Abstract
Background
Although cytotoxic T lymphocytes (CTLs) store perforin within cytoplasmic secretory granules for immediate use, perforin is synthesized anew within hours of TCR stimulation. Previously, we observed new perforin protein at an immunologic synapse independent of secretory lysosomes; herein we aimed to determine how new perforin transits to the synapse if not via lytic granules.
Results
We analyzed antigen-specific human CTLs via imaging flow cytometry and high-resolution confocal microscopy, with attention to intracellular trafficking components and new perforin. The recycling endosome compartments identified by rab8, rab11a, rab4, and rab37 co-localized with new perforin, as well as the SNAREs vti1b and VAMP4. After ablating the function of the recycling endosome pathway, we observed a relative accumulation of new perforin in rab8 vesicles.
Conclusions
The recycling endosome pathway may serve as an auxiliary intracellular route for the delivery of new perforin to an immunologic synapse in order to perpetuate a cytotoxic response.
Keywords: Cytotoxic T Lymphocytes, Perforin, Trafficking, Recycling Endosomes
INTRODUCTION
One mechanism by which human cytotoxic T lymphocytes (CTLs) kill compromised host cells is the targeted exocytosis of specialized secretory lysosomes (lytic granules) that contain perforin and granzyme proteins, the chief mediators of cytotoxicity. Perforin is indispensable for this killing response: CTLs from both humans and mice with defective perforin protein or perforin production experience profound immunodeficiency[1, 2]. While lytic granule release is an immediate response to stimulation, CTLs can sustain this function to eliminate multiple target cells serially[3, 4]. The mechanism by which CTLs continue to kill in the face of a persistent antigenic burden is not understood given that the supply of lytic granules in the cytoplasm available for immediate deployment is limited.
It is known that human antigen-specific CTLs produce new perforin protein within hours of stimulation[5]. During this response, new perforin is concentrated at the immunologic synapse independent of secretory lysosomes[6]. This observation begs the question of how new perforin traffics to a synapse if not via the canonical secretory granule route. There has been conjecture of a granule-independent trafficking pathway for perforin previously: it was postulated from biochemical studies that some perforin may transit via the constitutive secretory pathway[7], however this was never proven formally. While the authors concluded this phenomenon was strictly a non-specific byproduct of perforin regeneration, such a scenario would be inefficient and could produce significant collateral damage – a result that is inconsistent with validated literature of directed lymphocyte killing. We contend that new perforin is targeted immediately and specifically to the immunologic synapse to sustain an antigen-specific cytotoxic response[6]. In support, it was demonstrated that while glycosylation of perforin is necessary for its delivery to lytic granules, non-glycosylation-dependent, alternative trafficking of perforin occurred as well; this mode was responsible for approximately 20% of the observed killing by CTL and NK cells[8]. Thus, while the secretory granule pathway is the main destination of perforin protein, an alternative intracellular trafficking pathway is available to deliver perforin to the immunological synapse in times of duress. Our objective was to define the specific components of this auxiliary highway in human CD8 T cells.
T cells possess a variety of intracellular trafficking pathways to exocytose newly synthesized proteins. Rab GTPases are membrane proteins that direct this process by conferring orientation and specificity to intracellular transport vesicles[9, 10]. Similarly, Soluble NSF (N-ethylmaleimide-sensitive fusion) Attachment Protein Receptor (SNAREs) proteins mediate proper tethering and fusion of vesicles to their target compartments[11]. Unique Rab and SNARE proteins regulate discrete trafficking pathways for cargo with specialized functions: Activated CD4 T cells secrete IL-2 and IFN-g into the immunologic synapse for direct, local effects, whereas they release TNF-a and CCL3 in multiple directions away from the synapse in order to coordinate a general inflammatory response. The former process is associated with Rab3d, Rab19, and Rab37, whereas the latter pathway involves the SNARE protein syntaxin6[12]. In cytotoxic lymphocytes, trafficking of lytic granules to the immunologic synapse involves the fusion of a recycling endosome bearing rab11 and munc13-4 with a late endosomal compartment containing rab27a and rab7. The resulting vesicle then fuses with the immature lytic granule, providing the granule with the molecular machinery needed to dock at, and fuse with, the immunological synapse[13, 14].
