Abstract
Gastric cancer (GC) is one of the most common malignancies worldwide and has high morbidity and mortality rates. It is essential to elucidate the molecular events of GC proliferation and invasion, which will provide new therapeutic targets for GC. The inactivation of transforming growth factor‐β receptor 2 (TGFβR2) correlates with cancer cell growth and metastasis, but the mechanisms underlying the downregulation of TGFβR2 expression remain unknown. MicroRNAs (miRNAs) act as post‐transcriptional regulators and play a key role in the development of cancers. Bioinformatics analysis and luciferase reporter assays have shown that miR‐155 directly binds to the 3′‐UTR of TGFβR2 mRNA. In this study, we found that the TGFβR2 protein levels, but not mRNA levels, were downregulated in GC tissues, and the levels of miR‐155 were significantly increased in GC tissues. We deduced that miR‐155 was inversely correlated with TGFβR2 in GC cells. In vitro studies showed that overexpression of miR‐155 in SGC7901 inhibited the expression of TGFβR2 and then promoted GC cell proliferation and migration, whereas miR‐155 inhibitor showed opposite effects. In addition, the tumor‐suppressing function of TGFβR2 was verified by using siRNA and TGFβR2 overexpressing plasmids. The results showed that miR‐155 promotes cell growth and migration by negatively regulating TGFβR2. Thus, miR‐155‐regulated TGFβR2 as a potential therapeutic target in GC.
Keywords: gastric cancer, growth, invasion, miR‐155, transforming growth factor‐β receptor 2
Abbreviations
- CT
cycle threshold
- EdU
5‐ethynyl‐2‐deoxyuridine
- GC
gastric cancer
- miRNA
microRNA
- NC
negative control
- qRT‐PCR
quantitative RT‐PCR
- TGFβR2
transforming growth factor‐β receptor 2
1. INTRODUCTION
Gastric cancer is one of the most common malignancies worldwide, with the fourth highest morbidity rate and the third highest mortality rate.1, 2 The incidence rate of GC in Asia is above the worldwide average; furthermore, China accounts for nearly half of the cases in Asia.3 Gastric cancer is the third most common cancer in China.4 Despite important progress in the diagnosis and treatment of GC in recent years, the poor prognosis for GC patients remains because of the cancer's capacity to proliferate and metastasize.3 Our aim is to find effective methods to prevent the growth and metastasis of GC. To do so, it is necessary to identify the neoplastic etiopathogenesis of GC and clarify the molecular mechanisms underlying tumor metastasis. Analysis of the signaling pathways in GC cells provides novel biomarkers for valid molecular targeted therapy.
Transforming growth factor‐β receptor 2 is involved in many biological processes, such as cell differentiation, cell growth, apoptosis, and cell migration.5 Acting as a tumor suppressor, TGFβR2 participates in tumor growth and migration because of genetic mutation or promoter methylation, which has been reported in colorectal cancer, esophageal, and prostate cancers.6, 7, 8, 9 Furthermore, TGFβR2 as a cancer inhibitor is downregulated in GC, which is associated with increased tumorigenicity.10 Although the inactivation of TGFβR2 has been discovered in cancer, the molecular mechanisms connected with TGFβR2 in GC have not been investigated.
MicroRNAs are small non‐coding RNAs of 19‐25 nucleotides in length that post‐transcriptionally regulate gene expression.11, 12, 13 They play vital roles in regulating the translation and degradation of mRNAs by binding the 3′‐UTRs of mRNAs. The biological functions of miRNAs that regulating gene expression include cell proliferation, cell apoptosis, stress tolerance, and fat metabolism.14 Aberrant expression of miRNAs has been found in several cancers, which play novel roles as anti‐oncogenes or oncogenes, depending on the function of the target genes.15, 16, 17 Previous studies showed that aberrant expression of many miRNAs promote proliferation and metastases in GC.18, 19, 20 However, the molecular events are not fully understood.
In the present study, we identified specific targeting sites for miR‐155 in the 3′‐UTR of the TGFβR2 gene by using bioinformatics prediction. Experimental confirmation studies then illustrated that the negative regulation TGFβR2 by miR‐155 promoted the growth and invasion of GC.
Our findings indicate that miR‐155, as a novel therapeutic target, provides a potential new treatment for GC.
2. MATERIALS AND METHODS
2.1. Patients and tissue samples
This study was approved by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital (Tianjin, China). All patients provided written informed consent to participate in the study. We obtained 5 pairs of human GC tissue and matched adjacent normal gastric tissues from GC patients who were undergoing gastrostomy at the Tianjin Medical University Cancer Institute and Hospital. None of the patients received antitumor therapy before surgery. All GC tissue and normal gastric tissues were confirmed histologically. The pathological type of GC was confirmed to be adenocarcinoma. Tissue fragments were frozen in liquid nitrogen immediately after surgical excision. Total protein and RNA from tissue samples were extracted and stored at −80°C.
2.2. Animals
Male nude mice (BALB/c‐nu, 6‐8 weeks old) were purchased from the Model Animal Center of Nanjing University (Nanjing, China). They were housed in a pathogen‐free animal facility with access to water and food and allowed to eat and drink ad libitum. All of the experimental procedures were carried out in accordance with protocols approved by the Institutional Animal Care and Research Advisory Committee of Tianjin Medical University Cancer Institute and Hospital.
2.3. Cell lines and cell cultures
The human gastric adenocarcinoma cell lines SGC7901 and MGC803 were obtained from the Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). The cells were cultured in DMEM (Gibco, Carlsbad, CA, USA), supplemented with 10% FBS (Gibco) and 1% penicillin/streptomycin (Solarbio, Beijing, China). All of the cells were incubated in an atmosphere of 5% CO2 at 37°C.
2.4. Immunohistochemistry assay
Gastric cancer tissues and matched adjacent normal gastric tissues were fixed in 4% paraformaldehyde and embedded in paraffin. The tissues were sliced into 4‐μm‐thick sections. All slides were dewaxed twice with dimethylbenzene and rehydrated in a graded series of ethanol. Antigen retrieval was achieved by soaking the sections in 10 mmol/L citrate buffer (pH 6.0), then heating them to 220°C at high pressure for 3 minutes. To block endogenous peroxidase activity, the slides were soaked in 3% hydrogen peroxide for 20 minutes. The tissue slides were treated with anti‐human TGFβR2 mAb (1:50, sc‐400; Santa Cruz Biotechnology, CA, USA), then incubated overnight in a humidified chamber at 4°C. On day 2, the slides were washed in PBS 3 times and secondary antibodies were added dropwise to slides for 40 minutes at 37°C. After washing again with PBS 3 times, sections were stained by diaminobenzidine and counterstained with hematoxylin. The sections were then dehydrated and coverslipped. Quantitative analysis was carried out by quantifying the fluorescence intensity from 6 sections.
