The alarming global rise of antibiotic resistance among bacterial pathogens necessitates not only urgent discovery of new antibiotics but also practical strategies to preserve those that are currently used (1, 2). A system-level understanding of the killing mechanisms of bactericidal antibiotics holds the promise of shortening treatment time courses and decreasing the chances that sensitive bacteria will evolve resistance during treatment (3). As different classes of bactericidal antibiotics have different primary cellular target(s), it is not surprising that a significant aspect of the bacterial killing mechanism is specific to the essential function targeted by each antibiotic class: β-lactams induce a futile cycle of cell wall synthesis (4), aminoglycosides induce mistranslation of peptides that compromise inner membrane integrity (5), and fluoroquinolones cause DNA breaks by inhibiting DNA gyrase or topoisomerase IV (6). However, regardless of the primary cellular target, different bactericidal antibiotics all induce a similar cascade of events that contribute to their lethality in Escherichia coli: accelerated respiration (7); changes in iron and redox metabolism (8); formation of reactive oxygen species (9); oxidative damage of DNA (10), RNA, and proteins (11); and cell death. Interestingly, the same killing cascade is seen in a number of antibiotic-independent killing phenomena, for example, T6SS-effector dependent killing (12) and expression of a toxic fusion protein (13).
In PNAS, Fan et al. (14) explore whether different classes of bactericidal antibiotics used to treat Mycobacterium tuberculosis, the causative agent of tuberculosis, share a common killing cascade. They used bactericidal antibiotics of the aminoglycoside and fluoroquinolone classes that were also used in previous bactericidal antibiotic lethality studies in E. coli. They discovered that lethal oxidative DNA damage does occur in nongrowing M. tuberculosis cells treated with different bactericidal antibiotics. They also used rifampicin, an RNA polymerase inhibitor that is not bactericidal in E. coli, but rather bacteriostatic. Rifampicin does not show any of the hallmarks of the common killing mechanism of bactericidal antibiotics in E. coli, but is lethal in mycobacteria. The basis for this lethality is unknown (15), but nevertheless, it is fascinating evidence that shows how deeply embedded the common oxidative-damage–dependent killing mechanism is in the lethality of most, if not all, bactericidal antibiotics.
Reaction of hydrogen peroxide with reduced iron (Fe2+) produces hydroxyl radicals that damage both the ribose sugar and the nitrogen-containing base of each of the four nucleotides (16). As one might expect, various DNA damage repair mutants are hypersensitive to bona fide oxidants (hydrogen peroxide or ionizing radiation) (16). Because deoxynucleotide triphosphates chelate reduced iron, it is the free nucleotide pool and not the genomic DNA that is mostly susceptible to oxidative damage by hydroxyl radicals (17). Bacterial “sanitizer” enzymes are constantly monitoring the nucleotide pool and hydrolyze various oxidized nucleotide derivatives. In E. coli, an oxidized derivative of guanine, 8-Oxo-G, accumulates during oxidative stress as well as during antibiotic treatment (11). 8-Oxo-G is promutagenic by virtue of its base-pairing capacity with adenine.
In antibiotic-treated E. coli, overexpression of the MutT sanitizer protein (of the Nudix hydrolase family) partially protects growing cultures from killing, as do mutations that minimize error-prone incorporation of 8-Oxo-G (10). Once 8-Oxo-G is incorporated to genomic DNA, the fate of the bacterial cell is dependent on the oxidized guanine (GO) system. This system is composed of two DNA repair proteins: 8-Oxo-G–specific DNA glycosylase (MutM of the Fpg family) and DNA glycosylase (MutY), which is specific for adenine paired with either guanine or 8-Oxo-G. Of note, MutM cannot remove 8-Oxo-G paired with adenine, and this property of the enzyme prevents mutagenic repair of 8-Oxo-G lesions. The glycosylase activity of both enzymes removes the nitrogen base of the damaged nucleotide, thereby producing an abasic site. In addition, these enzymes have a weak DNA lyase catalytic activity toward abasic sites, although this nicking activity does not produce a polymerase-competent 3′-OH end in the same manner as do dedicated abasic site nucleases (18). Ultimately, antibiotic-treated E. coli accumulate lethal double-strand breaks (DSBs) in DNA and, strikingly, a mutM-mutY double mutant is more resistant to antibiotic treatment and accumulates less DSBs. Collectively, the results appear consistent with a mechanism of antibiotic killing involving the incomplete base excision repair of incorporated 8-Oxo-G by the GO system, which apparently contributes to the formation of lethal DSBs. It seems that the activity of the GO system is harmful under conditions of oxidative DNA damage, and future research will hopefully address in more detail this surprising observation. On one hand, then, 8-Oxo-G poses a real threat to bacterial viability by virtue of its contribution to DSB formation, but on the other hand its promutagenic potential (19) increases the chances of antibiotic-treated bacteria to evolve antibiotic resistance.
