Abstract
Background
Staphylococcus aureus, a common cause of bovine mastitis, is known for its ability to acquire to antimicrobial resistance and to secrete numerous virulence factors that can exacerbate inflammation. In addition, alpha-hemolysin has an important role in S. aureus infections, diversity of the hla gene (that produces alpha-hmolysin) in S. aureus isolated from bovine mastitis has not been well characterized. The objective was, therefore, to determine diversity of virulence genes, hla gene sequences, and clonal profiles of S. aureus from bovine mastitis in Chinese dairy herds, and to evaluate inter-relationships.
Results
The antimicrobials resistance varies from as low as 1.9% (2/103) for CTX to as high as 76.7% (79/103) for penicilin in the 103 isolates and 46 (44.7%) S. aureus were determined as multi-resistant isolates with diverse resistance patterns. Thirty-eight virulence gene patterns (with variable frequencies) were identified in the 103 isolates and correlated with MLST types, indicating a great diversity. Although the hla gene also had great diversity (14 genotypes), Hla peptides were relatively more conserved. With 7 clonal complexes identified from 24 spa types and 7 MLST types. Regarding the letter, ST 97 was the dominant type in S. aureus from bovine mastitis in China. Furthermore, based on phylogenetic analysis, there was a distinct evolutionary relationship between the hla gene and MLST.
Conclusion
Multi-resistant S. aureus occurred in bovine mastitis with diverse resistance patterns. The diversity of virulence gene profiles, especially the hla gene and, their relationship with molecular types were reported for the first time in S. aureus from bovine mastitis, which will be useful for future studies on immunogenicity and vaccine development. In addition, based on the distinct evolutionary relationship between the hla gene and MLST types, we inferred that the hla gene has potential role for molecular typing of S. aureus.
Electronic supplementary material
The online version of this article (10.1186/s12917-018-1374-7) contains supplementary material, which is available to authorized users.
Keywords: Staphylococcus aureus; Virulence gene; Hla; MLST; spa, Bovine mastitis
Background
Bovine mastitis causes huge economic losses in the global dairy industry by decreasing the quality and quantity of milk produced, as well as compromising dairy cow health and welfare. Staphylococcus aureus, one of the most prevalent etiologic agents, has an important role in clinical and subclinical mastitis, characterized by persistent and recurrent infections with low cure rates in response to antimicrobial therapy [1–3].
Staphylococcus aureus is known for the ability to develop resistance to antimicrobial agents (e.g. methicillin-resistant S. aureus, vancomycin-intermediate S. aureus, and vancomycin-resistant S. aureus) and to secrete numerous virulence factors to exacerbate inflammation. As an alternative to antimicrobials, anti-virulence therapies interfere with bacterial toxins or virulence factors and/or pathways that regulate their production [4]. Allen et al. [5] proposed that a combination of anti-virulence compounds targeting various virulence factors would be a more effective solution than conventional treatments. Therefore, characterization of virulence gene profiles and clonal diversity among S. aureus populations are very important in development of anti-virulence therapies [6–9].
Alpha-hemolysin (Hla) toxin is the most emphasized and characterized virulence factor [10] in S. aureus. Changes in key amino acid residues may affect Hla activity. For example, a H35L substitution had no hemolytic or lethal activity, whereas a C259T substitution resulted in a premature stop codon and a significant reduction in Hla production [11–13]. Additionally, promising results have been obtained using Hla as a candidate for developing a vaccine to prevent S. aureus infections [14–16]. However, variation in Hla peptide sequences could affect vaccine efficacy. It is therefore important to characterize genetic polymorphism of the hla gene.
To date, several studies on diversity of hla gene of S. aureus originating from humans have been reported [13, 17, 18]; however, there is paucity of research regarding sequence variability of this gene in S. aureus isolates from bovine mastitis. The objective was to determine virulence gene patterns and hla gene diversity in S. aureus from bovine mastitis and link them with potential molecular clones, as determined by multilocus sequence type (MLST) and spa typing.
Methods
A total of 103 S. aureus isolates from 1021 clinical mastitis samples collected from 2013 to 2016 on 19 dairy farms in 9 provinces of China were used in this study (Table 1). Isolates were identified as S. aureus according to conventional methods, including Gram staining, colony morphology, hemolysis, catalase and tube coagulase test, as well as 16S rRNA, coa and nuc gene sequence analysis, as described [6, 10].
Table 1.
Distribution of the 103 Staphylococcus aureus clinical mastitis isolates according to region and herd
| Region | Herd | No. of milk samples | No. of isolates | Total isolates of region | |
|---|---|---|---|---|---|
| Beijing | a | 407 | 41 | 48 | |
| b | 53 | 5 | |||
| c | 28 | 2 | |||
| Ningxia | d | 46 | 5 | 16 | |
| e | 31 | 4 | |||
| f | 29 | 3 | |||
| g | 28 | 3 | |||
| h | 28 | 1 | |||
| Heilongjiang | i | 56 | 11 | 11 | |
| Hebei | j | 38 | 6 | 11 | |
| k | 23 | 4 | |||
| l | 22 | 1 | |||
| Inner Mongolia | m | 25 | 3 | 5 | |
| n | 22 | 2 | |||
| Liaoning | o | 15 | 2 | 4 | |
| p | 15 | 2 | |||
| Guangdong | q | 98 | 3 | 3 | |
| Shandong | r | 40 | 3 | 3 | |
| Henan | s | 17 | 2 | 2 | |
| Total | 19 | 1021 | 103 | 103 |
Extraction of genomic DNA
Genomic DNA of all 103 isolates was extracted using Bacteria Genomic DNA Kit (CW, Beijing, China) according to the manufacturer’s instructions and stored at − 20 °C. In addition, quantity and quality of DNA were assessed using a Nanodrop ND-1000 spectrophotometer (Thermoscientific, Wilmington, DE).
Antimicrobial susceptibility testing
To analyze the antibiotic susceptibility profiles of the 103 isolates, minimum inhibitory concentrations (MICs) of a panel of 11 different antibiotics were determined using the broth microdilution method and resistance breakpoints for ampicillin (AMP, 0.5 mg/L), augmentin (AMX/CA, 32 mg/L), cefotaxime (CTX, 8 mg/L), ceftriaxone (CRO, 8 mg/L), ciprofloxacin (CIP, 4 mg/L), clindamicin (CL, 8 mg/L), erythromycin (E, 8 mg/L), gentamicin (GM, 16 mg/L), oxacillin (OX, 0.5 mg/L), penicillin (P, 0.25 mg/L), tetracycline (TE, 16 mg/L) as described in CLSI (2013) [19]. S. aurues ATCC 29213 was used as uality control in this study.
Antimicrobial agents above were selected either according to their availability in commercial products or working as representative of an antibiotic family. And those which were found to be resistant to at least three or more of antimicrobial agents were defined as multidrug resistant isolates.
Detection of virulence genes
All S. aureus isolates were screened by polymerase chain reaction (PCR) for the following virulence genes: fibronectin binding proteins A and B (fnbA and fnbB), clumping factors A and B (clfA and clfB), α-, β-, γ-, and δ-hemolysin (hla, hlb, hlc and hld), intracellular adhesion A and D (icaA and icaD), toxic shock syndrome toxin (tsst), and enterotoxins (sea, seb, sec, see, seg and sei). Primer sequences and PCR methods have been described [8, 20, 21]. Products amplified by PCR were sequenced by Beijing Sunbiotech Co. (Beijing, China). Virulence gene profiles were analyzed as binary data using the Maximum Parsimony tree by MEGA6 [22, 23]. Evolutionary distances were computed using the p-distances method and were in units of the number of base differences per site [24].