2.5. RNA extraction and RT‐PCR
Total RNA was extracted from the cultured cells and tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). In order to detect the quantity of the miRNA, we used TaqMan miRNA probes (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's protocol. RNA was reverse transcribed into cDNA by using AMV reverse transcriptase (TaKaRa, Dalian, China) and a stem‐loop RT primer (Applied Biosystems). The reaction conditions were: 16°C for 30 minutes, 42°C for 30 minutes, and 85°C for 5 minutes. Using a TaqMan PCR kit, RT‐PCR was carried out on ABI 7300 Sequence Detection System (Applied Biosystems). After the reactions were complete, the CT data were collected by using fixed threshold settings, and the mean CT was calculated from triplicate PCRs. A comparative CT method was used to compare each condition to the control reactions. U6 snRNA was used as an endogenous control; the mRNA levels of TGFβR2 were normalized to the endogenous control gene GAPDH. The relative amount of gene normalized to control was calculated with the equation 2−ΔCT, in which ΔCT = CT gene−CT control. All of the reactions were run in triplicate. The sequences of the primers for TGFβR2 and GAPDH were as follows: GAPDH sense, 5′‐AGAAGGCTGGGGCTCATTTG‐3′; GAPDH antisense, 5′‐AGGGGCCATCCACAGTCTTC‐3′; TGFβR2 sense, 5′‐GTAGCTCTGATGAGTGCAATGAC‐3′; and TGFβR2 antisense, 5′‐CAGATATGGCAACTCCCAGTG‐3′.
2.6. MicroRNA target prediction and Luciferase reporter assay
The miRNA target prediction and analysis were performed with the algorithms from TargetScan (http://www.targetscan.org/), PicTar (http://pictar.mdc-berlin.de/), and miRanda (http://www.microrna.org/). To test the binding specificity of miR‐155 to the target gene TGFβR2, luciferase reporter assays were carried out as follows. The entire 3′‐UTRs of human TGFβR2 were amplified from human genomic DNA using PCR. The PCR products were inserted into the luciferase reporter plasmid, which were designed by Genescript (Nanjing, China). Two micrograms of firefly luciferase reporter plasmid, 2 mg β‐galactosidase vector, and equal doses (200 pmol) of mimics, inhibitors, or scrambled negative control RNA were transfected into the prepared cells. At 24 hour post‐transfection, cells were analyzed using the Dual Luciferase Assay Kit (Promega, Madison, WI, USA) following the manufacturer's instructions. Each sample was prepared in triplicate, and the entire experiment was repeated independently 3 times on different days.
2.7. Cell transfection
SGC‐7901 cells were seeded in a 6‐well or other sized plate, and transfection was carried out after 24 hours. The TGFβR2 overexpression plasmid (pcDNA TGFβR2) and corresponding negative control plasmid (pcDNA NC) were designed and synthesized by GenePharma (Shanghai, China), and 2 μg plasmid were transfected into every single well. Cells were transfected with miR155‐5p mimics or miR155‐5p inhibitors as scrambled negative control, using Lipofectamine 2000 (Invitrogen, Life Technologies) and Opti‐MEM Reduced Serum Medium (Gibco, Life Technologies) following the manufacturer's instructions. For overexpression and knockdown of miRNA, 100 pmol miRNA mimics and inhibitors were transfected into each well, respectively. Then 100 pmol siRNAs (Santa Cruz Biotechnology) and scrambled negative control RNA were transfected into each well. The cells were harvested 24 or 48 hours after transfection for isolation of total RNA or at 48 hours for Western blot analysis.
2.8. Protein isolation and Western blot analysis
The expression of TGFβR2 was assessed by Western blot analysis and GAPDH was used as an internal control. Total proteins were extracted from tissues and cultured cells that were solubilized in lysis buffer supplemented with a protease and phosphatase inhibitor cocktail on ice. Proteins were separated by 10% SDS‐PAGE and then transferred to PVDF membranes (Roche, Mannheim, Germany). The membranes were incubated for 2 hour with 2% BSA at room temperature for 1 hour and incubated overnight at 4°C with primary anti‐TGFβR2 (1:200; Santa Cruz Biotechnology) and anti‐GAPDH (1:5000; Santa Cruz Biotechnology). Subsequently, the membranes were washed and incubated with appropriate secondary antibodies. The immune complexes were visualized with an ECL Prime Western Blot Detection System (Thermo, EHVEGFACL, Carlsbad, CA, USA).
2.9. Cell proliferation assay
SGC‐7901 cells were seeded on 24‐well plates and transfected with miR‐155 mimics, inhibitors, TGFβR2 siRNA, and the relevant negative controls. A Cell‐light EdU DNA cell kit (Ribobio, Guangzhou, China) was used to measure the cell proliferation ability at 24 hour post‐transfection, following the manufacturer's instructions. Cells were incubated with 50 μmol/L EdU for 5 hours and fixed within 4% paraformaldehyde for 30 minutes at room temperature. The cells were washed with PBS twice and treated with 0.5% Triton X‐100 for 10 minutes. Subsequently, the cells were incubated in the dark with Apollo (Ribobio) for 30 minutes and in Hoechst 33342 (Ribobio) for a further 30 minutes. All of the staining was carried out in triplicate.
2.10. Cell migration assay
The migration ability of SGC‐7901 cells transfected with miR‐155 mimics, inhibitors, TGFβR2 siRNA, and the relevant negative controls were measured by wound healing assays and Transwell assays. SGC‐7901 cells were seeded on 12‐well plates. When densities of the transfected cells were approximately 90%, each well was scratched with a 10‐μL pipette tip to create 2 empty linear regions. The cells were then cultured in DMEM without FBS in the humidified incubator. We observed and photographed the cells at 0 hour, 6 hours, 18 hours, and 24 hours after scraping. The distance of the wound zone was measured in at least 3 randomly selected fields using a phase contrast microscope and the cells were digitally photographed and compared with the control group.
