.Le S, Davis C, Konopka JB, Sternglanz R. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast. 1997;13:1029–1042. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1029::AID-YEA160>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 63.Kanellis P, Agyei R, Durocher D. Elg1 forms an alternative PCNA-interacting RFC complex required to maintain genome stability. Curr Biol. 2003;13:1583–1595. doi: 10.1016/s0960-9822(03)00578-5. [DOI] [PubMed] [Google Scholar]
- 64.Georgescu R, Langston L, O’Donnell M. A proposal: Evolution of PCNA’s role as a marker of newly replicated DNA. DNA Repair (Amst) 2015;29:4–15. doi: 10.1016/j.dnarep.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Recht J, et al. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc Natl Acad Sci USA. 2006;103:6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 67.Goldstein AL, Pan X, McCusker JH. Heterologous URA3MX cassettes for gene replacement in Saccharomyces cerevisiae. Yeast. 1999;15:507–511. doi: 10.1002/(SICI)1097-0061(199904)15:6<507::AID-YEA369>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 68.Lee ME, DeLoache WC, Cervantes B, Dueber JE. A highly characterized yeast toolkit for modular, multipart assembly. ACS Synth Biol. 2015;4:975–986. doi: 10.1021/sb500366v. [DOI] [PubMed] [Google Scholar]
- 69.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Janke R, Iavarone AT, Rine J. Oncometabolite D-2-Hydroxyglutarate enhances gene silencing through inhibition of specific H3K36 histone demethylases. eLife. 2017;6:e22451. doi: 10.7554/eLife.22451. [DOI] [PMC free article] [PubMed] [Google Scholar]