Abstract
With increasing uses of poly(ADP‐ribose) polymerase (PARP) inhibitors (PARPi) for cancer therapy, understanding their resistance is becoming urgent. However, acquired PARPi resistance in the phosphatase and tensin homolog (PTEN)‐deficient background is poorly understood. We generated 3 PARPi‐resistant PTEN‐deficient glioblastoma U251 variants separately with olaparib (U251/OP), talazoparib (U251/TP) and simmiparib (U251/SP). These variants displayed consistent resistance (2.46‐71.78‐fold) to all 5 PARPi, including niraparib and rucaparib, and showed higher degrees of resistance to the PARPi to which the parental cells were more sensitive. The resistance was characteristic of fast emergence and high stability. However, the resistance acquirement did not cause an increasingly aggressive phenotype. The resistance was not correlated to various factors, including PTEN mutations. The PARPi‐treated variants produced less γH2AX and G2/M arrest. Consistently, loss of 53BP1 occurred in all variants and its compensation enhanced their sensitivity to PARPi by approximately 76%. The variants revealed slightly different cross‐resistance profiles to 13 non‐PARPi anticancer drugs. All were resistant to Ara‐C (6‐8‐fold) but showed differential resistance to 5‐fluorouracil, gemcitabine and paclitaxel. Almost no resistance was observed to the rest drugs, including cisplatin. SAMHD1 was overexpressed in all the variants and its knockout completely restored their sensitivity to Ara‐C but did not affect their PARPi sensitivity. The present study demonstrates a consistent resistance profile to PARPi and a unique cross‐resistance profile to non‐PARPi drugs in different PARPi‐resistant U251 cells and reveals 53BP1 loss and SAMHD1 overexpression as the primary mechanisms responsible for their resistance to PARPi and Ara‐C, respectively. These effects probably result from heritable gene change(s) caused by persistent PARPi exposure.
Keywords: 53BP1, acquired resistance, phosphatase and tensin homolog deficiency, poly(ADP‐ribose) polymerase inhibitors, SAMHD1
1. INTRODUCTION
Poly(ADP‐ribose) polymerases (PARP) are important DNA repair enzymes. PARP inhibitors (PARPi), including olaparib, rucaparib and niraparib, have been used to treat ovarian cancers defective in DNA homologous recombination repair (HRR) in the clinic.1, 2, 3, 4, 15 Cancer cells have been shown to develop drug resistance to PARPi,1 suggesting that their clinical resistance might inevitably emerge from persistent/repeated treatments. Several mechanisms of PARPi resistance have been reported, including restoration of HRR function due to secondary mutation on BRCA alleles, stabilization of BRCA1 mutant protein, loss of 53BP1, RIF1 or REV7, loss of PARP‐1 and increased drug efflux.1 However, many issues on tumor drug resistance to PARPi remain to be clarified.
The present knowledge about PARPi resistance comes primarily from the studies on the olaparib‐induced resistance in BRCA or ATM‐defective cancer cells.1 We have only very limited understanding regarding the characteristics of drug resistance of the same cancer cells to different PARPi and their cross‐resistance profiles. Moreover, little is known about the characteristics and mechanisms of PARPi resistance in non‐BRCA/ATM‐defective cancer cells. There have been 3 PARPi approved for clinical cancer therapy and many others in clinical trials.1, 4, 5 Moreover, PARPi will probably be used to treat non‐BRCA‐defective cancers in the future. Therefore, the answers to these issues will be critical to understanding, monitoring, delaying and even preventing PARPi resistance in different clinical settings.
Phosphatase and tensin is one of the most frequently mutated genes in a wide range of hereditary and sporadic human tumors, including glioblastoma. Up to 70% of glioblastoma patients are defective for PTEN due to loss of 1 allele of the long arm of chromosome 10 on which PTEN is located.6 PTEN is important for the expression of RAD51, a critical HRR factor, and, thus, PTEN deficiency results in defective HRR.7 PTEN‐deficient tumor cells are sensitive to PARPi.8, 9, 10, 11 For example, PTEN‐deficient glioblastoma U87MG and U251 cells show different degrees of sensitivity to olaparib,12, 13, 14 simmiparib13 and mefuparib hydrochloride.14 The U251 cell line has been used extensively and its sensitivity to PARPi, including olaparib and simmiparib, is higher than that of the U87MG cell line.13 Olaparib is the first approved PARPi,1 talazoparib has been shown to be the most potent PARPi so far10 and is undergoing phase III clinical trials,15 and simmiparib is one of new PARPi developed by our institute and is currently in phase I clinical trials.13 These 3 inhibitors may represent the existing PARPi that possess different characteristics and are in different clinical stages.
In the present study, we separately exposed U251 cells to olaparib, talazoparib and simmiparib, and after approximately 4 months, obtained their corresponding drug‐resistant variants that were denoted by U251/OP, U251/TP and U251/SP, respectively. Then we examined their features of morphology, growth, migration and invasion, resistance to different PARPi and cross‐resistance to conventional anticancer drugs. After that, we investigated the possible mechanisms responsible for the drug resistance of PTEN‐deficient glioblastoma U251 cells to PARPi and non‐PARPi drugs. Our results reveal some important new aspects of PARPi resistance, which provide new insight into this promising class of anticancer drugs.
2. MATERIALS AND METHODS
2.1. Cell culture
U251 cells were purchased from the Institute of Cell Biology (Shanghai, China). KB/VCR cells were from the Sun Yat‐Sen University of Medical Sciences (Guangzhou, China). HUVEC cells were from American Type Culture Collection (ATCC, Manassas, VA, USA). All cell lines were cultured at the conditions specified by the suppliers.
2.2. Drugs
2.3. Proliferation inhibition assays
Proliferation inhibition was determined by sulforhodamine B (SRB) assay, as described previously.17
2.4. Growth rate assays
The growth rate of both U251 parental and resistant cells was determined using SRB assays.18
2.5. Transwell migration and invasion assays
Details are provided in Document S1.
2.6. DNA sequencing
Details are provided in Document S1.
2.7. Western blotting
Western blotting was performed using the standard procedure. Details are provided in Document S1.
2.8. Cell cycle assays
Cell cycle arrest was analyzed by propidium iodide‐staining‐based flow cytometry as described previously.13
2.9. Immunofluorescence
Details are provided in Document S1.
2.10. Transfection with 53BP1 plasmids
Details are provided in Document S1.
2.11. Quantitative real‐time PCR
Details are provided in Document S1.
2.12. Stable knockout of SAMHD1 with the CRISPR/Cas9 technique
Details are provided in Document S1.