Transwell migration assays were undertaken using Costar Transwell chambers (8‐μm pore size; Corning, Corning, NY, USA). Approximately 105 cells were seeded in the upper chamber of the wells with 200 μL serum‐free growth medium (105 cells/well of 8.0‐μm pore polycarbonate membrane insert). The lower chambers were filled with 700 μL medium containing 10% FBS in order to induce cell migration. After incubation at 37°C in 5% CO2 for 24 hours, non‐migratory cells on the upper surface of the membrane were removed slightly by cotton swabs. The membranes were fixed with methanol and stained with a two‐step staining set. All assays were carried out in triplicate. To minimize the bias, at least 3 randomly selected fields were counted with 200× magnification under a microscope, and the average number was taken.
2.11. Establishment of tumor xenograft in nude mice
SGC7901 cells treated with control lentivirus or miR‐155 overexpressing lentivirus or TGFβR2 overexpressing lentivirus were injected s.c. into nude mice (1 × 107 cells/mouse). Mice were killed after 4 weeks, and the weight and diameter of tumors were recorded.
2.12. Statistical analyses
All statistical analyses were undertaken using spss 20.0 (IBM, Chicago, IL, USA) to analyze the data. The results are described as the means ± SE and analyzed by using Student's t‐test. Statistical significance is shown by *P < .05, **P < .01, and ***P < .001. Transwell migration assay images are representative of at least 3 experiments. Quantitative RT‐PCRs, Western blot analyses, luciferase reporter assays, and cell viability, proliferation, and migration assays were carried out in triplicate, and each experiment was repeated several times.
3. RESULTS
3.1. Transforming growth factor‐β receptor 2 expression decreases in human GC tissue specimens
It has been reported that TGFβR2 is downregulated in multiple cancer types, but the expression pattern in GC has not been fully elucidated. In order to analyze the expression characteristics of TGFβR2 protein in GC, 5 pairs of GC patient tissues and corresponding adjacent non‐cancerous tissues were subjected to immunohistochemistry, Western blot, and qRT‐PCR analyses. As shown in θ Figure 1, the TGFβR2 protein was dramatically upregulated in GC tissues compared with the normal adjacent non‐cancerous tissues. Immunohistochemical assays showed that the expression characteristic of TGFβR2 is positive cytoplasm staining, and results revealed higher expression levels in GC tissues than in non‐cancerous tissues (Figure 1F), which agreed with the results of Western blot analysis. However, there was a slight decline in TGFβR2 mRNA levels in GC tissues compared to non‐cancerous tissues (Figure 1C). These results indicate that the expression of TGFβR2 is mainly dependent on post‐transcriptional regulators in GC.
Figure 1.
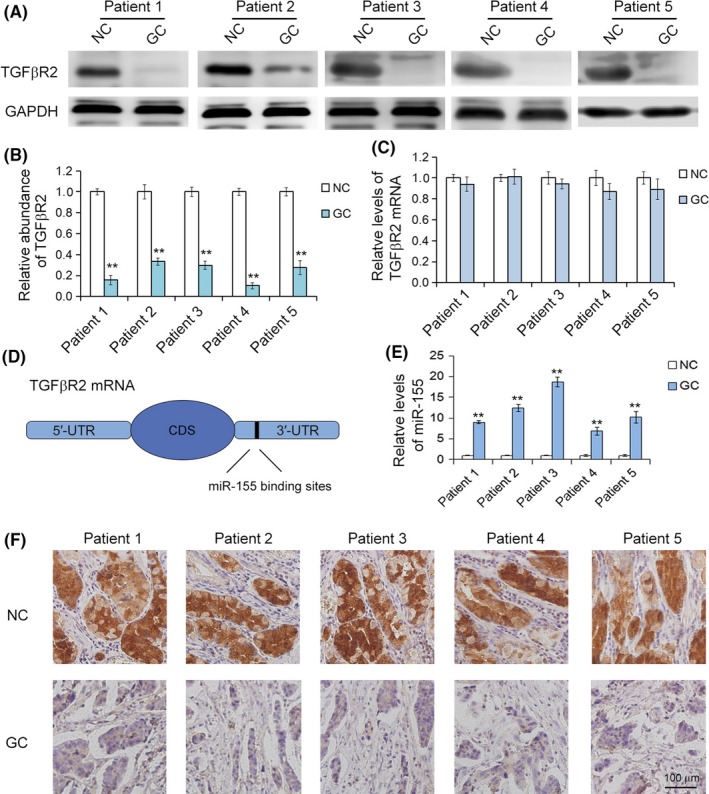
Inverse correlation between transforming growth factor‐β receptor 2 (TGFβR2) and microRNA‐155 (miR‐155) in gastric cancer (GC) tissues. A, Western blot analysis of TGFβR2 protein expression in GC tissues and paired non‐cancerous tissues (NC) (n = 5). B, Quantitative analysis of (A). C, Quantitative RT‐PCR analysis of TGFβR2 mRNA levels in GC and NC tissues (n = 5). D, Predicted binding region of miR‐155 in the mRNA of TGFβR2. CDS, coding sequence. E, Relative levels of miR‐155 in GC and NC tissues (n = 5). F, Immunohistochemistry assays of TGFβR2 expression in GC and NC tissues (n = 6). **P < .01
3.2. Identification of TGFβR2 as a potential downstream target of miR‐155
The underlying molecular mechanisms of downregulation of TGFβR2 protein expression are largely unknown in GC. MicroRNAs play a significant role in post‐transcriptional genes by cleaving specific mRNA or translational repression. We utilized 3 public bioinformatics tools to predict the theoretical potential target of miR‐155 and the binding sites. Then we found that miR‐155 can potentially bind a region in the 3′‐UTR of TGFβR2 mRNA (Figure 1D). Furthermore, the region that miR‐155 directly targets is highly conserved, and the complementary base‐pairing reactions are shown in Figure 2A. One predicted hybridization is observed between miR‐155 and the 3′‐UTR of TGFβR2.
Figure 2.