Fan et al. (14) focus on the oxidized derivative of dCTP, 5-OH-dCTP, rather than on the oxidized derivative of guanine, 8-Oxo-G. The presence of four mutT-like genes in mycobacteria and the absence of the mutagenic signature associated with 8-Oxo-G (GC→TA) in mutational analysis studies suggest that mycobacteria are efficient in removing 8-Oxo-G from its genome. That 5-OH-dCTP is a major oxidized nucleotide derivative resulted from previous studies on the mycobacteria MazG sanitizer protein (of the all-α NTP pyrophosphatase family) by the same group (20, 21). While MazG of M. tuberculosis can hydrolyze all four canonical nucleotides in vitro, it does so with very high Km values. The sensitivity of the mazG mutant to oxidative stress provided evidence that its preferred substrate is likely to be an oxidized derivative of one of the canonical nucleotides. The increased mutation rates of CG→TA substitutions together with an earlier study describing the HPLC-based characterization of oxidized derivatives of dCTP in E. coli (22) led the group to successfully identify 5-OH-dCTP as the preferred substrate for the enzymatic activity of MazG.
Fan et al. (14) show that the mazG mutant of M. tuberculosis and of Mycobacterium smegmatis is more sensitive than the respective wild-type strain to bactericidal antibiotics in stationary phase and during macrophage infection. This sensitivity occurs presumably because of the incorporation of 5-OH-dCTP into DNA. Unfortunately, while incorporation of 8-Oxo-G into DNA can be detected with the use of antibodies, detection of 5-OH-dCTP in DNA from cell lysates proves to be difficult. Fan et al. reason that if 5-OH-dCTP is indeed incorporated into DNA in stationary phase, an error-prone polymerase is more likely to incorporate it and, therefore, a mutant in such a polymerase will incorporate a significantly lower amount of 5-OH-dCTP into the genome. In support of this idea, double mutants of mazG and dnaE2, the predominant error-prone polymerase of M. tuberculosis (23), were as resistant as wild-type cells to bactericidal antibiotic treatment in stationary phase. Differential expression of dnaE2 also suggests a growth phase-specific contribution of 5-OH-dCTP to antibiotic lethality; mRNA levels of dnaE2 are greatly increased in stationary phase.
Does a dedicated system for the removal of 5-OH-dCTP from DNA (analogous to the GO system for base excision repair of 8-Oxo-G) exist? Perhaps. In mycobacteria, Nth (endonuclease III) is a glycosylase/lyase dedicated to the removal of oxidized pyrimidines from DNA that nicks the abasic site and thereby produces a 3′
The current work of Fan et al. highlights both shared and distinctive features of the mechanism of antibiotic killing through oxidative DNA damage between E. coli and M. tuberculosis.
blocked nick, which is then repaired by the AP nuclease (24). However, it does not act in concert with a MutY-like partner. This difference in processing of oxidized lesions in DNA between 8-Oxo-G and 5-OH-dCTP might indicate that, unlike 8-Oxo-G, 5-OH-dCTP is not readily base-pairing with a single preferred nucleotide.