2.13. Statistical analysis
All the data, if applicable, were presented as mean ± SD. Comparison between 2 groups was performed with Student's t‐test. P < .05 was considered statistically significant.
3. RESULTS
3.1. Modeling acquired resistance of phosphatase and tensin homolog‐deficient glioblastoma U251 cells to poly(ADP‐ribose) polymerase inhibitors
To investigate the acquired resistance of PTEN‐deficient glioblastoma cells to PARPi, we generated a series of PARPi‐resistant cells by separately exposing PTEN‐deficient glioblastoma U251 cells to olaparib, talazoparib and simmiparib. Their concentrations started from 5 μmol L−1 (olaparib), 0.5 μmol L−1 (talazoparib) and 0.1 μmol L−1 (simmiparib) and then stepwisely increased up to 40, 2 and 2 μmol L−1, respectively (Figure1A). After 112‐day (olaparib), 117‐day (talazoparib) and 88‐day (simmiparib) exposure, the cells acquired 10.07, 24.62 and 30.05‐fold resistance to the corresponding PARPi and were denoted by U251/OP, U251/TP and U251/SP, respectively (Figures 1A,B and S1A). Moreover, all 3 variants showed cross‐resistance to other PARPi that were not used to establish the corresponding resistant cells, including olaparib, talazoparib, simmiparib, niraparib and rucaparib, with resistance factor ranges of 8.32‐9.85, 6.37‐27.28, 51.09‐71.78, 7.38‐14.72 and 2.46‐4.35, respectively (Figures 1C and S1A). The averaged IC50 values of talazoparib and simmiparib are 0.43 and 1.27 μmol L−1, respectively, lower than those of olaparib (7.74 μmol L−1), rucaparib (4.21 μmol L−1) and niraparib (3.30 μmol L−1) in U251 cells; in contrast, the averaged resistance factors of the former two (19.42 for talazoparib and 50.97 for simmiparib) are greater than those of the latter three (9.41 for olaparib, 3.41 for rucaparib and 10.61 for niraparib) in the resistant variants. The data show that the resistant variants are more resistant to the PARPi to which the parental U251 cells are more sensitive. Notably, once generated, the resistant cells did not need to be maintained with PARPi and did not lose their drug resistance. This result indicates the stability of their PARPi resistance, which, once acquired, will become independent of their corresponding PARPi used to generate them.
Figure 1.
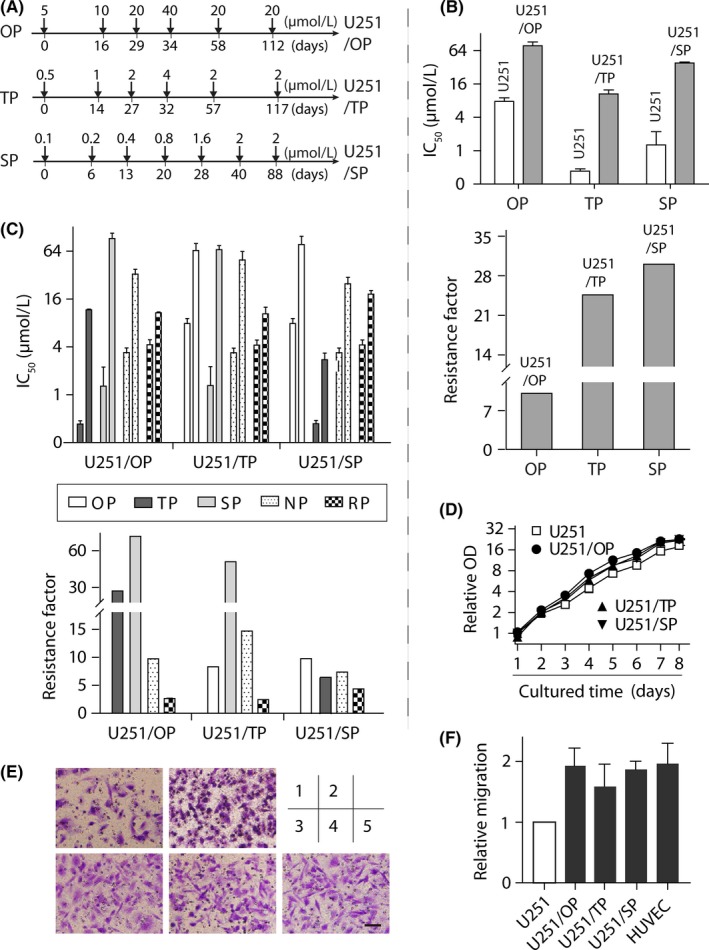
Generation of poly(ADP‐ribose) polymerase inhibitors (PARPi)‐resistant variants and their resistance profiles to PARPi. A, workflow flowcharts used to generate PARPi‐resistant cells. Phosphatase and tensin homolog (PTEN)‐deficient glioblastoma U251 cells were separately exposed to increasing concentrations of olaparib (OP), talazoparib (TP) and simmiparib (SP). The resulting PARPi‐resistant variants were denoted by U251/OP, U251/TP and U251/SP, respectively. B,C, PARPi‐resistance profiles. Cells were exposed to olaparib (OP), talazoparib (TP), simmiparib (SP), niraparib (NP) or rucaparib (RP) for 7 days and then subjected to sulforhodamine B (SRB) assays. The IC 50 value was expressed as mean ± SD from 3 separate experiments and the resistance factor was calculated as the ratio of the averaged IC 50 value of the indicated PARPi in the given resistant cells to that of the same PARPi in the parental U251 cells. B shows the resistance of a given PARPi‐resistant variant to the PARPi that was used to establish the variant while C indicates the cross‐resistance to the PARPi that were not used to establish the indicated variant. D, Growth curves of the resistant and parental U251 cells. Samples were determined by the SRB assay and their optic density (OD) values were normalized using that of the parental U251 group on day 1 (relative OD). E, representative images of cell migration from 3 independent Transwell migration assays. 1, U251; 2, HUVEC; 3, U251/OP; 4, U251/TP; 5, U251/SP. Scale bar: 50 μm. F, Quantiffication results of E. Relative migration was calculated in relation to the migration of parental U251 cells. HUVEC cells served as the positive control
There were no apparent differences in cell morphology observed between parental and PARPi‐resistant U251 cells (data not shown but readers may refer to Figures 1E and S1B). However, all resistant cells grew slightly faster than the parental cells with the doubling time of 17.94, 17.80, 18.22 and 20.17 hour for U251/OP, U251/TP, U251/SP and U251 cells, respectively (Figure 1D), although they were not statistically different. In addition, the migration abilities of the resistant cells were increased by 1.93 (U251/OP), 1.59 (U251/TP) and 1.87 (U251/SP)‐fold relative to that of their parental cells (Figure 1E,F), which had no statistical difference either. The invasion capacity of the resistant cells was similar to that of the parental cells (Figure S1C).