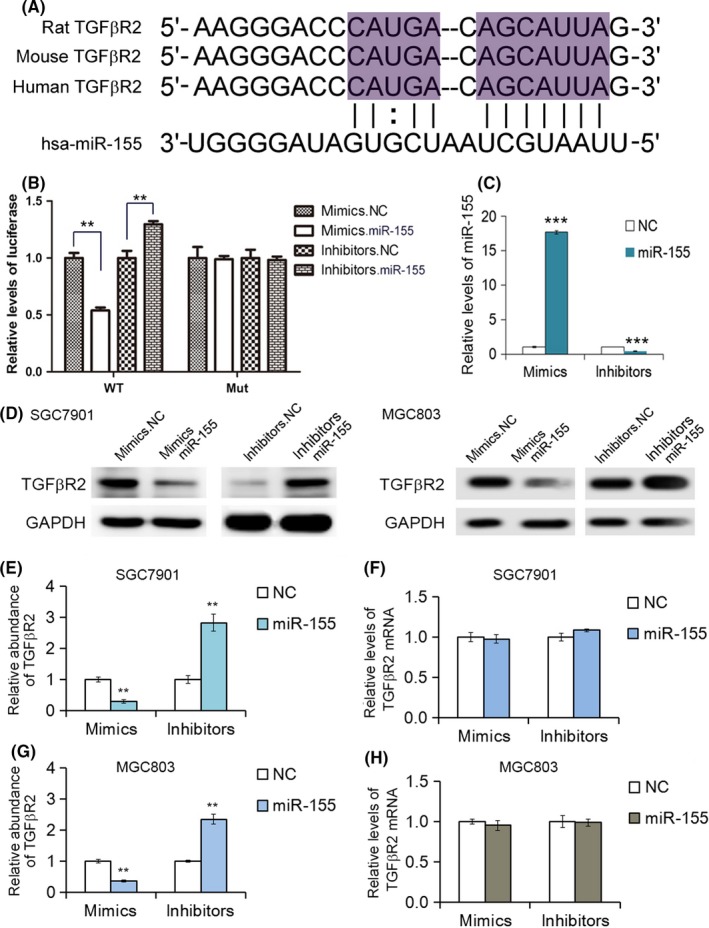
MicroRNA‐155 (miR‐155) regulates transforming growth factor‐β receptor 2 (TGFβR2) expression in gastric cancer (GC) cells. A, Description of the base‐pairing interaction between miR‐155 and TGFβR2 mRNA. B, Luciferase reporter assay. SGC7901 cells were cotransfected with firefly luciferase reporters containing either WT or mutant TGFβR2 3′‐UTR with miR‐155 mimics and inhibitors (n = 3). inhibitors.NC, control inhibitors; mimics.NC, control mimics. C, Relative levels of miR‐155 in SGC7901 cells transfected with mimics or inhibitors by quantitative RT‐PCR analysis (n = 3). D, Suppression of TGFβR2 expression by miR‐155 in SGC7901 and MGC803 cells (n = 3). E, Quantitative analysis of (D) (n = 3). F, Quantitative RT‐PCR analysis of TGFβR2 mRNA in SGC7901 cells transfected with miR‐155 mimics or inhibitors by quantitative RT‐PCR analysis (n = 3). G, Quantitative analysis of (D) (n = 3). H, Quantitative RT‐PCR analysis of TGFβR2 mRNA in MGC803 cells transfected with miR‐155 mimics or inhibitors by quantitative RT‐PCR analysis (n = 3). **P < .01; ***P < .001
3.3. Upregulation of miR‐155 in GC tissues and validation of TGFβR2 as a direct target of miR‐155
Low expression of TGFβR2 proteins were explored in GC tissues. In order to validate the actual relationship between miR‐155 and TGFβR2 in GC, we further analyzed the levels of miR‐155 in GC and NC tissues by real‐time PCR. Compared with the adjacent non‐cancerous tissues, a significant decrease of miR‐520b/e was observed in GC tissues. As expected, miR‐155 was clearly increased in GC tissues and decreased in the paired NC tissues (Figure 1E). Therefore, miR‐155 is most likely the directly regulator of TGFβR2 in GC cells.
In order to investigate whether miR‐155 could bind to the putative binding sites in the 3′‐UTR of TGFβR2, we undertook a luciferase assay. As shown in Figure 2B, the relative luciferase activity of the reporter gene in SGC7901 cells cotransfected with p‐MIR‐TGFβR2 and miR‐130 mimics was markedly inhibited compared with the control (cotransfected with p‐MIR‐TGFβR2 and miRNA NC). There was no difference in the relative luciferase activity of the reporter gene between SGC7901 cells cotransfected with mutated p‐MIR‐TGFβR2 and miR‐155 mimics or miRNA NC. These data verified that miR‐155 directly binds the 3′‐UTR of TGFβR2 mRNA and suppresses TGFβR2 expression.
3.4. MicroRNA‐155 regulates TGFβR2 expression in GC cells
There was an inverse relationship between the miR‐155 and TGFβR2 expression levels in GC tissue. To further clarify the relationship between miR155 and TGFβR2 in GC, miR‐155 mimics or inhibitors were transfected into the SGC7901 cell line. After 24 hours, the relative levels of miR‐155 and the expression of TGFβR2 protein and mRNA were detected using qRT‐PCR or Western blot analysis. Transfection of miRNA mimics significantly increases miR‐155 levels, while inhibitors clearly down‐regulate miR‐155 in cells (Figure 2C). As shown in Figure 2D,E, the overexpression of miR‐155 by transfection of mimics led to the clear suppression of TGFβR2 protein, and the transfection of miR‐155 inhibitors enhanced the expression of TGFβR2 in SGC7901 cells. In contrast, TGFβR2 mRNA was not changed with the transfection of mimics or inhibitors (Figure 2F). Moreover, we repeated the above experiments in another GC cell line (MGC803) to validate the robustness of the test. As expected, miR‐155 also repressed TGFβR2 expression (Figure 2D,G), and the TGFβR2 mRNA was not changed with the transfection of mimics or inhibitors (Figure 2H). These data indicated that miR‐155 is an important post‐transcriptional regulator that negatively regulates TGFβR2 expression in GC cells. The EdU proliferation incorporation assay was used to measure the proliferation of SGC‐7901 cells.
3.5. MicroRNA‐155 promotes cell proliferation and migration
We next analyzed the biological consequences of the miR‐155‐driven repression of TGFβR2 expression in vivo. MicroRNA‐155 mimics, miR‐155 inhibitors, or NCs were transfected into the SGC7901 cell line. The EdU proliferation assay was used to measure the proliferation of SGC‐7901 cells. The results showed high levels of proliferation of SGC7901 cells transfected with miR‐155 mimics and low levels of proliferation of SGC7901 cells transfected with miR‐155 inhibitors compared with the NC groups. In contrast, the proliferation rate in SGC7901 cells transfected with miR‐155 inhibitors showed a sharp decrease compared to the corresponding NC inhibitor group (Figure 3). Based on these results, we concluded that miR‐155 as an oncogene promotes the proliferation of SGC7901 cells.
Figure 3.