Genotyping of hla gene and phylogenetic analysis
Primers used for amplification of the complete 960 bp hla gene were: hlaF1 (5′- TTAGCCGAAAAACATCATTTC -3′) and hlaR1 (5′- TTATTCCCGACGAAATTCCAA -3′), as described Mocca et al. (2014) [15]. The PCR was performed with initial denaturation at 95 °C for 5 min, followed by 33 cycles of 94 °C for 1 min, 55 °C for 1 min, 72 °C for 1 min, and a final extension at 72 °C for 10 min. Products were sent to Beijing Sunbiotech Co. (Beijing, China) for bidirectional sequencing using both primers: hlaF2 (5′- GAAGTTATCGGCTAAAGTTATAA -3′) and hlaR2 (5′- CATAATTAATACCCTTTTTCTC -3′ [15]. All PCR-amplified products were sequenced twice.
Staphylococcus aureus strain WOOD46 (GenBank accession no. X01645) was selected as the reference strain, as reported [13, 16, 25–27], and hla gene sequences were aligned using BioEdit v7.0.9 (Micro Focus, Newbury, UK) to designate genotypes. Corresponding peptide sequences were deduced and aligned to determine the presence of amino acid substitutions. NetPhosBac 1.0 Server (http://www.cbs.dtu.dk/services/NetPhosBac-1.0) was used to predict serine and threonine phosphorylation sites in Hla peptide sequences, which were identified with non-synonymous amino acid substitutions [28]. In addition, phylogenetic trees of the hla gene and 7 MLST alleles in the collection were generated by MEGA6 (http://www.megasoftware.net/) using the neighbor-joining method by the Kimura 2-parameter model with 1000 bootstrap replicates [22].
Molecular typing
Staphylococcus aureus isolates that harbored the hla gene were genotyped by staphylococcal protein A (spa) typing (http://spa.ridom.de/index.shtml) using SPATypeMapper software (download at http://www.clondiag.com/fileadmin/Media/Downloads/SPATypeMapper_0_6.zip) and multilocus sequence typing (MLST; http://saureus.mlst.net/) [9, 29]. In addition, the goeBURST algorithm (http://goeBURST.phyloviz.net) was used to infer evolutionary associations of MLST types (STs).
Statistical analyses
Associations between virulence gene profiles, Hla peptide types and clonal background were analyzed using a Chi-square or Fisher’s exact test, as appropriate, on contingency tables (virulence types or Hla peptide types vs clonal backgrounds or geography; virulence types vs Hla peptide types) using SPSS 23.0 (SPSS Inc., Chicago, IL). Statistical significance was defined as P < 0.05.
Results
Antimicrobial resistance patterns
Overall, there were only 5 S. aureus isolates susceptible to all tested antimicrobial compounds, as shown in Table 2. And a total of 46 (44.7%) S. aureus were determined as multi-resistant isolates with diverse resistance patterns. In addition, the antimicrobials resistance varies from as low as 1.9% (2/103) for CTX to as high as 76.7% (79/103) for penicillin.
Table 2.
Antimicrobial resistance patterns of S. aureus strains isolated from clinical mastitis in cows
| Number of types of resistance | Patterns of resistance | S. aurues (n = 103) |
|---|---|---|
| 0 (5) | 5 | |
| 1 (8) | CL | 2 |
| CRO | 1 | |
| E | 3 | |
| AMP | 2 | |
| 2 (45) | P, AMP | 39 |
| CIP, AMP | 1 | |
| OX, E | 1 | |
| E, P | 1 | |
| CIP, P | 1 | |
| OX, AMP | 1 | |
| OX, CIP | 1 | |
| 3 (28) | OX, P, AMP | 2 |
| CIP, AMP, P | 4 | |
| P, AMP, CL | 3 | |
| E, P, AMP | 14 | |
| P, AMP, CRO | 1 | |
| AMX/CA, P, AMP | 1 | |
| OX, E, CL | 1 | |
| TE, AMP, P | 2 | |
| 4 (13) | OX, E, CIP, CL | 1 |
| OX, AMX/CA, P, AMP | 1 | |
| E, AMX/CA, P, AMP | 1 | |
| OX, E, AMP, CL | 1 | |
| OX, TE, E, AMP | 1 | |
| OX, CIP, P, AMP | 1 | |
| E, CIP, AMP, CL | 3 | |
| TE, E, P, AMP | 1 | |
| OX, E, P, AMP | 1 | |
| E, P, AMP, CL | 2 | |
| ≥5 (5) | OX, E, P, CTX, AMP | 1 |
| GM, E, P, AMP, CL | 1 | |
| GM, E, CIP, AMP, CL | 1 | |
| OX, E, CIP, P, AMP, CL | 1 | |
| OX, TE, GM, E, CIP, P, CTX, AMP, CL, CRO | 1 |
AMP ampicillin, AMX/CA augmentin, CFX/K cephalexin/kanamycin, CIP ciprofloxacin, CL clindamycin, CRO ceftriaxone, CTX cefotaxime, E erythromycin, GM gentamicin, OX oxacillin, P penicillin, TE tetracycline
Virulence gene profiles
The PCR amplification results of 17 virulence genes are shown in Additional file 1: Figure S1, Additional file 2: Figure S2, Additional file 3: Figure S3 and Additional file 4: Figure S4, and their relative frequency in 103 clinical mastitis S. aureus are shown (Fig. 1). Among them, clfB and icaA genes were detected in 100% of isolates, followed by clfA (n = 99; 96%), hld (n = 98; 95%), fnbA (n = 97; 94%), hla (n = 93; 90%), hlc (n = 91; 88%), fnbB (n = 88; 85%) and hlb (n = 82; 80%), tsst (n = 31; 30%), icaD (n = 26; 25%), sec (n = 22; 21%), sei (n = 5; 5%), seg (n = 3; 3%), seb and see (n = 2; 2%). None of the 103 S. aureus isolates had the sea gene.
Fig. 1.