3.2. Acquired resistance is not associated with poly(ADP‐ribose) polymerase inhibition, phosphatase and tensin homolog deficiency and drug transporters
The majority of present PARPi can inhibit multiple members in the PARP family, among which PARP1, PARP2, PARP3 and Tankyrase1/2 have been revealed to participate in DNA repair and integrity maintenance.1, 2 In the PARPi‐resistant variants, however, the base protein levels of these PARP members were not found to change (Figure2A). In resistant and parental U251 cells, moreover, there were no significant differences in the time‐dependent PAR formation driven by either MMS or H2O2 and in the suppression of PAR formation by the corresponding PARPi used to establish the resistant variants (Figures 2B,C and S2A‐C). The data suggest that these PARP family members do not contribute to the acquired resistance in these PARPi‐resistant variants.
Figure 2.
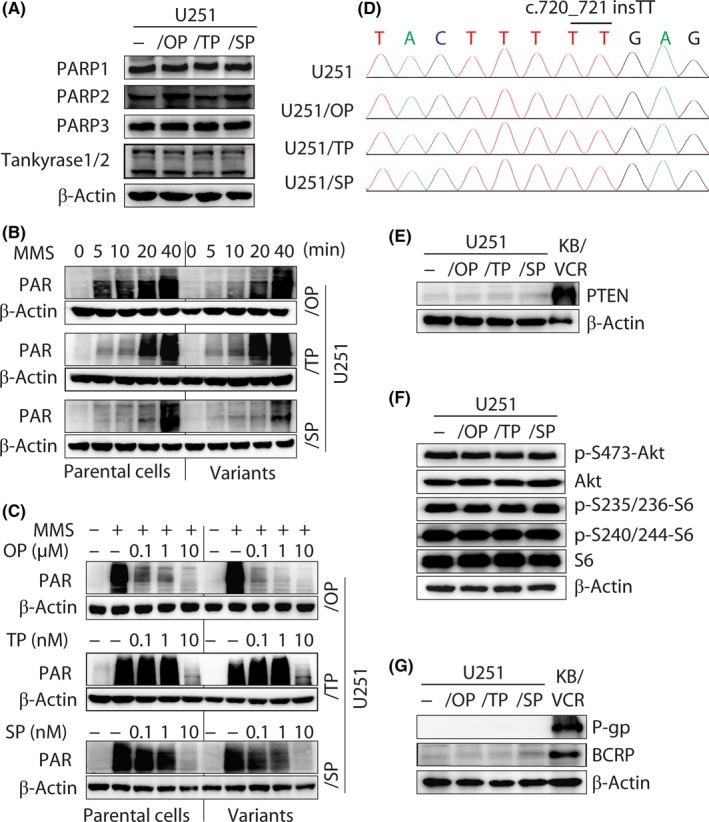
Poly(ADP‐ribose) polymerase (PARP) expression and inhibition, phosphatase and tensin homolog (PTEN) deficiency and drug‐transporter expression in PARP inhibitor (PARPi)‐resistant variants. A, protein levels of PARP1, PARP2, PARP3 and Tankyrase1/2 detected by western blotting in PARPi‐resistant variants and parental U251 cells. B, PAR formation induced by the DNA damage agent MMS (0.01%). C, PARPi inhibited the formation of PAR triggered by MMS (0.01%). D, DNA sequencing revealed the same homozygous c.720_721 insTT of PTEN existing in all 3 PARPi‐resistant variants and parental U251 cells. E,F, Protein levels of PTEN (E) and its downstream signaling factors (F) determined by western blotting. KB/VCR cells served as the positive control of PTEN expression (E). g, protein levels of drug transporters P‐gp and BCRP were detected by western blotting. KB/VCR cells served as the positive control. OP, olaparib; SP, simmiparib; TP, talazoparib
Secondary mutations on mutated BRCA1 or BRCA2 have been shown to be potential PARPi‐resistant mechanisms.1, 19, 20 The U251 cell line harbors a protein‐truncating c.720_721 insTT frameshift mutation, which results in lack of PTEN expression.21 This frameshift mutation on PTEN (c.720_721 insTT) was retained in all the 3 PARPi‐resistant variants revealed by whole‐exome sequencing (Figure 2D). Indeed, the resistant and parental cells did not express the PTEN protein (Figure 2E). In these cells, moreover, all the exons of BRCA1, BRCA2, ATM and TP53 showed no differences (Table S1). PTEN regulates the Akt‐mTOR‐ribosomal protein S6 signaling axis and phosphorylation of S6 has been reported to participate in PARPi resistance.22 However, the levels of the components in this signaling axis, including Akt, p‐S473‐Akt and S6, p‐S235/236‐S6 or p‐S240/244‐S6, showed no differences in the resistant and parental cells (Figure 2F). The result indicates that PARPi resistance is independent of the PTEN‐Akt‐mTOR‐S6 signaling axis in these variants.
In addition, overexpression of drug transporters such as P‐gp might contribute to olaparib resistance.1 However, all the PARPi‐resistant variants, just as their parental cells, did not express P‐gp or BCRP (Figure 2G).
3.3. Poly(ADP‐ribose) polymerase inhibitors resistance leads to decreased cell cycle arrest and DNA damage induced by poly(ADP‐ribose) polymerase inhibitors
Olaparib elicited G2/M arrest in the parental U251 cells. In contrast, the PARPi caused reduced G2/M arrest in the corresponding resistant variants (Figure 3A,B). Consistently, the treatment with olaparib led to more apparent increase in the levels of γH2AX, a marker of DNA double‐strand breaks (DSB), in the parental cells than in U251/OP cells (Figure 3C). Olaparib also promoted the formation of γH2AX foci in the parental and resistant cells with significant differences. At 1 μM olaparib, γH2AX‐positive cells accounted for 37.63% in the parental group but only 7.75% in the resistant group (Figure 3D,E). Moreover, when exposed to ionizing irradiation, U251/OP cells accumulated much less γH2AX over time than U251 cells (Figure 3F). Similar results including G2/M arrest and γH2AX accumulation were obtained in the other 2 PARPi‐resistant variants (U251/TP and U251/SP) when they were treated separately with talazoparib, simmiparib or ionizing irradiation (Figures S3 and S4).
Figure 3.