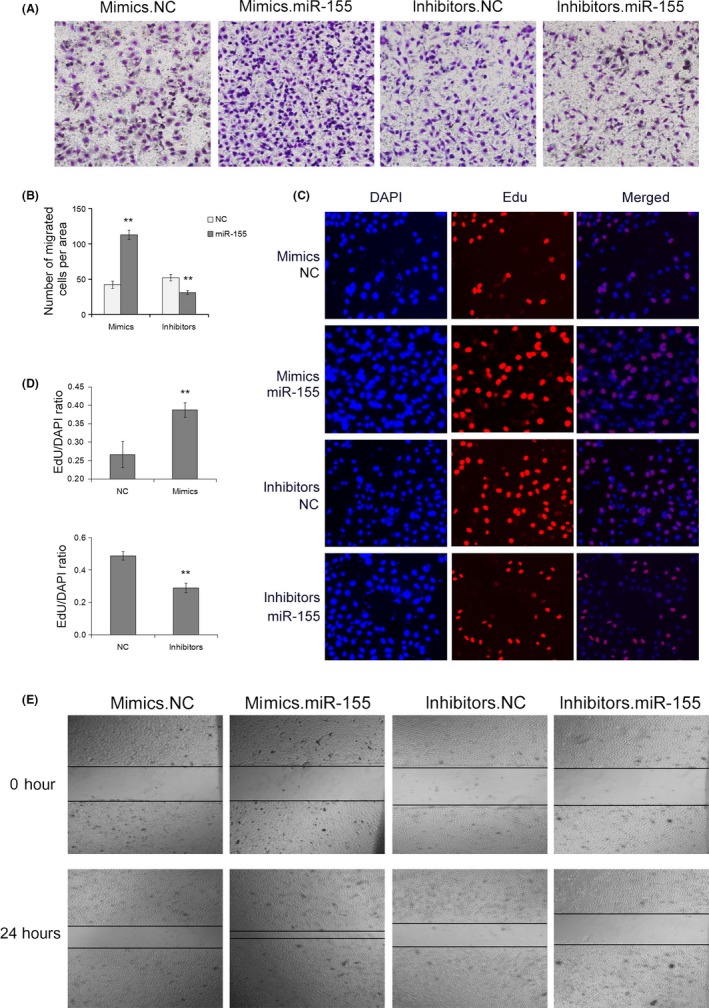
Oncogenic microRNA‐155 (miR‐155) promotes cell proliferation and migration of SGC7901 gastric cancer cells. A, Transwell assays show that miR‐155 promotes cell migration in SGC7901 cells (n = 3). B, Quantitative analysis of (A) (n = 3). C, 5‐Ethynyl‐2‐deoxyuridine (EdU) assays show that miR‐155 enhances cell proliferation of SGC7901 cells (n = 3). D, Quantitative analysis of (C) (n = 3). E, Validation of miR‐155‐mediated cell migration by wound healing assay (n = 3). **P < .01
To detect whether miR‐155 affects cell migration, we undertook wound healing and Transwell invasion assays (Figure 3A) using SGC‐7901 cells transfected with miR‐155 mimics, inhibitors, or NC. In the wound healing assay, overexpression of miR‐155 markedly accelerated cell migration at the edges of the scratch wound of SGC7901 cells, whereas downregulation of miR‐155 suppressed cell migration (Figure 3E). In accordance with the results of the wound healing assay, the Transwell invasion assay showed a higher ratio in migration of SGC7901 cells transfected with miR‐155 mimics, whereas inhibition of miR‐155 caused a decrease in cell migration. Taken together, results suggested that the function of miR‐155 promotes GC cell proliferation and migration.
3.6. Effects of TGFβR2 overexpression and silencing in SGC7901 cells
The biological function of TGFβR2 was explored by overexpression or silencing of TGFβR2 using plasmid or siRNA, respectively. The siRNA sequence that targets human TGFβR2 cDNA was used to knock down the expression of TGFβR2, and the plasmid that expresses the ORF of TGFβR2 was designed to overexpress TGFβR2. As shown in Figure 4, the expression levels of TGFβR2 protein was dramatically decreased in SGC7901 cells transfected with TGFβR2 siRNA compared with the control. Conversely, transfection with plasmid increased TGFβR2 protein expression (Figure 4C). Then the silencing and overexpressing efficiencies were assessed by qRT‐PCR for TGFβR2 protein and mRNA (Figure 4B,D). To further investigate the function of TGFβ2 in cell proliferation and migration, EdU proliferation, wound healing, and Transwell invasion assays were used to assess SGC7901 cells transfected with TGFβR2 siRNA or plasmid. The EdU assay results (Figure 5) showed a higher proliferation rate of SGC7901 cells transfected with TGFβR2 plasmid than NC control, whereas siRNA targeting TGFβR2 promoted proliferation. Subsequently, the downregulation of TGFβR2 significantly promoted SGC7901 cell migration (Figure 5A,E). The results indicated that TGFβR2 acts as a tumor suppressor; its downregulation can promote the proliferation and migration in GC cells.
Figure 4.
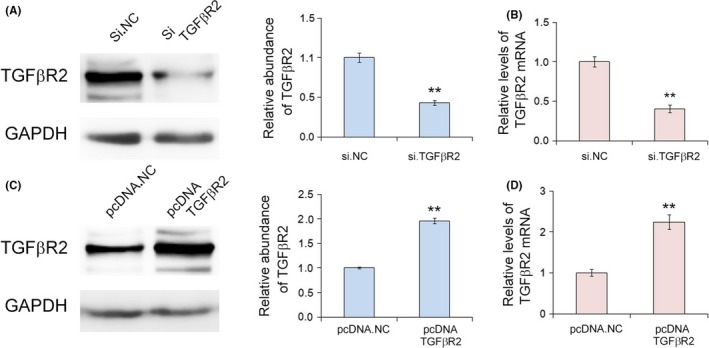
Effects of transforming growth factor‐β receptor 2 (TGFβR2) silencing and overexpression in SGC7901 gastric cancer cells. A, Western blot assay of TGFβR2 protein expression in SGC7901 cells transfected with TGFβR2 siRNA or control (si.NC). B, Quantitative RT‐PCR analysis of the relative levels of TGFβR2 protein and TGFβR2 mRNA in SGC7901 cells transfected with siRNA. C, Western blot assay of TGFβR2 protein expression in SGC7901 cells transfected with plasmid (n = 3). D, Quantitative RT‐PCR analysis of the relative levels of TGFβR2 protein and TGFβR2 mRNA in SGC7901 cells transfected with plasmid (n = 3) **P < .01
Figure 5.