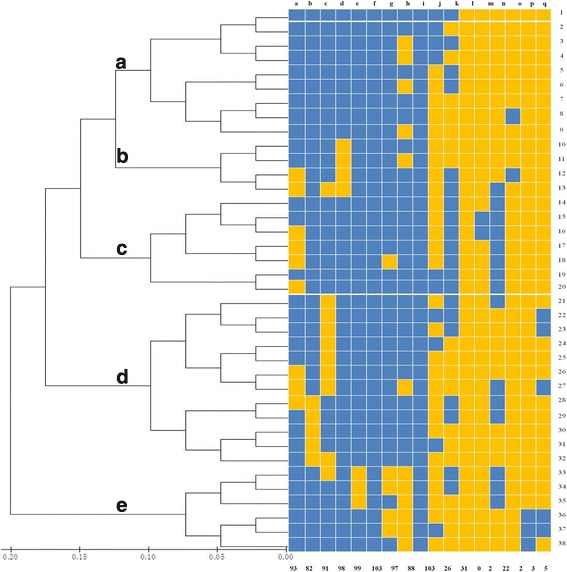
Analysis of virulence gene profiles of 103 Chinese Staphylococcus aureus isolates from bovine clinical mastitis. This dendrogram was generated based on the presence/absence of virulence genes with the sum of branch length = 0.91629109. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. (a-e) refer to different virulence gene clusters. Blue boxes indicate the presence and yellow boxes the absence of the corresponding virulence genes, and the number on the right refer to the virulence profile patterns, whereas the number under the box refer to the corresponding number of positive isolates in each virulence gene. (a-q), refers to the virulence genes in order: hla, hlb, hlc, hld, clfA, clfB, fnbA, fnbB, icaA, icaD, tsst, sea, seb, sec, see, seg, sei
The 103 S. aureus isolates comprised 38 virulence gene profiles, which were then categorized into 5 clusters (A to E; Fig. 1). Geographic distribution of these clusters are shown (Table 3). Prevalence of virulence clusters differed among herds from different provinces. Different virulence cluster combinations appeared in herds from different provinces with only 2 exceptions (Herds m and q and Herds s and l; Table 3). Isolates from herds in same province usually had similar cluster combinations. Predominant clusters therefore differed among provinces (P < 0.01): Cluster A was the most frequently detected in Beijing herds, Cluster D in herds from Ningxia, and Cluster C in herds from Hebei and Heilongjiang.
Table 3.
Distribution of virulence gene clusters and Hla peptide sequence types in Staphylococcus aureus from 103 cases of clinical mastitis
| Province | Herd | Virulence cluster | Hla peptide type | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | I | II | III | IV | V | VI | VII | VIII | IX | X | ||
| Beijing | a | 36 | 2 | – | 3 | – | 35 | – | 3 | – | – | – | – | 1 | 1 | 1 |
| b | 4 | – | – | – | 1 | 3 | – | – | 2 | – | – | – | – | – | – | |
| c | 1 | 1 | – | – | – | 2 | – | – | – | – | – | – | – | – | – | |
| Ningxia | d | – | – | – | 5 | – | 5 | – | – | – | – | – | – | – | – | – |
| e | – | – | – | 4 | – | 4 | – | – | – | – | – | – | – | – | – | |
| f | – | – | – | 3 | – | 3 | – | – | – | – | – | – | – | – | – | |
| g | – | – | – | 3 | – | 3 | – | – | – | – | – | – | – | – | – | |
| h | – | – | – | 1 | – | 1 | – | – | – | – | – | – | – | – | – | |
| Heilongjiang | i | 3 | – | 4 | 4 | – | 7 | – | – | – | – | 1 | – | – | – | – |
| Hebei | j | 1 | 1 | 1 | 3 | – | 4 | – | – | – | – | – | – | – | – | – |
| k | – | – | 4 | – | – | 2 | – | – | – | – | – | – | – | – | – | |
| l | 1 | – | – | – | – | 1 | – | – | – | – | – | – | – | – | – | |
| Inner Mongolia | m | – | – | – | – | 3 | – | 2 | – | – | – | – | 1 | – | – | – |
| n | – | – | 1 | 1 | – | 2 | – | – | – | – | – | – | – | – | – | |
| Liaoning | o | 1 | – | – | 1 | – | 2 | – | – | – | – | – | – | – | – | – |
| p | 1 | – | – | 1 | – | 1 | – | – | – | – | – | – | – | – | – | |
| Guangdong | q | – | – | – | – | 3 | 1 | 2 | – | – | – | – | – | – | – | – |
| Shandong | r | 1 | 1 | 1 | – | – | 1 | – | – | – | – | – | – | – | – | – |
| Henan | s | 2 | – | – | – | – | – | – | – | – | 2 | – | – | – | – | – |
- No isolates
Genotyping based on hla gene
Using S. aureus strain WOOD 46 as a reference sequence, 22 single nucleotide mutations were detected, 4 in the signal peptide encoding portion (nucleotide positions − 42, − 22, − 6 and − 5) and 18 on the mature hla encoding sequence (nucleotide positions 47, 144, 214, 237, 255, 262, 399, 438, 453, 498, 499, 519, 606, 686, 765, 797, 824, 833) (Table 4). As a result 14 hla genotypes were identified based on nucleotide sequence analysis and 10 peptide sequence types (I to X) among 93 isolates with the hla gene (Tables 4 and 5). Amongst the 14 hla genotypes, Genotype 1 contained 65 (70%) isolates (Table 4).
Table 4.
Nucleotide mutations and corresponding amino acid substitutions of Staphylococcus aureus from cases of clinical mastitis
| Geno type | No. | Nucleotide mutation position/corresponding amino acid substitutiona | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −42 | −22 | −6 &-5 | 47 | 144 | 214 | 237 | 255 | 262 | 399 | 438 | 453 | 498 | 499 | 519 | 606 | 686 | 765 | 797 | 824 | 833 | ||
| C | G | GG | G | C | G | C | G | T | C | G | T | C | T | T | T | C | T | A | C | T | ||
| 1 | 65 | −b | A | AA | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Sb | G(−2)N | |||||||||||||||||||||
| 2 | 5 | – | A | AA | – | – | – | – | – | C | T | C | G | – | – | – | C | – | C | – | – | – |
| S | G(−2)N | S | S | S | S | S | S | |||||||||||||||
| 3 | 4 | T | A | AA | – | – | – | – | – | – | T | C | G | – | – | – | C | – | C | – | – | – |
| S | S | G(− 2)N | S | S | S | S | S | |||||||||||||||
| 4 | 4 | – | A | AA | – | – | – | – | – | – | – | – | – | T | – | – | – | – | – | – | – | – |
| S | G(−2)N | S | ||||||||||||||||||||
| 5 | 3 | – | A | AA | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | A |
| S | G(−2)N | S278Y | ||||||||||||||||||||
| 6 | 2 | – | A | AA | – | – | – | – | A | – | – | – | – | – | – | – | – | – | – | – | – | – |
| S | G(−2)N | S | ||||||||||||||||||||
| 7 | 2 | – | A | AA | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | T |
| S | G(−2)N | S278F | ||||||||||||||||||||
| 8 | 2 | – | A | AA | – | T | – | – | – | – | – | – | – | – | – | – | C | – | C | C | T | – |
| S | G(−2)N | S | S | S S T275I | ||||||||||||||||||
| 9 | 1 | – | A | AA | – | – | A | – | A | – | – | – | – | – | – | – | – | – | – | – | – | – |
| S | G(−2)N | Q72S | S | |||||||||||||||||||
| 10 | 1 | – | A | AA | T | – | – | – | – | – | T | C | G | – | – | – | C | – | C | – | – | – |
| S | G(−2)N | S16I | S | S | S | S | S | |||||||||||||||
| 11 | 1 | – | A | AA | – | – | – | A | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| S | G(−2)N | S | ||||||||||||||||||||
| 12 | 1 | – | A | AA | – | – | – | – | – | – | – | – | – | – | – | – | – | G | – | – | – | A |
| S | G(−2)N | A229G | S278Y | |||||||||||||||||||
| 13 | 1 | – | A | AA | – | – | – | – | – | – | – | – | – | – | – | A | – | – | – | – | – | – |
| S | G(−2)N | N173 K | ||||||||||||||||||||
| 14 | 1 | – | A | AA | – | – | – | – | – | – | – | – | – | – | Db | – | – | – | – | – | – | – |
| S | G(−2)N | W167Gc | ||||||||||||||||||||
aNucleotide/peptide positions were designated relative to the first nucleotide/amino acid of the mature 훂-hemolysin (S. aureus WOOD46)
bS synonymous mutation, D deletion mutation, − = no mutation;
cThe deletion at nucleotide position 499 of Genotype 14 resulted in peptide termination at residue position 169
Table 5.