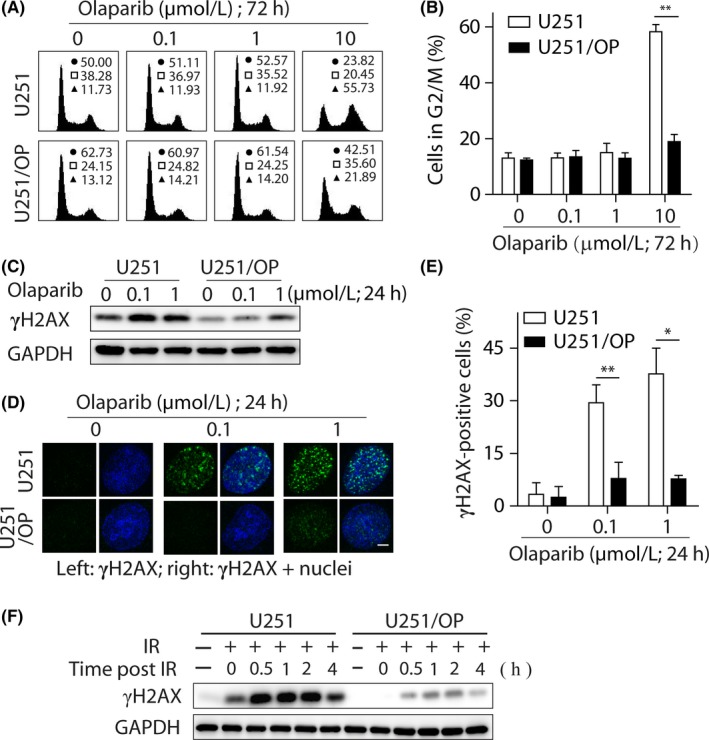
Cell cycle arrest and γH2AX accumulation in U251/OP cells exposed to olaparib. A,B, G2/M arrest induced by olaparib was decreased determined by FACS. A, Representative histograms; in the inserts, ●, □ and ▲ show the percentage of cells in G0/G1, S and G2/M phases, respectively. B, Percentage of cells in the G2/M phase expressed as mean ± SD from 3 independent experiments. **P < .01. C‐E, Levels of cellular γH2AX were reduced in U251/OP cells exposed to olaparib. Both U251 and U251/OP cells were treated with olaparib for 24 hours and then subjected to western blotting (C) and confocal microscopy (D). Scale bar in d: 5 μm. Cells that contained 5 or more γH2AX foci per nucleus were considered γH2AX‐positive cells. At least 50 cells were analyzed in each group. The data were expressed as mean ± SD from 3 independent experiments (E). *P < 0.05; **P < .01. F, Levels of cellular γH2AX induced by irradiation (IR; 10 Gy) were decreased in U251/OP cells determined by western blotting
3.4. Levels of 53BP1 are reduced in poly(ADP‐ribose) polymerase inhibitor‐resistant variants and its exogenous expression partially restored poly(ADP‐ribose) polymerase inhibitor sensitivity
Previous studies have shown that loss of 53BP1, REV7, KU70 or PTIP is implicated in PARPi resistance.1 Other proteins that separately participate in repairing different forms of DNA damage, such as BRCA1, Rad51, SMC1, RPA32, Ku80, ATM, ATR, MSH2 and MLH1, might also contribute to PARPi resistance.1 In the 3 PARPi‐resistant variants, however, we found that only the levels of 53BP1 were reduced consistently while the levels of all the other examined proteins basically remained unchanged (Figure 4A). Transfection with the full‐length 53BP1 cDNA completely restored the levels of 53BP1 in the resistant variants up to that in the parental U251 cells (Figure 4B). The exogenous 53BP1 expression also enhanced the levels of γH2AX in U251/OP+53BP1 cDNA cells when exposed to olaparib (Figure 4C). Consistently, the ectopic expression of 53BP1 restored, significantly although not completely, the sensitivity of U251/OP cells to PARPi, including olaparib, talazoparib, simmiparib and niraparib (Figure 4D,E). Their resistance factors reduced from 10.07, 27.28, 71.78 and 9.71‐3.60, 6.68, 23.91 and 2.68, respectively; in other words, 53BP1 compensation caused a resistance reduction up to 64.25%‐75.51%. Similar results were obtained in the other 2 resistant variants (U251/TP and U251/SP) (Figure S5 and S6).
Figure 4.
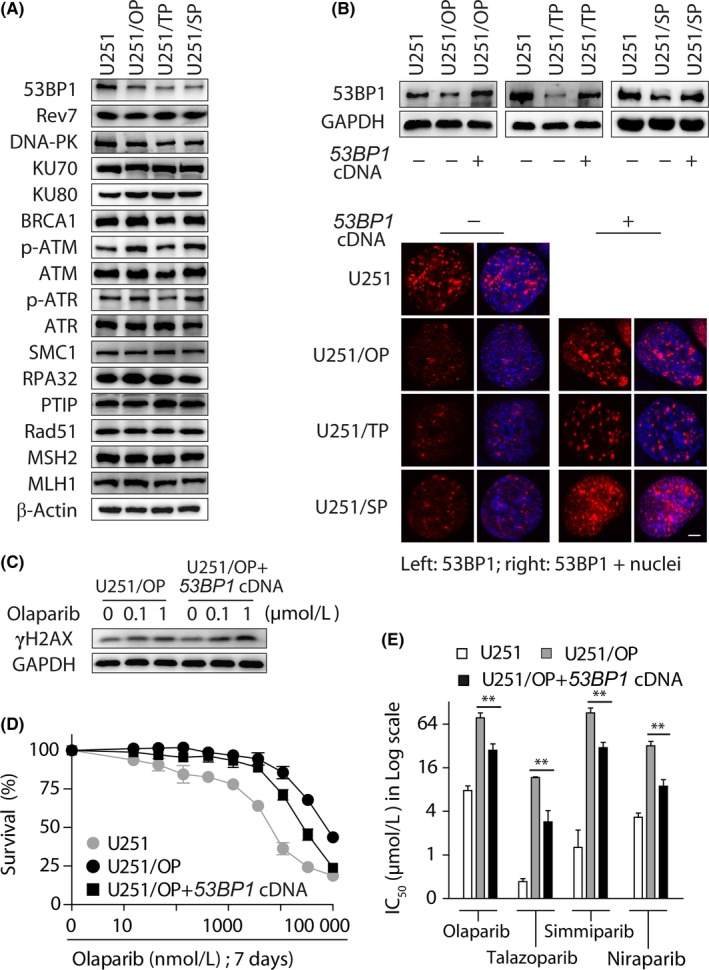
Exogenous expression of 53BP1 partially restored the sensitivity of poly(ADP‐ribose) polymerase inhibitors (PARPi)‐resistant variants to PARPi. A, Levels of DNA repair‐related proteins in the parental and resistant U251 cells determined by western blotting. B, After U251/OP, U251/TP and U251/SP cells were transfected with the full‐length 53BP1 cDNA, the levels of 53BP1 protein were determined by western blotting (upper) and immunofluorescence (lower). Scale bar: 5 μm. C, Accumulation of γH2AX triggered by olaparib was increased in U251/OP+53BP1 cDNA cells relative to U251/OP cells. D,E, Exogenous expression of 53BP1 in U251/OP cells partially restored olaparib sensitivity. D, Survival curves of olaparib‐treated U251, U251/OP and U251/OP+53BP1 cDNA cells. E, IC 50 in the log scale. **P < .01
3.5. Poly(ADP‐ribose) polymerase inhibitor‐resistant variants are cross‐resistant to Ara‐C, which is correlated to the increased SAMHD1