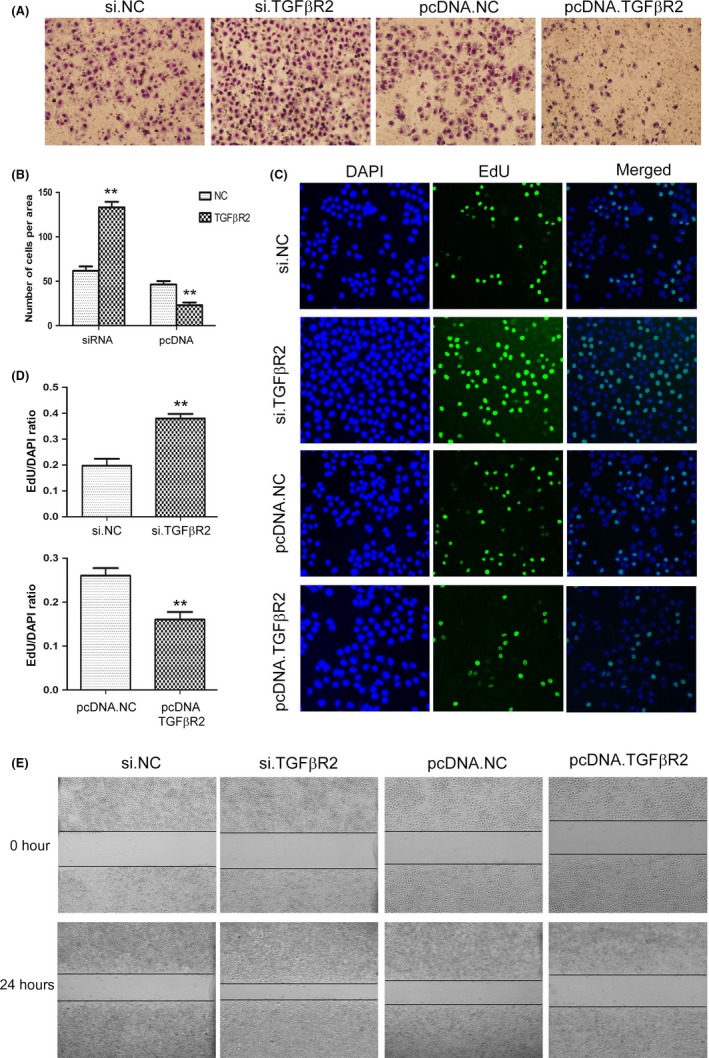
Overexpression or silencing of transforming growth factor‐β receptor 2 (TGFβR2) regulates cell proliferation and migration in gastric cancer (GC) cells. A, Transwell assays show that silencing of TGFβR2 enhances cell migration, whereas overexpression of TGFβR2 inhibits cell migration in SGC7901 cells (n = 3). B, Quantitative analysis of (A) (n = 3). C, 5‐Ethynyl‐2‐deoxyuridine (EdU) assays show that silencing of TGFβR2 promotes cell proliferation of GC cells, whereas overexpression of TGFβR2 inhibits cell proliferation (n = 3). D, Quantitative analysis of (C) (n = 3). E, Validation of TGFβR2‐mediated cell migration by wound healing assay. pcDNA.NC, control plasmid; pcDNA TGFβR2, overexpression plasmid (n = 3). **P < .01
3.7. In vivo role of miR‐155 and TGFβR2 in gastric cancer
We evaluated the function of miR‐155 and TGFβR2 in the growth of human GC cell xenografts in nude mice. The SGC7901 cells were treated with lentivirus particles to rapidly produce high intracellular levels of mature miR‐155 or TGFβR2, and cells were harvested and injected s.c. in the armpit of mice (Figure 6A). It was observed that the tumor sizes and weights were clearly increased in the miR‐155‐OE group compared with the control group, whereas the tumor growth was strongly inhibited in the TGFβR2‐overexpressing group (Figure 6B,C). The expression of miR‐155 and TGFβR2 in tumors was measured. MicroRNA‐155 was raised 10‐fold and TGFβR2 mRNA was raised 7‐fold in the corresponding groups (Figure 6D‐F). Immunohistochemical staining revealed lower TGFβR2 levels in tumors from the miR‐155‐overexpressing group, whereas tumors from the TGFβR2‐overexpressing group showed increased TGFβR2 (Figure 6G). The tumor‐implanted experiment offers a strong confirmation that the miR‐155–TGFβR2 pathway effectively regulates tumor growth in GC, and implies that inhibition of miR‐155 is a potential novel method for anti‐GC tumor therapy.
Figure 6.
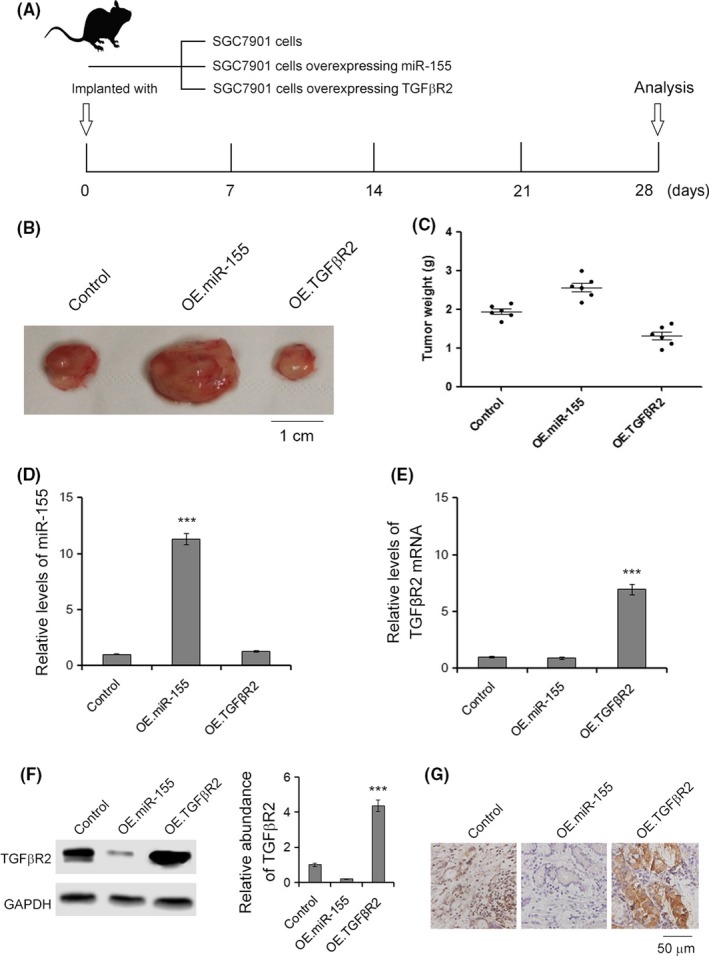
Effects of the microRNA‐155–transforming growth factor‐β receptor 2 (miR‐155/TGFβR2) pathway on gastric cancer tumor growth in vivo. A, Graphic description of the in vivo experiments. B, Morphology of the tumors from tumor‐implanted nude mice (n = 6). Scale bar = 1 cm. C, Weight of tumors excised from mice implanted with control SGC7901 cells, miR‐155‐overexpressing (OE.miR‐155) or TGFβR2‐overexpressing (OE.TGFβR2) SGC7901 cells (n = 6). D, Relative levels of miR‐155 in tumors (n = 6). E, Relative levels of TGFβR2 in tumors (n = 6). F, Western blot analysis of TGFβR2 in tumors (n = 6). G, Immunohistochemistry assays of TGFβR2 expression in tumors (n = 6). ***P < .001
4. DISCUSSION
MicroRNAs are involved in a series of tumorigenic processes, including cell proliferation, apoptosis, migration, and invasion. As reported in previous studies, many miRNAs are aberrantly expressed in GC tissues, serum, and cells, and these alterations are connected with the occurrence, progression, and prognosis of cancer.21, 22 As post‐transcriptional regulators, miRNAs affect the expression of a large number of targeted mRNAs that encode proteins. Thus, it is essential to identify the pathway connecting miRNAs with their target genes and regulated proteins. Novel potential molecular therapy can then be developed to target cancer.