Association between hla peptide types and virulence gene profiles of 93 Staphylococcus aureus isolated from clinical mastitis
| Prediction of phosphorylation sites (n) | Peptide type | hla | Virulence gene profile cluster | ||||
|---|---|---|---|---|---|---|---|
| A | B | C | D | E | |||
| A (85) | I (77) | 1 | 37 | 3 | 6 | 18 | 1 |
| 2 | 0 | 0 | 0 | 5 | 0 | ||
| 4 | 3 | 0 | 0 | 1 | 0 | ||
| 6 | 0 | 0 | 1 | 0 | 1 | ||
| 11 | 1 | 0 | 0 | 0 | 0 | ||
| II (4) | 3 | 4 | 0 | 0 | 0 | 0 | |
| V (2) | 8 | 2 | 0 | 0 | 0 | 0 | |
| VII (1) | 10 | 0 | 0 | 0 | 0 | 1 | |
| IX (1) | 13 | 1 | 0 | 0 | 0 | 0 | |
| B (6) | III (3) | 5 | 2 | 0 | 0 | 1 | 0 |
| IV (2) | 7 | 2 | 0 | 0 | 0 | 0 | |
| VIII (1) | 12 | 1 | 0 | 0 | 0 | 0 | |
| C (1) | VI (1) | 9 | 1 | 0 | 0 | 0 | 0 |
| X (1) | 14 | 1 | 0 | 0 | 0 | 0 | |
Hla peptide type I, composed of hla genotypes 1, 2, 4, 6 and 11, was present in 18 herds, peptide sequence type II was in 2 herds, whereas others were identified in only 1 herd with 1 or 2 isolates, indicating that Hla peptide were conserved among herds (Table 3).
Three results were obtained from the prediction of serine and threonine phosphorylation sites among the 92 complete peptide sequences (Fig. 2). Figure 2a presents the prediction results of peptide sequence type I, II, V, VII and IX, which were same as the reference strain. Conversely, there were variations at position 300 in Fig. 2b (results of peptide sequence types III, IV and VIII) and position 100 in Fig. 2c (result of peptide sequence type VI) when comparing with that of the reference strain. Type A was the most dominant in the 93 isolates (n = 86, 93%), indicating that the Hla peptide sequence was conserved to an extent (Table 5)
Fig. 2.
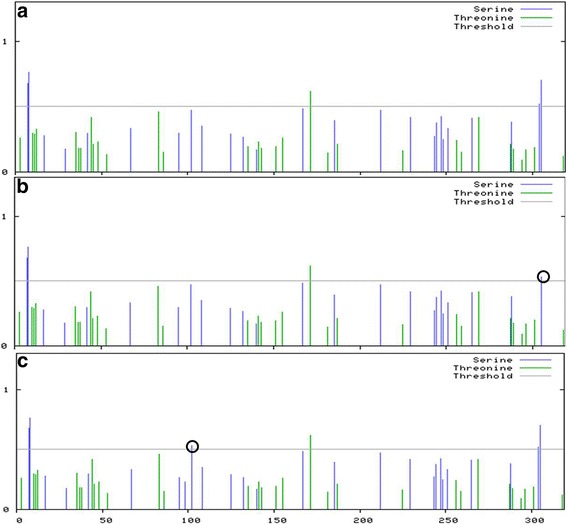
Phosphorylation sites prediction of Hla peptide types with non-synonymous substitutions. The score for each serine or threonine residue is plotted against the sequence position of that residue. When the score is > 0.5, the residue is a predicted phosphorylation site. Variations from the reference sequence are indicated by black circles. a) Phosphorylation sites prediction of reference strain WOOD46 and Hla peptide I, II, V, VII, IX; b) Phosphorylation sites prediction of Hla peptide type III, IV and VIII; c) Phosphorylation sites prediction of Hla peptide type VI and X
Molecular typing
The analysis of 103 S. aureus isolates by spa typing revealed 24 spa types, of which 19 were known types (t189, t127, t2699, t359, t237, t4682, t521, t730, t224, t6297, t2756, t131, t1234, t267, t034, t529, t518 and t528), 3 others were reported for the first time (t16314, t16315, and t17095), and 2 were unassigned types with repeats r07r23r12r21r17r34r34r33r34r13 (UK-1) and r07r16r34r33r13r26 (UK-2) (Table 6).
Table 6.
Distribution of spa typing and MLST types of 103 Staphylococcus aureus isolated from clinical mastitis in China
| Clonal complex (CC) | MLST type | Spa type | aBJ | NX | HJ | HB | IM | LN | GD | SD | HN | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a | b | c | d | e | f | g | h | i | j | k | l | m | n | o | p | q | r | s | |||
| CC1 (90) | ST188 | t189 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| ST1 | t127 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | |
| t2699 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | ||
| t17095 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | ||
| UK02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | ||
| ST97 | t359 | 25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| t4570 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| t237 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| t4682 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| t521 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| t730 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 4 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | ||
| t16314 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| t16315 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| t224 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| t6297 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| t2756 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| t131 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| t1234 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| t267 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| UK01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| CC7(1) | ST398 | T034 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| CC12(5) | ST705 | t529 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 3 | 0 | 0 |
| CC22(6) | ST50 | t518 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| CC30(1) | ST479 | t528 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
aBJ Beijing, NX Ningxia, HJ Heilongjiang, HB Hebei, IM Inner Mongolia, GD Guangdong, SD Shandong, HN Henan, LN Liaoning
In addition, 7 MLST types were obtained, which clustered into 7 clonal complexes. Of them, ST97, composed of 82 isolates, was detected in 12 herds from 7 provinces and shared by 15 spa types, whereas other MLST types were present in isolates from only 1 or 2 provinces (Table 6). In addition, there was great diversity among isolates within or between herds. Nine of the 19 herds contained > 1 spa type (Table 6).
Phylogenetic tree of hla gene
According to nucleotide diversity, the 93 hla sequences branched into 3 distinct major clusters (Fig. 3a). Furthermore, the corresponding ST types were also grouped into various clusters: ST97 and ST1 were in Cluster A, ST118 was in Cluster B, whereas ST 50, ST705 and ST479 were grouped into Cluster C (Fig. 3a). The position of STs in the tree constructed by the concatenated sequences of the 7 housekeeping genes used in MLST was almost the same as that of hla tree, with only ST705 as an exception (Fig. 3b).
Fig. 3.