Some PARPi‐resistant cells have been reported to be cross‐resistant to conventional anticancer drugs such as cisplatin.20 Therefore, to further understand the cross‐resistance profile of PARPi‐resistant U251 variants to non‐PARPi anticancer drugs, we evaluated the proliferative inhibition of these drugs, including the antimetabolite drugs Ara‐C, methotrexate, hydroxyurea, 6‐mercaptopurine, gemcitabine and 5‐fluorouracil, the RNA synthesis inhibitor actinomycin D, the topoisomerase inhibitors doxorubicin, etoposide and SN‐38, the alkylating agent temozolomide, cisplatin and the microtubulin inhibitor paclitaxel in the resistant and parental cells. The results showed that all 3 resistant U251 variants displayed similar cross‐resistance to Ara‐C with 6.81‐7.12‐fold reduced sensitivity relative to the parental cells (Figure 5A). The resistance of U251/OP cells to 5‐fluorouracil and paclitaxel and U251/SP cells to gemcitabine and paclitaxel reached more than 3‐fold; however, U251/TP cells were almost not resistant to the 3 drugs with the resistance factors of 0.57 for paclitaxel, 1.55 for 5‐fluorouracil and 2.44 for gemcitabine. In contrast, the resistance of all the 3 resistant variants to the rest tested drugs including cisplatin was lower than 2‐fold (Figure 5A). In contrast to the resistance profile of the PARPi‐resistant variants to PARPi, therefore, our data showed a unique cross‐resistance profile to those non‐PARPi anticancer drugs.
Figure 5.
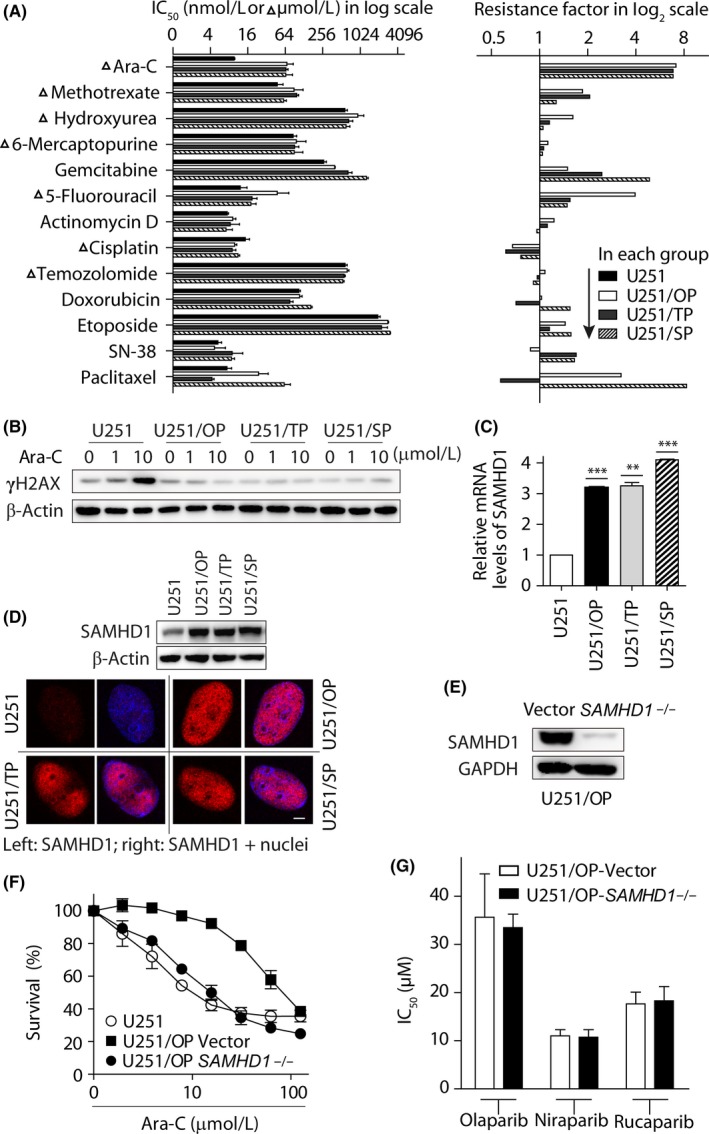
Upregulation of SAMHD1 was associated with the resistance of poly(ADP‐ribose) polymerase inhibitors (PARPi)‐resistant variants to Ara‐C. A, Cells were treated with the indicated drugs for 72 hours and then subjected to sulforhodamine B (SRB) assays. IC 50 values (left panel) were calculated from 3 independent experiments and expressed as mean ± SD. The resistance factor (right panel) is the ratio of the averaged IC 50 value of the indicated drug in the given resistant cells to that of the same drug in the parental U251 cells. b, Levels of γ‐H2AX induced by Ara‐C reduced in the resistant cells determined by western blotting. C, mRNA level of SAMHD1 was detected by qRT‐PCR and normalized using that of the parental U251 cells. **P < .01; ***P < .001. D, Protein level (upper) and distribution (lower) of SAMHD1 detected by western blotting and immunofluorescence, respectively. Scale bar: 5 μm. E, CRISPR/Cas9‐mediated knockout of SAMHD1 in U251/OP cells was confirmed by western blotting. f,g, cells were treated with Ara‐C for 3 days (F) or PARPi for 7 days (G) and then subjected to SRB assays. Data from 3 independent experiments were expressed as mean ± SD
Consistent with its drug resistance, Ara‐C induced much less γH2AX in the 3 resistant variants than in the parental U251 cells (Figure 5B). SAMHD1, a deoxynucleoside triphosphate (dNTP) triphosphohydrolase, has been shown to be a biomarker for Ara‐C sensitivity in acute myeloid leukemia.23, 24 Interestingly, all 3 resistant variants expressed much more SAMHD1 mRNA (Figure 5C) and protein (Figure 5D) than the parental cells. Importantly, depletion of SAMHD1 by using the CRISPR/Cas9 technique (Figure 5E) almost completely restored the sensitivity of U251/OP to Ara‐C (Figure 5F). Similar results were obtained in both U251/TP and U251/SP cells (Figure S7A). However, depletion of SAMHD1 did not sensitize the PARPi‐resistant cells to PARPi (Figures 5G and S7B).