MicroRNA‐155 has been found to be upregulated and act as a tumor promoter in cancer tissues and cell lines,23, 24 including gastric cancer;25, 26 however, its function and the relevant pathways concerned with TGFβR2 have not been sufficiently elucidated. In this study, an inverse correlation was found between miR‐155 and TGFβR2 after measuring the expression levels of miR‐155 and TGFβR2 in GC tissues and paired peritumoral gastric tissues. However, the mRNA level of TGFβR2 only slightly declined in GC tissues. Subsequently, we found that miR‐155 directly binding the region in the 3′‐UTR of TGFβR2 was highly conserved. Furthermore, overexpression of miR‐155 led to a decrease in the protein levels of TGFβR2, while miR‐155 inhibitors enhanced TGFβR2 expression in vitro. These results illustrated that miR‐155 acts as a direct upstream regulator of TGFβR2. Biological functional experiments showed that elevated expression of miR‐155 or silencing of TGFβR2 enhanced proliferation and migration of SGC7901 cells. Suppression of miR‐155 expression or plasmid‐mediated TGFβR2 overexpression inhibited proliferation and migration in vitro. Therefore, miR‐155 as an oncogene directly regulated expression of TGFβR2 and promoted cancer progression in GC. Our findings could serve to identify new molecular alterations while enriching the miRNA profiling of human GC. This novel pathway provided potential targets by repressing miR‐155 or enhancing TGFβR2 expression for the treatment of GC. MicroRNA‐155 is primarily expressed within lymphocytes and functions as an important regulator of the immune system.27 In the immune system, miR‐155 is unique in its ability to shape the transcriptome of activated myeloid and lymphoid cells controlling diverse biological functions ranging from inflammation to immunological memory. Previous studies have illustrated that miR‐155 played an oncogene role in GC, which is in accord with the present study. We also noticed that several miRNA profiling studies reported a downregulated alteration of miR‐155 in GC.28
The function of TGFβR2 in tumorigenesis is controversial. Previous research showed that TGFβR2 acted as a tumor inhibitor and was downregulated in cancer.29 In this study, TGFβR2 also functioned as a tumor suppressor and its levels were decreased in GC tissue. However, the other research showed that TGFβR2 acted as a proto‐oncogene; its expression was dramatically increased in non‐small‐cell lung cancer tissues compared with non‐neoplastic tissues, and overexpression of TGFβR2 was a significant risk factor for decreased overall survival and disease‐free survival in non‐small‐cell lung cancer patients.30 The inconsistent expression of miR‐155 and TGFβR2 might be explained by the differences in tumor classification, tumor staging, or immune functions. We should further address the biological functions of miR‐155 and TGFβR2 in GC in future studies.
It was validated that TGFβR2 was a direct target of miR‐155 in this study. Previous research reported that multiple downstream targets are regulated by miR‐155 in GC, such as c‐myc, SMAD1, signal transducer and activator of transcription 1, calcium binding protein 39, CXC chemokine receptor 4, and carbonic anhydrase 9,26, 31 all of which played vital roles in the evolution and progression of GC. These pathways indicated that one miRNA could target multiple genes, and 1 gene could also be regulated by more than 1 miRNA.
In conclusion, the present study showed that the upregulation of miR‐155 promoted GC progression by suppressing the expression of TGFβR2, and promoted proliferation and migration of GC cells. Furthermore, interferon regulatory transcription factor 2 is a direct downstream target of miR‐155 in GC. This novel pathway could provide a potential target for the treatment of GC.
CONFLICT OF INTEREST
The authors have no conflict of interest.
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81772629, 81602158, 81602156, 81702437, 81702431, 81702275, 81772843) and Tianjin health and family planning commission foundation of science and technology (15KG142). This work was also supported by Tianjin Science Foundation (No. 16PTSYJC00170). The funders had no role in study design; collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit this article for publication.
Qu Y, Zhang H, Sun W, et al. MicroRNA‐155 promotes gastric cancer growth and invasion by negatively regulating transforming growth factor‐β receptor 2. Cancer Sci. 2018;109:618–628. https://doi.org/10.1111/cas.13472
Funding Information
National Natural Science Foundation of China; Tianjin Health and Family Planning Commission Foundation of Science and Technology.
Yajing Qu, Haiyang Zhang, Wu Sun and Yueting Han contributed equally to this work.
Contributor Information
Guoguang Ying, Email: yingguoguang163@163.com.
Yi Ba, Email: bayi@tjmuch.com.