Phylogenetic trees of hla gene (a) and concatenated sequences of MLST alleles (b) in Staphylococcus aureus isolates. The percentage of the trees in which the associated hla sequence clustered together is shown next to the branches. The tree is drawn to scale, with branch lengths measured in number of substitutions per site. Evolutionary analyses were conducted in MEGA6. Note: BJ = Beijing; NX = Ningxia; SD = Shandong; HB = Hebei; LN = Liaoning; HN = Henan; GD = Guangdong; IM = Inner Mongolia; HJ = Heilongjiang. The number after regions refers to the corresponding hla type
Relationships among virulence gene profiles, hla genotypes and clonal typing
Isolates in hla type 1/peptide I were present in 5 virulence gene clusters in various proportions, revealing great diversity of virulence genes, whereas isolates in other hla genotypes and peptide types demonstrated simplex virulence gene profile (Table 5). Nevertheless, there was no association between hla gene genotypes and virulence gene profiles (P = 0.12), and no association between Hla peptide types and virulence gene profiles (P = 0.80).
Isolates belonging to the same ST may comprise same virulence gene profile. Isolates from ST705 were grouped into Cluster E, whereas ST50 isolates were included in Cluster D. With the exception ST97 and ST1, in which isolates were sprinkled in more than 3 clusters (Table 7). Hence, there was a correlation between virulence gene profiles and molecular clones (P < 0.01).
Table 7.
Association between virulence gene clusters and Hla peptide types, and MLST types and spa types
| MLST type | Spa type | Virulence gene cluster | Hla peptide sequence type | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | I | II | III | IV | V | VI | VII | VIII | IX | X | ||
| ST97 | t359, t237, t4682, t521, t730, t16314, t16315, t224, t6297, t2756, t131, t1234, t267, UK-1 | 45 | 5 | 11 | 20 | 1 | 66 | 0 | 0 | 0 | 1 | 3 | 2 | 1 | 1 | 1 |
| ST705 | t529 | 0 | 0 | 0 | 0 | 5 | 1 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ST50 | t518 | 0 | 0 | 0 | 6 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ST1 | t127, t2699, t17095, UK-2 | 4 | 0 | 1 | 1 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ST479 | t528 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ST188 | t189 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ST398 | t034 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Each Hla peptide sequence type was only present in 1 ST type, except for peptide I, which had isolates in ST97, ST705, ST50, and ST1 (Table 7). Similarly, isolates belonging to the same ST harbored same peptide type, except for ST97, in which isolates were clustered into 7 peptide sequence types (I, V, VI, VII, VIII, IX and X) (Table 5). Therefore, Hla peptide types and ST types were correlated (P < 0.01).
Discussion
It is important to monitor antimicrobial resistance in veterinary medicine. As a frequent and major contagious pathogen in bovine mastitis, S. aureus readily becomes resistant to antimicrobials and causes persistent noncurable intramammary infection. Recent investigations on antimicrobial resistance of S. aureus are available [30–33]. However, lacking of a common clinical breakpoint for antibiotics which are frequently used to treat mastitis make it difficult to comparing with other data. Overall, the percentage of antimicrobial resistant S. aureus in this study is much higher (76.7% vs 25%) in resistant to penicillin according to the work of de Jong et al. [31].
In the present study, the 17 virulence genes were distributed with varying frequencies among S. aureus isolates. Consistent with previous research, none of the 103 isolates contained the pvl gene [34, 35], whereas adhesin genes (clfA, clfB, fnbA and fnbB) were the most prevalent. In contrast, Gogoi-Tiwari et al. (2015) reported that fnbB was only present in 1.3% of S. aureus isolates, which was much lower than in our study (85%) [36]. Thompson et al. (2014) reported a lower occurrence of sec gene in MSSA than the current study (14 vs 21%, respectively) [37]. In the present study, 30% clinical isolates of S. aureus contained the tsst gene, whereas in another study [35], none of 47 S. aureus mastitis isolates were positive for this gene. In agreement with a previous study, none of the isolates harbored the sea gene; this was the gene most frequently encountered from food poisoning in humans and subclinical mastitis cases in cattle [6, 38]. Li et al. (2017) reported that no enterotoxin gene was present in ST97, ST188 or ST398 isolates, which was different from the current study, where some ST97 isolates had seb, sec and see genes [38]. Overall, virulence gene patterns of S. aureus isolates had variable distributions among herds from different regions.
There were many reports regarding contribution of α-hemolysin to the pathogenesis of S. aureus infection, including cell signaling pathways that govern cell proliferation, cytokine secretion, inflammatory responses and cell-cell interactions [39–42]. Although polymorphisms in the hla promoter region have been described [43], the range of genetic diversity and evolution of this toxin was only assessed in human [43–45], with no report of a large representative collection of S. aureus from dairy cows. Among the 22 single nucleotide mutations, 14 (64%) mutations were the same as those isolates from humans [13], Among the 22 single nucleotide mutations, 14 (64%) were the same as reported earlier from human isolates, indicating that transmission of S. aureus might occur between cows and humans, which would be of great relevance to public health security. The high degree of hla gene diversity was in accordance with other reports [17, 23]. However, with the analysis of Hla peptide sequences in this study, we predicted that most (n = 86) isolates carried the same phosphorylation site as in our reference strain; therefore, we inferred that Hla peptide sequences were conserved. Although influence of the different phosphorylation sites was uncertain with regards to their influence on the 3-dimensional structure stability of the protein [23], Burnside et al. (2015) reported that a serine/threonine kinase and phosphatase regulated expression of hemolysin in S. aureus [46]. Therefore, the exact influence of disparate phosphorylation sites here remains to be studied. Sharma-Kuink et al. (2015) co-related peptide polymorphism to random mutations in nature and selective pressure from the immune system, due to generation of antibodies that bind to alpha-toxins [17]. In this study, the correlation between hla gene genotype/ peptides types and geography partly supported the above conclusion. In addition, variations in peptide sequences may influence Hla functionality and may change antigenic epitopes and potentially cause vaccine failure, as reported [15, 16].
Analysis of hla-based phylogenetic tree revealed 3 clusters with distinct nucleotide diversity and the ST types of isolates in Cluster A were ST97 and ST1, Cluster B were ST118 and Cluster C were ST 50, ST705 and ST479. Therefore, we inferred that the hla gene in S. aureus is evolving at varying rates in various genetic backgrounds. This was very similar to a previous study [23]. Interestingly, the tree based on MLST sequences was almost as same as the hla tree, revealing that the hla gene evolved with MLST background.
Based on analysis of clonal diversity, the 103 S. aureus isolates had 24 spa types and 7 MLST types, with t359, t730 and ST97 present in most isolates, consistent with previously reports in which ST97 was also the dominant ST type in S. aureus from bovine mastitis from China [38, 47]. Conversely, ST239 was reported to be the dominant ST type among 608 hospital-acquired S. aureus isolates recovered from human respiratory specimens, sterile body fluids, S. aureus bacteremia and non-S. aureus bacteremia patients in China [47, 48]. Importantly, 5 new spa types (t16314 from Beijing, t16315 from Beijing, t17095 from Liaoning, UK-1 from Hebei and UK-2 from Liaoning and Hebei) were reported for the first time in this study, indicating evolutionary occurrence of unique clones in the region. Most herds harbored at least 3 spa types, indicating diversity of the clonal background. When comparing spa/ ST types with distribution of isolates, there was a correlation, despite the presence of disparate molecular types.
Notably, there was no correlation between hla gene/peptide types and virulence gene clusters. Both virulence gene profile and hla gene/peptide types were associated with the molecular clone background, consistent with previous studies in which there was a strong correlation between hla gene/ peptide types and clonal background of isolates from human [13]. In addition, there were also differences in virulence genes of various molecular types of strains [38]. Therefore, clone background should be taken into consideration when using Hla as a candidate for vaccines.