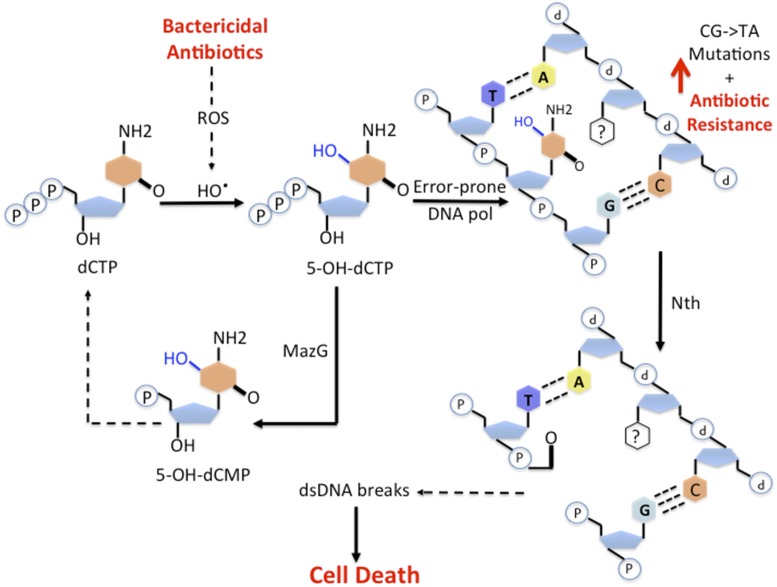
Fan et al. provide further evidence that in an attempt to repair the oxidized lesion, Nth seems to exacerbate oxidative DNA damage and to increase the amount of DSBs, a striking resemblance to the mechanism of bactericidal antibiotic-induced lethality in E. coli (Fig. 1). Whereas a mazG mutant accumulates a greater number of DSBs than do wild-type cells during bactericidal antibiotic treatment, the mazG nth double mutant restores DSBs to the wild-type level. Further evidence for the accumulation of DSBs during bactericidal antibiotic treatment results from the simultaneous treatment with bactericidal antibiotics and an inhibitor of homologous recombination, which is the major mycobacteria DSB repair pathway. Low levels of the RecA-inhibitor suramin sensitize the bactericidal antibiotic killing of the mazG mutants even further. Moreover, a high concentration of suramin also sensitized wild-type cells to bactericidal antibiotic killing.
Fig. 1.
Oxidative DNA damage is a common killing mechanism of bactericidal antibiotics in mycobacteria. Different classes of bactericidal antibiotics increase generation of hydroxyl radicals and other reactive oxygen species. In this intracellular environment, dCTP is oxidized to 5-OH-dCTP. Error-prone DNA polymerase then incorporates the damaged nucleotide into DNA, unless it is hydrolyzed first by MazG. Once in DNA, 5-OH-dCTP is mutagenic. Moreover, incomplete repair of this lesion by the Nth glycosylase/lyase can potentially lead to the formation of DSBs and cell death.
The current work of Fan et al. highlights both shared and distinctive features of the mechanism of antibiotic killing through oxidative DNA damage between E. coli and M. tuberculosis. Also, the strong mutator phenotype of sanitizer protein mutants in both E. coli and M. tuberculosis suggests that oxidized derivatives of nucleotides drive much of the mutagenesis detected under conditions of stress.
Future translational studies will hopefully reveal strategies to disable bacterial defense systems that protect bacteria against antibiotic-mediated oxidative damage (25–28). Within the realm of basic research, it will be interesting to determine whether it is possible to develop high-throughput methodologies for the identification of the true substrates of novel sanitizer proteins, as well as to identify the types of DNA damage responsible for unknown mutational signatures that appear in sequencing data (29).
Acknowledgments
This work was supported by the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
See companion article on page 2210.
References
- 1.Ling LL, et al. A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King AM, et al. Aspergillomarasmine A overcomes metallo-β-lactamase antibiotic resistance. Nature. 2014;510:503–506. doi: 10.1038/nature13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levin-Reisman I, et al. Antibiotic tolerance facilitates the evolution of resistance. Science. 2017;355:826–830. doi: 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 4.Cho H, Uehara T, Bernhardt TG. Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell. 2014;159:1300–1311. doi: 10.1016/j.cell.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis BB. The lethal action of aminoglycosides. J Antimicrob Chemother. 1988;22:1–3. doi: 10.1093/jac/22.1.1. [DOI] [PubMed] [Google Scholar]
- 6.Chen CR, Malik M, Snyder M, Drlica K. DNA gyrase and topoisomerase IV on the bacterial chromosome: Quinolone-induced DNA cleavage. J Mol Biol. 1996;258:627–637. doi: 10.1006/jmbi.1996.0274. [DOI] [PubMed] [Google Scholar]