4. DISCUSSION
As PARPi are increasingly being used for cancer therapy, understanding their resistance is becoming more and more significant and urgent. In fact, clinical resistance to PARPi, including olaparib,25 rucaparib26 and veliparib,27 has been reported. In addition, PARPi will probably extend their application to non‐BRCA/ATM‐mutated cancers.1 Although different PARPi, such as olaparib, rucaparib, niraparib and talazoparib, have similar PARP1 inhibition activity, they display differential capacity in trapping PARP1‐DNA complexes.1, 28 Moreover, different from other PARPi, niraparib elicits its anticancer effects appearing in a manner of relatively low dependency on HR deficiency in either animal models29 or patients.30 However, current studies primarily focus on the resistance to only one PARPi, for example, olaparib. Here, we present the first report on the resistance of PTEN‐deficient glioblastoma cells to three different PARPi simultaneously.
In this study, we generated 3 different PARPi‐resistant variants of U251 cells (U251/OP, U251/TP and U251/SP) with olaparib, talazoparib and simmiparib by concentration‐increased induction. In addition to the PARPi that was used to establish the corresponding resistant cells, all these variants were cross‐resistant to the other tested PARPi; that is, olaparib, rucaparib, niraparib, talazoparib and simmiparib. Notably, the PARPi resistance profile reveals that the resistant variants appear to possess higher degrees of resistance to the PARPi to which the parental U251 cells are more sensitive. However, this might not indicate that the cancers resistant to the former can be treated with the latter because anticancer drugs are generally used clinically at their maximum tolerated doses. It is also worthwhile noting that the PARPi resistance in our case is characteristic of fast emergence (approximately 4 months) and high stability and, once acquired, it is persistent and independent of the resistance‐induced PARPi. This characteristic indicates that while acquiring the PARPi resistance, the cells are likely to develop heritable changes at their gene levels that contribute to their resistance to PARPi. Fortunately, however, the acquirement of PARPi resistance seems not to confer an increasingly aggressive phenotype to the U251 variants because the resistant and parental cells showed no significant differences in cell morphology, growth rate, migration or invasion in vitro.
In contrast to their similar resistance to PARPi, the 3 PARPi‐resistant U251 variants display slightly different cross‐resistance profiles to 13 tested non‐PARPi anticancer drugs. All 3 variants show consistent resistance to the antimetabolite drug Ara‐C but no apparent resistance to methotrexate, hydroxyurea, 6‐mercaptopurine, actinomycin D, doxorubicin, etoposide, SN‐38, temozolomide and cisplatin. However, U251/OP cells are resistant to 5‐fluorouracil and paclitaxel, and U251/SP cells are resistant to gemcitabine and paclitaxel but U251/TP cells are almost not resistant to the 3 drugs. Understanding the non‐PARPi cross‐resistance profiles and their differences might be meaningful not only for clinical drug selections for patients with PARPi‐resistant cancers but also for explorations on the drug resistance mechanisms. In contrast, some drugs in our test such as cisplatin have been reported to be cross‐resistant to PARPi in both clinical27 and preclinical studies.31, 32, 33, 34 However, cisplatin was shown not to be cross‐resistant to rucaparib in ovarian cancers defective for nonhomologous end joining.35 All our PARPi‐resistant PTEN‐deficient variants are not resistant to cisplatin (resistance factor: 0.62‐0.76) either. Such inconsistent results further suggest that different genetic backgrounds of cancers and different PARPi might have unique cross‐resistance profiles.
Mechanistic studies reveal that all the PARPi‐resistant variants, when compared with the parental U251 cells, display no significant changes in the levels of PARP1–3, and tankyrase1/2, the formation of PAR, mutations on PTEN, BRCA1, BRCA2, ATM and TP53 genes, the levels of PTEN and the members in the related Akt‐mTOR‐S6 signaling axis and the expression of drug transporters P‐gp and BCRP. The data indicate that these factors, some of which have been reported to be correlated to PARPi resistance,1, 36 do not contribute to the acquired PARPi resistance of U251 cells. Consistent with their reduced proliferation inhibition, the PARPi‐resistant variants, relative to the parental cells, produce decreased DSB and G2/M arrest when exposed to PARPi or irradiation. However, except 53BP1, the other 15 examined DNA repair and damage response factors are comparable at their protein levels in both resistant and parental U251 cells. The protein levels of 53BP1 are consistently reduced in the 3 PARPi‐resistant variants. The exogenous expression of 53BP1 completely restores its cellular protein levels and enhances the sensitivity of the PARPi‐resistant variants to PARPi by approximately 76%. Previously, loss of 53BP1 was primarily shown to cause PARPi resistance in BRCA37, 38, 39, 40 or ATM41‐deficient breast37, 39, 41 or ovarian cancer cells.40 For the first time, our results reveal that loss of 53BP1 also dominantly mediates the resistance of PTEN‐deficient glioblastoma cells to different PARPi, strongly suggesting that loss of 53BP1 is a relatively universal mechanism responsible for PARPi resistance at different conditions. The data also indicate a possibility of the combination of 53BP1 levels with PTEN deficiency, just as with BRCA1 deficiency, as a biomarker to predict PARPi sensitivity.37
In addition, we further demonstrate that SAMHD1 is responsible for the resistance of all 3 PARPi‐resistant U251 variants to Ara‐C evidenced by its knockout completely restoring the Ara‐C sensitivity. However, SAMHD1 seems not to be correlated to their PARPi resistance because its knockout did not change the sensitivity of the variants to PARPi. However, the exogenous expression of 53BP1 in the PARPi‐resistant variants did not affect their sensitivity to Ara‐C (Supplementary Figure S7C). This is interesting because the loss of 53BP1 and the increased expression of SAMHD1 coexist in a persistent and stable mode in the PARPi‐resistant variants that are not maintained with PARPi. Together with the stability of PARPi resistance, this further suggests that PARPi‐resistant variants develop heritable genetic change(s), which regulate the expression of 53BP1 and SAMHD1 in both negative (53BP1) and positive (SAMHD1) manners. However, several important issues, such as what the heritable genetic changes are, how the persistent PARPi exposure causes them and how they lead to the changes in the expression of 53BP1 and SAMHD1 need further investigation in the future.