The distribution on peptide types, virulence gene profile and molecular clone according to geographical location was not determined, due to the large variation in numbers of S. aureus isolates in herds and provinces. However, based on the difference of virulence cluster and spa types among herds, we inferred that geography might be an important factor when developing effective treatment strategies for bovine mastitis.
Conclusions
Multi-resistant S. aureus occurred in bovine mastitis with diverse resistance patterns. A great diversity of virulence gene patterns and spa typing was determined; hla gene evolved with MLST types; ST97 was the dominant types in S. aureus from bovine mastitis in China and virulence gene patterns were correlated with MLST types. All of these finding will be useful for future studies on anti-virulence therapies, immunogenicity and vaccine development. In addition, the similarity in diversity of hla gene from both humans and bovines make great significance in public health security.
Additional files
Figure S1. Original picture of PCR result-1: hla; 2: hlb; 3: hlc; 4: icaD; 5: sec; 6: sei; 7: seg; 8: icaA; 9: tsst; 10: coa; 11: nuc; 12: clfA; 13: clfB; 14: fnbA; 15: fnbB. (ZIP 31 kb)
Figure S2. Original picture of PCR result of hld gene. (JPEG 39 kb)
Figure S3. Original picture of PCR result of seb gene. (JPEG 7 kb)
Figure S4. Original picture of PCR result of see gene. (JPEG 5 kb)
Acknowledgments
The authors thank all the milking technicians, farm veterinarians and the commercial farms supporting the samples collection.
Funding
This research was financially supported by the National Natural Science Foundation of China (No. 31772813, 31572587 and No. 31550110200) and the National Key R&D Project (No. 2016YFD0501203), which beared the expenses, and High-end Foreign Experts Recruitment Program (No. GDT20171100013), with which we got the English native speakers revise the manuscript.
Availability of data and materials
The FASTA sequences of virulence genes in this study are not publicly available to avoid breaching owner confidentiality. However, they are available from the corresponding author on reasonable request. And the phylogenetic data was deposited into TreeBase (www.treebase.org) and is publicly available at http://purl.org/phylo/treebase/phylows/study/TB2:S22292. The study ID on Treebase is 22,292.
Abbreviations
- AMP
Ampicillin
- AMX/CA
augmentin
- CIP
Ciprofloxacin
- CL
Clindamicin
- clfA
clumping factors A
- clfB
clumping factors B
- CRO
Ceftriaxone
- CTX
Cefotaxime
- E
Erythromycin
- fnbA
fibronectin binding proteins A
- fnbB
fibronectin binding proteins B
- GM
Gentamicin
- Hla
Alpha-hemolysin
- hla
α- hemolysin
- hlb
β-hemolysin
- hlc
γ-hemolysin
- hld
δ-hemolysin
- icaA
intracellular adhesion A
- icaD
intracellular adhesion D
- MICs
Minimum inhibitory concentrations
- MLST
Multilocus sequence type
- OX
Oxacillin
- P
penicillin
- PCR
Polymerase chain reaction
- S. aureus
Staphylococcus aureus
- sea
Staphylococcus aureus enterotoxin a
- sec
Staphylococcus aureus enterotoxin c
- see
Staphylococcus aureus enterotoxin e
- seg
Staphylococcus aureus enterotoxin g
- sei
Staphylococcus aureus enterotoxin i
- spa
staphylococcal protein A
- STs
MLST types
- TE
tetracycline
- tsst
toxic shock syndrome toxin
Authors’ contributions
BH conceived and designed the experiment. LZ performed the research and wrote the manuscript, GL and YD collected the S. aureus isolates from milk samples, JG assisted in the result analysis. HWB, TA, SN and JPK revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The current work was carried out according to the ethical guide lines of China Agricultural University (CAU), Beijing. Prior to the initiation of this study, proper approval was obtained from the ethics committee of College of Veterinary Medicine, China Agricultural University, Beijing. Furthermore, sampling was performed in accordance to the standard protocols of National Mastitis Council (NMC). There are no vulnerable populations involved, and no endangered species was used in the experiments. Farm managers provided verbal consent to collect the milk samples.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12917-018-1374-7) contains supplementary material, which is available to authorized users.
Contributor Information
Limei Zhang, Email: 20073175@163.com.
Jian Gao, Email: gaojian2016@cau.edu.cn.
Herman W. Barkema, Email: barkema@ucalgary.ca
Tariq Ali, Email: tariq_ali@cau.edu.cn.
Gang Liu, Email: liugang_0402@126.com.
Youtian Deng, Email: idyt@outlook.com.
Sohail Naushad, Email: sohail.naushad@ucalgary.ca.
John P. Kastelic, Email: j.kastelic@ucalgary.ca
Bo Han, Phone: 86-10-62733801, Email: hanbo@cau.edu.cn.
References
- 1.Barkema HW, Schukken YH, Zadoks RN. Invited review: the role of cow, pathogen, and treatment regimen in the therapeutic success of bovine Staphylococcus aureus mastitis. J Dairy Sci. 2006;89:1877–1895. doi: 10.3168/jds.S0022-0302(06)72256-1. [DOI] [PubMed] [Google Scholar]
- 2.Schukken YH, Günther J, Fitzpatrick J, Fontaine MC, Goetze L, Holst O, Leigh J, Petzl W, Schuberth HJ, Sipka A, Smith DGE, Quesnell R, Watts J, Yancey R, Zerbe H, Gurjar A, Zadoks RN, Seyfert HM. Host-response patterns of intramammary infections in dairy cows. Vet Immun Immunopathol. 2011;144:270–289. doi: 10.1016/j.vetimm.2011.08.022. [DOI] [PubMed] [Google Scholar]
- 3.Gao J, Barkema HW, Zhang L, Liu G, Deng Z, Cai L, Shan R, Zhang S, Zou J, Kastelic JP, Han B. Incidence of clinical mastitis and distribution of pathogens on large Chinese dairy farms. J Dairy Sci. 2017;100:4797–4806. doi: 10.3168/jds.2016-12334. [DOI] [PubMed] [Google Scholar]
- 4.Kong C, Neoh HM, Nathan S. Targeting Staphylococcus aureus toxins: a potential form of anti-virulence therapy. Toxins. 2016;8:72. doi: 10.3390/toxins8030072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen RC, Popat R, Diggle SP, Brown SP. Targeting virulence: can we make evolution-proof drugs? Nat Rev Microbiol. 2014;12:300–308. doi: 10.1038/nrmicro3232. [DOI] [PubMed] [Google Scholar]
- 6.Yang FL, Li XS, Liang XW, Zhang XF, Qin GS, Yang BZ. Detection of virulence-associated genes in Staphylococcus aureus isolated from bovine clinical mastitis milk samples in Guangxi. Trop Anim Health Prod. 2012;44:1821–1826. doi: 10.1007/s11250-012-0143-z. [DOI] [PubMed] [Google Scholar]
- 7.Mitra SD, Velu D, Bhuvana M, Krithiga N, Banerjee A, Shome R, Rahman H, Ghosh SK, Shome BR. Staphylococcus aureus spa type t267, clonal ancestor of bovine subclinical mastitis in India. J Appl Microbiol. 2013;114:1604–1615. doi: 10.1111/jam.12186. [DOI] [PubMed] [Google Scholar]