REFERENCES
- 1. Ferro A, Peleteiro B, Malvezzi M, et al. Worldwide trends in gastric cancer mortality (1980‐2011), with predictions to 2015, and incidence by subtype. Eur J Cancer. 2014;50:1330‐1344. [DOI] [PubMed] [Google Scholar]
- 2. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:9. [DOI] [PubMed] [Google Scholar]
- 3. Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287‐314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen WQ, Zheng RS, Zhang SW, Zeng HM, Zou XN. The incidences and mortalities of major cancers in China, 2010. Chin J Cancer. 2014;33:402‐405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moustakas A, Heldin CH. The regulation of TGFbeta signal transduction. Development. 2009;136:3699‐3714. [DOI] [PubMed] [Google Scholar]
- 6. Zhang Q, Rubenstein JN, Jang TL, et al. Insensitivity to transforming growth factor‐beta results from promoter methylation of cognate receptors in human prostate cancer cells (LNCaP). Mol Endocrinol. 2005;19:2390‐2399. [DOI] [PubMed] [Google Scholar]
- 7. Dong Z, Guo W, Guo Y, Kuang G, Yang Z. Concordant promoter methylation of transforming growth factor‐beta receptor types I and II occurs early in esophageal squamous cell carcinoma. Am J Med Sci. 2012;343:375‐381. [DOI] [PubMed] [Google Scholar]
- 8. Duan J, Zhang H, Qu Y, et al. Onco‐miR‐130 promotes cell proliferation and migration by targeting TGFbetaR2 in gastric cancer. Oncotarget. 2016;7:44522‐44533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guo W, Dong Z, Guo Y, Kuang G, Yang Z, Shan B. Concordant repression and aberrant methylation of transforming growth factor‐beta signaling pathway genes occurs early in gastric cardia adenocarcinoma. Mol Biol Rep. 2012;39:9453‐9462. [DOI] [PubMed] [Google Scholar]
- 10. Nadauld LD, Garcia S, Natsoulis G, et al. Metastatic tumor evolution and organoid modeling implicate TGFBR2 as a cancer driver in diffuse gastric cancer. Genome Biol. 2014;15:014‐0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857‐866. [DOI] [PubMed] [Google Scholar]
- 12. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281‐297. [DOI] [PubMed] [Google Scholar]
- 13. Ambros V. The functions of animal microRNAs. Nature. 2004;431:350‐355. [DOI] [PubMed] [Google Scholar]
- 14. Ivey KN, Srivastava D. microRNAs as Developmental Regulators. Cold Spring Harb Perspect Biol. 2015;7:a008144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu R, Zhang H, Wang X, et al. The miR‐24‐Bim pathway promotes tumor growth and angiogenesis in pancreatic carcinoma. Oncotarget. 2015;6:43831‐43842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fan Q, Meng X, Liang H, et al. miR‐10a inhibits cell proliferation and promotes cell apoptosis by targeting BCL6 in diffuse large B‐cell lymphoma. Protein Cell. 2016;7:899‐912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yan X, Chen X, Liang H, et al. miR‐143 and miR‐145 synergistically regulate ERBB3 to suppress cell proliferation and invasion in breast cancer. Mol Cancer. 2014;13:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ni J, Yang Y, Liu D, Sun H, Jin S, Li J. MicroRNA‐429 inhibits gastric cancer migration and invasion through the downregulation of specificity protein 1. Oncol Lett. 2017;13:3845‐3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shi J, Chen P, Sun J, et al. MicroRNA‐1258: an invasion and metastasis regulator that targets heparanase in gastric cancer. Oncol Lett. 2017;13:3739‐3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang J, Li L, Jiang M, Li Y. MicroRNA‐195 inhibits human gastric cancer by directly targeting basic fibroblast growth factor. Clin Transl Oncol. 2017;12:017‐1668. [DOI] [PubMed] [Google Scholar]
- 21. Zhou J, Zhang Y, Qi Y, Yu D, Shao Q, Liang J. MicroRNA‐152 inhibits tumor cell growth by directly targeting RTKN in hepatocellular carcinoma. Oncol Rep. 2017;37:1227‐1234. [DOI] [PubMed] [Google Scholar]
- 22. Sekar D, Krishnan R, Thirugnanasambantham K, Rajasekaran B, Islam VI, Sekar P. Significance of microRNA 21 in gastric cancer. Clin Res Hepatol Gastroenterol. 2016;40:538‐545. [DOI] [PubMed] [Google Scholar]
- 23. Shen R, Wang Y, Wang CX, et al. MiRNA‐155 mediates TAM resistance by modulating SOCS6‐STAT3 signalling pathway in breast cancer. Am J Transl Res. 2015;7:2115‐2126. [PMC free article] [PubMed] [Google Scholar]
- 24. He F, Cai L. [Meta analysis of the association between the expression of microRNA‐155 and the outcome of patients with lung cancer]. Wei Sheng Yan Jiu. 2014;43:1004‐1008. [PubMed] [Google Scholar]
- 25. Zhang T, Liu C, Huang S, Ma Y, Fang J, Chen Y. A Downmodulated MicroRNA Profiling in Patients with Gastric Cancer. Gastroenterol Res Pract. 2017;1526981:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Feng JA. Target research on tumor biology characteristics of mir‐155‐5p regulation on gastric cancer cell. Pak J Pharm Sci. 2016;29:711‐718. [PubMed] [Google Scholar]
- 27. Vigorito E, Kohlhaas S, Lu D, Leyland R. miR‐155: an ancient regulator of the immune system. Immunol Rev. 2013;253:146‐157. [DOI] [PubMed] [Google Scholar]
- 28. Gu J, Lu Z, Ji C, et al. Melatonin inhibits proliferation and invasion via repression of miRNA‐155 in glioma cells. Biomed Pharmacother. 2017;93:969‐975. [DOI] [PubMed] [Google Scholar]
- 29. Zhang W, Zeng Z, Fan S, et al. Evaluation of the prognostic value of TGF‐beta superfamily type I receptor and TGF‐beta type II receptor expression in nasopharyngeal carcinoma using high‐throughput tissue microarrays. J Mol Histol. 2012;43:297‐306. [DOI] [PubMed] [Google Scholar]
- 30. Han Y, Jia C, Cong X, et al. Increased Expression of TGFbetaR2 Is Associated with the Clinical Outcome of Non‐Small Cell Lung Cancer Patients Treated with Chemotherapy. PLoS One. 2015;10:e0134682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sun S, Sun P, Wang C, Sun T. Downregulation of microRNA‐155 accelerates cell growth and invasion by targeting c‐myc in human gastric carcinoma cells. Oncol Rep. 2014;32:951‐956. [DOI] [PubMed] [Google Scholar]