- 7.Lobritz MA, et al. Antibiotic efficacy is linked to bacterial cellular respiration. Proc Natl Acad Sci USA. 2015;112:8173–8180. doi: 10.1073/pnas.1509743112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dwyer DJ, et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc Natl Acad Sci USA. 2014;111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 10.Foti JJ, Devadoss B, Winkler JA, Collins JJ, Walker GC. Oxidation of the guanine nucleotide pool underlies cell death by bactericidal antibiotics. Science. 2012;336:315–319. doi: 10.1126/science.1219192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belenky P, et al. Bactericidal antibiotics induce toxic metabolic perturbations that lead to cellular damage. Cell Rep. 2015;13:968–980. doi: 10.1016/j.celrep.2015.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong TG, et al. Generation of reactive oxygen species by lethal attacks from competing microbes. Proc Natl Acad Sci USA. 2015;112:2181–2186. doi: 10.1073/pnas.1425007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi N, et al. Lethality of MalE-LacZ hybrid protein shares mechanistic attributes with oxidative component of antibiotic lethality. Proc Natl Acad Sci USA. 2017;114:9164–9169. doi: 10.1073/pnas.1707466114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan X-Y, et al. Oxidation of dCTP contributes to antibiotic lethality in stationary-phase mycobacteria. Proc Natl Acad Sci USA. 2018;115:2210–2215. doi: 10.1073/pnas.1719627115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keren I, Mulcahy LR, Lewis K. Persister eradication: Lessons from the world of natural products. Methods Enzymol. 2012;517:387–406. doi: 10.1016/B978-0-12-404634-4.00019-X. [DOI] [PubMed] [Google Scholar]
- 16.Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat Rev Microbiol. 2013;11:443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rush JD, Koppenol WH. Reactions of Fe(II)-ATP and Fe(II)-citrate complexes with t-butyl hydroperoxide and cumyl hydroperoxide. FEBS Lett. 1990;275:114–116. doi: 10.1016/0014-5793(90)81452-t. [DOI] [PubMed] [Google Scholar]
- 18.Manuel RC, et al. Reaction intermediates in the catalytic mechanism of Escherichia coli MutY DNA glycosylase. J Biol Chem. 2004;279:46930–46939. doi: 10.1074/jbc.M403944200. [DOI] [PubMed] [Google Scholar]
- 19.Rotman E, Kuzminov A. The mutT defect does not elevate chromosomal fragmentation in Escherichia coli because of the surprisingly low levels of MutM/MutY-recognized DNA modifications. J Bacteriol. 2007;189:6976–6988. doi: 10.1128/JB.00776-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu LD, et al. Mycobacterial MazG is a novel NTP pyrophosphohydrolase involved in oxidative stress response. J Biol Chem. 2010;285:28076–28085. doi: 10.1074/jbc.M109.088872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyu L-D, Tang B-K, Fan X-Y, Ma H, Zhao G-P. Mycobacterial MazG safeguards genetic stability via housecleaning of 5-OH-dCTP. PLoS Pathog. 2013;9:e1003814. doi: 10.1371/journal.ppat.1003814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feig DI, Sowers LC, Loeb LA. Reverse chemical mutagenesis: Identification of the mutagenic lesions resulting from reactive oxygen species-mediated damage to DNA. Proc Natl Acad Sci USA. 1994;91:6609–6613. doi: 10.1073/pnas.91.14.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boshoff HIM, Reed MB, Barry CE, 3rd, Mizrahi V. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell. 2003;113:183–193. doi: 10.1016/s0092-8674(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 24.Moolla N, Goosens VJ, Kana BD, Gordhan BG. The contribution of Nth and Nei DNA glycosylases to mutagenesis in Mycobacterium smegmatis. DNA Repair (Amst) 2014;13:32–41. doi: 10.1016/j.dnarep.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Gusarov I, Shatalin K, Starodubtseva M, Nudler E. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science. 2009;325:1380–1384. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shatalin K, Shatalina E, Mironov A, Nudler E. H2S: A universal defense against antibiotics in bacteria. Science. 2011;334:986–990. doi: 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]
- 27.Vilchèze C, et al. Enhanced respiration prevents drug tolerance and drug resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2017;114:4495–4500. doi: 10.1073/pnas.1704376114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mironov A, et al. Mechanism of H2S-mediated protection against oxidative stress in Escherichia coli. Proc Natl Acad Sci USA. 2017;114:6022–6027. doi: 10.1073/pnas.1703576114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jee J, et al. Rates and mechanisms of bacterial mutagenesis from maximum-depth sequencing. Nature. 2016;534:693–696. doi: 10.1038/nature18313. [DOI] [PMC free article] [PubMed] [Google Scholar]