- 8.Martins KB, Faccioli-Martins PY, Riboli DF, Pereira VC, Fernandes S, Oliveira AA, Dantas A, Zafalon LF, da Cunha Mde L. Clonal profile, virulence and resistance of Staphylococcus aureus isolated from sheep milk. Braz J Microbiol. 2015;46:535–543. doi: 10.1590/S1517-838246220131164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roetzer A, Haller G, Beyerly J, Geier CB, Wolf HM, Gruener CS, Model N, Eibl MM. Genotypic and phenotypic analysis of clinical isolates of Staphylococcus aureus revealed production patterns and hemolytic potentials unlinked to gene profiles and source. BMC Microbiol. 2016;16:13. doi: 10.1186/s12866-016-0630-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang D, Zhang L, Zhou X, He Y, Yong C, Shen M, Szenci O, Han B. Antimicrobial susceptibility, virulence genes, and randomly amplified polymorphic DNA analysis of Staphylococcus aureus recovered from bovine mastitis in Ningxia, China. J Dairy Sci. 2016;99:9560–9569. doi: 10.3168/jds.2016-11625. [DOI] [PubMed] [Google Scholar]
- 11.Menzies BE, Kernodle DS. Site-directed mutagenesis of the alpha-toxin gene of Staphylococcus aureus: role of histidines in toxin activity in vitro and in a murine model. Infect Immun. 1994;62:1843–1847. doi: 10.1128/iai.62.5.1843-1847.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGavin MJ, Arsic B, Nickerson NN. Evolutionary blueprint for host- and niche-adaptation in Staphylococcus aureus clonal complex CC30. Front Cell Infect Microbiol. 2012;2:48. doi: 10.3389/fcimb.2012.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao M, Zhao R, Zhang Q, Fan X, O'Sullivan MV, Li DF. Genotypic diversity of Staphylococcus aureusα-hemolysin gene (hla) and its association with clonal background: implications for vaccine development. PLoS One. 2016;11:e0149112. doi: 10.1371/journal.pone.0149112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adhikari RP, Thompson CD, Aman MJ, Lee JC. Protective efficacy of a novel alpha hemolysin subunit vaccine (AT62) against Staphylococcus aureus skin and soft tissue infections. Vaccine. 2016;34:6402–6407. doi: 10.1016/j.vaccine.2016.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mocca CP, Brady RA, Burns DL. Role of antibodies in protection elicited by active vaccination with genetically inactivated alpha hemolysin in a mouse model of Staphylococcus aureus skin and soft tissue infections. Clin Vaccine Immunol. 2014;21:622–627. doi: 10.1128/CVI.00051-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oscherwitz J, Cease KB. Identification and validation of a linear protective neutralizing epitope in the β-pore domain of alpha toxin. PLoS One. 2015;10:e0116882. doi: 10.1371/journal.pone.0116882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma-Kuinkel BK, Wu Y, Tabor DE, Mok H, Sellman BR, Jenkins A, Yu L, Jafri HS, Rude TH, Ruffin F, Schell WA, Park LP, Yan Q, Thaden JT, Messina JA, Fowler VG, Jr, Esser MT. Characterization of alpha-toxin hla gene variants, alpha-toxin expression levels, and levels of antibody to alpha-toxin in hemodialysis and postsurgical patients with Staphylococcus Aureus bacteremia. J Clin Microbiol. 2015;53:227–236. doi: 10.1128/JCM.02023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tabor DE, Yu L, Mok H, Tkaczyk C, Sellman BR, Wu Y, Oganesyan V, Slidel T, Jafri H, McCarthy M, Bradford P, Esser MT. Staphylococcus aureus Alpha-toxin is conserved among diverse hospital respiratory isolates collected from a global surveillance study and is neutralized by monoclonal antibody MEDI4893. Antimicrob Agents Chemother. 2016;60:5312–5321. doi: 10.1128/AAC.00357-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial disk and dilution suscptibility tests for bacteria isolated from animals. Approved standard-fourth edition. 2013; CLSI document VET01-A4, Wayne, PA.
- 20.Lin Y, Barker E, Kislow J, Kaldhone P, Stemper ME, Pantrangi M, Moore FM, Hall M, Fritsche TR, Novicki T, Foley SL, Shukla SK. Evidence of multiple virulence subtypes in nosocomial and community-associated MRSA genotypes in companion animals from the upper midwestern and northeastern United States. Clin Med Res. 2011;9:7–16. doi: 10.3121/cmr.2010.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kot B, Szweda P, Frankowska-Maciejewska A, Piechota M, Wolska K. Virulence gene profiles in Staphylococcus aureus isolated from cows with subclinical mastitis in eastern Poland. J Dairy Res. 2016;83:228–235. doi: 10.1017/S002202991600008X. [DOI] [PubMed] [Google Scholar]
- 22.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tavares A, Nielsen JB, Boye K, Rohde S, Paulo AC, Westh H, Schonning K, de Lencastre H, Miragaia M. Insights into alpha-hemolysin (Hla) evolution and expression among Staphyloccus aureus clones with hospital and community origin. PLoS One. 2014;(7):e98634. [DOI] [PMC free article] [PubMed]
- 24.Li YL, Han GM, He SE, Zhang ZJA. New strategy for construction of phylogenetic tree based on DNA molecular marker data. China J Bioinformatics. 2007;4:168–170. [Google Scholar]
- 25.Gray GS, Kehoe M. Primary sequence of the alpha-toxin gene from Staphylococcus aureus wood 46. Infect Immun. 1984;46:615–618. doi: 10.1128/iai.46.2.615-618.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koziel J, Chmiest D, Bryzek D, Kmiecik K, Mizgalska D, Maciag-Gudowska A, Shaw LN, Potempa J. The Janus face of alpha-toxin: a potent mediator of cytoprotection in staphylococci-infected macrophages. J. Innate Immunity. 2015;7:187–198. doi: 10.1159/000368048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rauch S, Gough P, Kim HK, Schneewind O, Missiakas D. Vaccine protection of leukopenic mice against Staphylococcus aureus bloodstream infection. Infect Immun. 2014;82:4889–4898. doi: 10.1128/IAI.02328-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller ML, Soufi B, Jers C, Blom N, Macek B, Mijakovic I. NetPhosBac – a predictor for Ser/Thr phosphorylation sites in bacterial proteins. Proteomics. 2009;9:116–125. doi: 10.1002/pmic.200800285. [DOI] [PubMed] [Google Scholar]
- 29.Jamrozy DM, Fielder MD, Butaye P, Coldham NG. Comparative genotypic and phenotypic characterisation of methicillin-resistant Staphylococcus aureus ST398 isolated from animals and humans. PLoS One. 2012;7:e40458. doi: 10.1371/journal.pone.0040458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdi RD, Gillespie BE, Vaughn J, Merrill C, Headrick SI, Ensermu DB, D'Souza DH, Agga GE, Almeida RA, Oliver SP, Kerro Dego O. Antimicrobial resistance of Staphylococcus aureus isolates from dairy cows and genetic diversity of resistant isolates. Foodborne Pathog Dis. 2018; 10.1089/fpd.2017.2362. [DOI] [PubMed]
- 31.de Jong A, Garch FE, Simjee S, Moyaert H, Rose M, Youala M, Siegwart E, VetPath Study Group Monitoring of antimicrobial susceptibility of udder pathogens recovered from cases of clinical mastitis in dairy cows across Europe: VetPath results. Vet Microbiol. 2018;213:73–81. doi: 10.1016/j.vetmic.2017.11.021. [DOI] [PubMed] [Google Scholar]