At present, PARPi are being tested for the treatment of glioblastoma in clinical trials.42, 43 Recently, the PARPi olaparib was reported to be found in the core of the tumors in 27 of 35 glioblastoma patients. In 10 of the 27 patients, moreover, olaparib was found to reach tumor cells in the neighboring regions.44 The results suggest a great possibility of PARPi for the treatment of glioblastoma. In addition, other PTEN‐defective tumors, such as prostate and endometrial cancers, have been shown to be sensitive to PARPi.1, 8, 9, 10, 11 Therefore, our findings in this study could lead in a second step to ancillary studies in clinical research to evaluate PARPi in glioblastoma or other related tumor subtypes.
Together, our study demonstrates a consistent resistance profile to PARPi and a unique cross‐resistance profile to non‐PARP conventional anticancer drugs in 3 PARPi‐resistant variants generated from PTEN‐deficient glioblastoma U251 cells induced by different PARPi. The drug resistance is persistent and stable but does not cause other aggressive malignant phenotypes. The loss of 53BP1 and the enhanced expression of SAMHD1 in all the PARPi‐resistant variants are the primary mechanisms responsible for their resistance to PARPi and Ara‐C, respectively. These findings provide new insight into this promising targeted therapy.
DISCLOSURE
The authors have no conflicts of interest to declare.
Supporting information
Document S1
Wang Y‐T, Yuan B, Chen H‐D, et al. Acquired resistance of phosphatase and tensin homolog‐deficient cells to poly(ADP‐ribose) polymerase inhibitor and Ara‐C mediated by 53BP1 loss and SAMHD1 overexpression. Cancer Sci. 2018;109:821–831. https://doi.org/10.1111/cas.13477
Funding information
National Natural Science Foundation of China 81573450 to Z. H. Miao and 81603160 and 81773764 to J. X. He Chinese Academy of Sciences 29201731121100101 to J. X. He and the Hundred Talents Project XDA12020104 to Z. H. Miao Science and Technology Commission of Shanghai Municipality 16JC1406300 to Z. H. Miao State Key Laboratory of Drug Research SIMM1601ZZ‐03 to Z. H. Miao
Contributor Information
Jin‐Xue He, Email: jinxue_he@simm.ac.cn.
Ze‐Hong Miao, Email: zhmiao@simm.ac.cn.
REFERENCES
- 1. Wang YQ, Wang PY, Wang YT, Yang GF, Zhang A, Miao ZH. An update on poly(ADP‐ribose)polymerase‐1 (PARP‐1) inhibitors: opportunities and challenges in cancer therapy. J Med Chem. 2016;59:9575‐9598. [DOI] [PubMed] [Google Scholar]
- 2. He JX, Yang CH, Miao ZH. Poly(ADP‐ribose) polymerase inhibitors as promising cancer therapeutics. Acta Pharmacol Sin. 2010;31:1172‐1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ding J, Miao ZH, Meng LH, Geng MY. Emerging cancer therapeutic opportunities target DNA‐repair systems. Trends Pharmacol Sci. 2006;27:338‐344. [DOI] [PubMed] [Google Scholar]
- 4. Syed YY. Rucaparib: first global approval. Drugs. 2017;77:585‐592. [DOI] [PubMed] [Google Scholar]
- 5. Scott LJ. Niraparib: first global approval. Drugs. 2017;77:1029‐1034. [DOI] [PubMed] [Google Scholar]
- 6. Shen WH, Balajee AS, Wang J, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128:157‐170. [DOI] [PubMed] [Google Scholar]
- 7. Lester A, Rapkins R, Nixdorf S, Khasraw M, McDonald K. Combining PARP inhibitors with radiation therapy for the treatment of glioblastoma: is PTEN predictive of response? Clin Transl Oncol. 2017;19:273‐278. [DOI] [PubMed] [Google Scholar]
- 8. Mendes‐Pereira AM, Martin SA, Brough R, et al. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol Med. 2009;1:315‐322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dedes KJ, Wetterskog D, Mendes‐Pereira AM, et al. PTEN deficiency in endometrioid endometrial adenocarcinomas predicts sensitivity to PARP inhibitors. Sci Transl Med. 2010;2:53ra75. [DOI] [PubMed] [Google Scholar]
- 10. Shen Y, Rehman FL, Feng Y, et al. BMN 673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin Cancer Res. 2013;19:5003‐5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Janzen DM, Paik DY, Rosales MA, et al. Low levels of circulating estrogen sensitize PTEN‐null endometrial tumors to PARP inhibition in vivo. Mol Cancer Ther. 2013;12:2917‐2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Majuelos‐Melguizo J, Rodriguez MI, Lopez‐Jimenez L, et al. PARP targeting counteracts gliomagenesis through induction of mitotic catastrophe and aggravation of deficiency in homologous recombination in PTEN‐mutant glioma. Oncotarget. 2015;6:4790‐4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yuan B, Ye N, Song SS, et al. Poly(ADP‐ribose)polymerase (PARP) inhibition and anticancer activity of simmiparib, a new inhibitor undergoing clinical trials. Cancer Lett. 2017;386:47‐56. [DOI] [PubMed] [Google Scholar]
- 14. He JX, Wang M, Huan XJ, et al. Novel PARP1/2 inhibitor mefuparib hydrochloride elicits potent in vitro and in vivo anticancer activity, characteristic of high tissue distribution. Oncotarget. 2017;8:4156‐4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hidau MK, Kolluru S, Palakurthi S. Development and validation of a high‐performance liquid chromatography method for the quantiffication of talazoparib in rat plasma: application to plasma protein binding studies. Biomed Chromatogr. 2017. https://doi.org/10.1002/bmc.4046. PMID:28677821. [DOI] [PubMed] [Google Scholar]
- 16. Ye N, Chen CH, Chen T, et al. Design, synthesis, and biological evaluation of a series of benzo[de][1,7]naphthyridin‐7(8H)‐ones bearing a functionalized longer chain appendage as novel PARP1 inhibitors. J Med Chem. 2013;56:2885‐2903. [DOI] [PubMed] [Google Scholar]
- 17. Yi JM, Huan XJ, Song SS, Zhou H, Wang YQ, Miao ZH. Triptolide induces cell killing in multidrug‐resistant tumor cells via CDK7/RPB1 rather than XPB or p44. Mol Cancer Ther. 2016;15:1495‐1503. [DOI] [PubMed] [Google Scholar]
- 18. Miao ZH, Tong LJ, Zhang JS, Han JX, Ding J. Characterization of salvicine‐resistant lung adenocarcinoma A549/SAL cell line. Int J Cancer. 2004;110:627‐632. [DOI] [PubMed] [Google Scholar]