- 32.Kulangara V, Nair N, Sivasailam A, Sasidharan S, Kollannur JD, Syam R. Genotypic and phenotypic β-lactam resistance and presence of PVL gene in staphylococci fromdry bovine udder. PLoS One. 2017;12:e0187277. doi: 10.1371/journal.pone.0187277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haubert L, Kroning IS, Iglesias MA, da Silva WP. First report of the Staphylococcus Aureus isolate from subclinical bovine mastitis in the south of Brazil harboring resistance gene dfrG and transposon family Tn916-1545. Microb Pathog. 2017;113:242–247. doi: 10.1016/j.micpath.2017.10.022. [DOI] [PubMed] [Google Scholar]
- 34.Prashanth K, Rao KR, Reddy PV, Saranathan R, Makki AR. Genotypic characterization of Staphylococcus aureus obtained from humans and bovine mastitis samples in India. J Global Infect Dis. 2011;3:115–122. doi: 10.4103/0974-777X.81686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feltrin F, Alba P, Kraushaar B, Ianzano A, Argudín MA, Di Matteo P, Porrero MC, Aarestrup FM, Butaye P, Franco A, Battisti A. A livestock-associated, multidrug-resistant, methicillin-resistant Staphylococcus aureus clonal complex 97 lineage spreading in dairy cattle and pigs in Italy. Appl Environ Microbiol. 2016;82:816–821. doi: 10.1128/AEM.02854-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gogoi-Tiwari J, Waryah CB, Eto KY, Tau M, Wells K, Costantino P, Tiwari HK, Isloor S, Hegde N, Mukkur T. Relative distribution of virulence-associated factors among Australian bovine Staphylococcus aureus isolates: potential relevance to development of an effective bovine mastitis vaccine. Virulence. 2015;6:419–423. doi: 10.1080/21505594.2015.1043508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson T, Brown PD. Comparison of antibiotic resistance, virulence gene profiles, and pathogenicity of methicillin-resistant and methicillin-susceptible Staphylococcus aureus using a caenorhabditis elegans infection model. Pathog Glob Health. 2014;108:283–291. doi: 10.1179/2047773214Y.0000000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li T, Lu H, Wang X, Gao Q, Dai Y, Shang J, Li M. Molecular characteristics of Staphylococcus aureus causing bovine mastitis between 2014 and 2015. Front Cell Infect Microbiol. 2017;7:127. doi: 10.3389/fcimb.2017.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mairpady Shambat S, Haggar A, Vandenesch F, Lina G, van Wamel WJB, Arakere G, Svensson M, Norrby-Teglund A. Levels of alpha-toxin correlate with distinct phenotypic response profiles of blood mononuclear cells and with agr background of community-associated Staphylococcus aureus isolates. PLoS One. 2014;9:e106107. doi: 10.1371/journal.pone.0106107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maurer K, Reyes-Robles T, Alonzo F, 3rd, Durbin J, Torres VJ, Cadwell K. Autophagy mediates tolerance to Staphylococcus aureus Alpha-toxin. Cell Host Microbe. 2015;17:429–440. doi: 10.1016/j.chom.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Popov LM, Marceau CD, Starkl PM, Lumb JH, Shah J, Guerrera D, Cooper RL, Merakou C, Bouley DM, Meng W, Kiyonari H, Takeichi M, Galli SJ, Bagnoli F, Citi S, Carette JE, Amieva MR. The adherens junctions control susceptibility to Staphylococcus aureus Alpha-toxin. PNAS. 2015;112:14337–14342. doi: 10.1073/pnas.1510265112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tkaczyk C, Kasturirangan S, Minola A, Jones-Nelson O, Gunter V, Shi YY, Rosenthal K, Aleti V, Semenova E, Warrener P, Tabor D, Stover CK, Corti D, Rainey G, Sellman BR. Multimechanistic monoclonal antibodies (MAbs) targeting Staphylococcus aureus Alpha-toxin and clumping factor a: activity and efficacy comparisons of a MAb combination and an engineered bispecific antibody approach. Antimicrob Agents Chemother. 2017;61:e00629-17. doi: 10.1128/AAC.00629-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang X, Hall JW, Yang J, Yan M, Doll K, Bey R, Ji Y. Identification of single nucleotide polymorphisms associated with hyperproduction of alpha-toxin in Staphylococcus aureus. PLoS One. 2011;6:e18428. doi: 10.1371/journal.pone.0018428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeLeo FR, Kennedy AD, Chen L, Wardenburg JB, Kobayashi SD, Mathema B, Braughton KR, Whitney AR, Villaruz AE, Martens CA, Porcella SF, McGavin MJ, Otto M, Musser JM, Kreiswirth BN. Molecular differentiation of historic phage-type 80/81 and contemporary epidemic Staphylococcus aureus. PNAS. 2011;108:18091–18096. doi: 10.1073/pnas.1111084108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burnside K, Lembo A. De los Reyes M, lliuk a, BinhTran NT, Connelly JE. Regulation of hemolysin expression and virulence of Staphylococcus aureus by a serine/threonine kinase and phosphatase. PLoS One. 2015;5:e11071. doi: 10.1371/journal.pone.0011071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang D, Wang Z, Yan Z, Wu, J, Ali T, Li J, Han B. Bovine mastitis Staphylococcus aureus: antibiotic susceptibility profile, resistance genes and molecular typing of methicillin-resistant and methicillin-sensitive strains in China. Infect. Genet. Evol. 2015; 31: 9–16. [DOI] [PubMed]
- 47.Li T, Song Y, Zhu Y, Du X, Li M. Current status of Staphylococcus aureus infection in a central teaching hospital in shanghai, China. BMC Micorob. 2013;13:153. doi: 10.1186/1471-2180-13-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang B, Yin S, You B, Huang G, Yang Z, Zhang Y, Chen Y, Chen J, Yuan Z, Rao X, Hu X, Gong Y, Peng Y. A 5-year survey reveals increased susceptibility to glycopeptides for methicillin-resistant Staphylococcus aureus isolates from Staphylococcus aureus bacteremia patients in a Chinese burn center. Front Microbiol. 2017;8:2531. doi: 10.3389/fmicb.2017.02531. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Original picture of PCR result-1: hla; 2: hlb; 3: hlc; 4: icaD; 5: sec; 6: sei; 7: seg; 8: icaA; 9: tsst; 10: coa; 11: nuc; 12: clfA; 13: clfB; 14: fnbA; 15: fnbB. (ZIP 31 kb)
Figure S2. Original picture of PCR result of hld gene. (JPEG 39 kb)
Figure S3. Original picture of PCR result of seb gene. (JPEG 7 kb)
Figure S4. Original picture of PCR result of see gene. (JPEG 5 kb)
Data Availability Statement
The FASTA sequences of virulence genes in this study are not publicly available to avoid breaching owner confidentiality. However, they are available from the corresponding author on reasonable request. And the phylogenetic data was deposited into TreeBase (www.treebase.org) and is publicly available at http://purl.org/phylo/treebase/phylows/study/TB2:S22292. The study ID on Treebase is 22,292.