- 19. Norquist B, Wurz KA, Pennil CC, et al. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J Clin Oncol. 2011;29:3008‐3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Edwards SL, Brough R, Lord CJ, et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111‐1115. [DOI] [PubMed] [Google Scholar]
- 21. Furnari FB, Lin H, Huang HS, Cavenee WK. Growth suppression of glioma cells by PTEN requires a functional phosphatase catalytic domain. Proc Natl Acad Sci U S A. 1997;94:12479‐12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun CK, Zhang F, Xiang T, et al. Phosphorylation of ribosomal protein S6 confers PARP inhibitor resistance in BRCA1‐deficient cancers. Oncotarget. 2014;5:3375‐3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schneider C, Oellerich T, Baldauf HM, et al. SAMHD1 is a biomarker for Ara‐C response and a therapeutic target in acute myeloid leukemia. Nat Med. 2017;23:250‐255. [DOI] [PubMed] [Google Scholar]
- 24. Herold N, Rudd SG, Ljungblad L, et al. Targeting SAMHD1 with the Vpx protein to improve Ara‐C therapy for hematological malignancies. Nat Med. 2017;23:256‐263. [DOI] [PubMed] [Google Scholar]
- 25. Lheureux S, Bruce JP, Burnier JV, et al. Somatic BRCA1/2 recovery as a resistance mechanism after exceptional response to poly (ADP‐ribose) polymerase inhibition. J Clin Oncol. 2017;35:1240‐1249. [DOI] [PubMed] [Google Scholar]
- 26. Kondrashova O, Nguyen M, Shield‐Artin K, et al. Secondary somatic mutations restoring RAD51C and RAD51D associated with acquired resistance to the PARP inhibitor rucaparib in high‐grade ovarian carcinoma. Cancer Discov. 2017. https://doi.org/10.1158/2159-8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pishvaian MJ, Biankin AV, Bailey P, et al. BRCA2 secondary mutation‐mediated resistance to platinum and PARP inhibitor‐based therapy in pancreatic cancer. Br J Cancer. 2017;116:1021‐1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murai J, Huang SY, Renaud A, et al. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol Cancer Ther. 2014;13:433‐443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. AlHilli MM, Becker MA, Weroha SJ, et al. In vivo anti‐tumor activity of the PARP inhibitor niraparib in homologous recombination deficient and proficient ovarian carcinoma. Gynecol Oncol. 2016;143:379‐388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mirza MR, Monk BJ, Herrstedt J, et al. Niraparib maintenance therapy in platinum‐sensitive, recurrent ovarian cancer. N Engl J Med. 2016;375:2154‐2164. [DOI] [PubMed] [Google Scholar]
- 31. Anantha RW, Simhadri S, Foo TK, et al. Functional and mutational landscapes of BRCA1 for homology‐directed repair and therapy resistance. Elife 2017;6:pii: e21350 https://doi.org/10.7554/elife.21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Drost R, Dhillon KK, van der Gulden H, et al. BRCA1185delAG tumors may acquire therapy resistance through expression of RING‐less BRCA1. J Clin Invest. 2016;126:2903‐2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ray Chaudhuri A, Callen E, Ding X, et al. Replication fork stability confers chemoresistance in BRCA‐deficient cells. Nature. 2016;535:382‐387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Johnson SF, Cruz C, Greifenberg AK, et al. CDK12 inhibition reverses de novo and acquired PARP inhibitor resistance in BRCA wild‐type and mutated models of triple‐negative breast cancer. Cell Rep. 2016;17:2367‐2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McCormick A, Donoghue P, Dixon M, et al. Ovarian cancers harbor defects in nonhomologous end joining resulting in resistance to rucaparib. Clin Cancer Res. 2017;23:2050‐2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lord CJ, Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355:1152‐1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang ZM, Liao XM, Chen Y, et al. Combining 53BP1 with BRCA1 as a biomarker to predict the sensitivity of poly(ADP‐ribose) polymerase (PARP) inhibitors. Acta Pharmacol Sin. 2017;38:1038‐1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Johnson N, Johnson SF, Yao W, et al. Stabilization of mutant BRCA1 protein confers PARP inhibitor and platinum resistance. Proc Natl Acad Sci U S A. 2013;110:17041‐17046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jaspers JE, Kersbergen A, Boon U, et al. Loss of 53BP1 causes PARP inhibitor resistance in Brca1‐mutated mouse mammary tumors. Cancer Discov. 2013;3:68‐81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pennington KP, Wickramanayake A, Norquist BM, et al. 53BP1 expression in sporadic and inherited ovarian carcinoma: relationship to genetic status and clinical outcomes. Gynecol Oncol. 2013;128:493‐499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hong R, Ma F, Zhang W, et al. 53BP1 depletion causes PARP inhibitor resistance in ATM‐deficient breast cancer cells. BMC Cancer. 2016;16:725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Olaparib and temozolomide in treating patients with relapsed glioblastoma; Cancer Research UK. https://clinicaltrials.gov/ct2/show/NCT01390571?term=PARP+inhibitor&cond=Glioblastoma&rank=1. Accessed April 14, 2016.
- 43. Cediranib maleate and olaparib compared to bevacizumab in treating patients with recurrent glioblastoma; National Cancer Institute (NCI). https://clinicaltrials.gov/ct2/show/NCT02974621?term=PARP+inhibitor&cond=Glioblastoma&rank=4. Accessed: 27 November 2017.
- 44. Werntraub A. Could AstraZeneca's ovarian cancer drug also treat glioblastoma? 6 November 2017. https://www.fiercebiotech.com/research/could‐astrazeneca‐s‐ovarian‐cancer‐drug‐also‐treat‐glioblastoma?mkt_tok=eyJpIjoiTVRZMllXUmtaV0V3TlRoayIsInQiOiJOWm5Vc2F2TlRpcVphT2pqWnUybmxzTnMyM2ZNVDRcL0hXMFwvQ0FCNFZTUHVyVFhRREtuTDlJTnNRNjBBbHhVSGozVnkxdHZHcEpWb1FFMEdcL0dnbktjV2s4aTYySFU3Y2prUE5JSHZjZ0NSTUk1WkdrME90cUZrODRhV0labm9OSSJ9&mrkid=23680060&utm_medium=nl&utm_source=internal.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Document S1