New perforin synthesis is critical to replenish the cytoplasmic store of lytic granules, however in human CTLs new perforin protein appears rapidly after stimulation at the immunologic synapse independent of secretory lysosomes [6]. Given this specialized function to sustain a cytotoxic response, an auxiliary pathway likely exists to expedite the delivery of new perforin to the synapse in times of need by bypassing the granule intermediate. We hypothesized that new perforin may travel through recycling endosomes, as they are known to transport cargo to and from the plasma membrane, as well as the immunologic synapse. Here, we define essential components to the trafficking pathway utilized by newly synthesized perforin in activated human CD8 T cells.
RESULTS
High-throughput screening of perforin localization in antigen-specific human CTL
Previously, via confocal microscopy, new perforin was visualized at the immunologic synapse (IS) between human CMV-specific CD8 T cells and antigen-presenting cells (APCs) bearing HLA-restricted cognate peptide: The new perforin did not co-localize with rab7-labeled late endosomes – the immediate precursors to secretory granules -- nor Lysotracker-labeled secretory lysosomes[6]. This observation begs the question: If not via the canonical lytic granule secretory pathway, how does new perforin transit from the Golgi to an immunological synapse? We hypothesized a trafficking pathway for new perforin protein, complimentary to the traditional secretory granule route, that utilizes the cell’s recycling endosomes, as they are known conduits for newly synthesized, rapidly secreted proteins[15].
Finding a renewable source of perforin-upregulating human CTLs for extensive study is challenging, as the ability of CTLs to upregulate perforin varies from person-to-person[5]. Previous studies of newly synthesized perforin were done with ex vivo human CTL and anti-human antibodies, and no suitable cell line or mouse model exists. To study the intracellular trafficking of new perforin, we aimed to screen 3–6 human donors per protein trafficking marker (rabs and SNAREs), with attention to the co-localization of newly synthesized perforin with those markers. To accomplish this, we screened human CTL for perforin upregulation in response to various antigen pools—CMV pp65, Influenza non-structural protein, Influenza matrix protein, CEF peptide pool representing immunodominant epitopes from CMV, EBV, and influenza viruses, and staphylococcal enterotoxin B (SEB), a super-antigen—via flow cytometry. CTL from donors with positive responses were then expanded with the appropriate peptide pool ex vivogenerating a semi-renewable source of perforin-upregulating human CTL. Detection of new, granule-independent perforin is made possible through the tandem use of two anti-perforin antibody clones: the D48 anti-perforin clone detects new and granule-associated perforin while the δG9 clone detects only granule-associated perforin. Therefore, new perforin is D48+ δG9-, while granule-associated perforin is D48+ δG9+[5, 6]. The reliance on these antibodies necessitates using static microscopy rather than live cell imaging. The combination of these issues would have made our initial colocalization screening process, consisting of 16 different rabs and SNAREs, impractical by traditional confocal microscopy. Therefore, we aimed to find a high-throughput means of performing our initial screening, with the idea that positive results could be followed up by traditional confocal microscopy. We utilized imaging flow cytometry (IFC)—IFC combines the statistical robustness of flow cytometry with the imaging power of confocal microscopes, allowing the user to acquire thousands of cells for traditional flow cytometric analysis, while also obtaining images of each cell.
To analyze the IFC results, we used IDEAS software (Amnis) to create custom masks to distinguish newly synthesized perforin from granule-associated perforin and assess its co-localization with rabs and SNAREs. Masks are analysis tools that identify all areas with a signal above background for a particular antibody. Using the existing knowledge that the D48 anti-perforin clone detects new and granule-associated perforin while the δG9 clone detects only granule-associated perforin, masks were created that recognized all spots of either perforin D48 or δG9, as well as masks for the rabs and SNAREs. Boolean logic was then used to create combination masks that would identify and distinguish new perforin—spots that are covered by the D48 mask but not the δG9 mask (D48+ δG9-), and granule perforin—spots covered by both the D48 and δG9 masks (D48+ δG9+). A custom co-localization analysis feature was then created, which measures the degree of overlap of the D48+ δG9- mask with a mask created for each trafficking marker. The co-localization features in IDEAS generate Bright Detail Similarity (BDS) scores—BDS scores are log-transformed Pearson’s correlation coefficients, and are a measure of the degree to which the images of two probes are correlated within a defined area. Analysis of co-localization was performed exclusively on the areas covered by our custom masks. Acquired cells were gated to eliminate out-of-focus cells, debris and dead cells, and NK (CD56+) cells; CD8+ Perforin+ cells were selected (gating strategy shown in Figure S1). For comparison, we included extracellular CD8 as a negative co-localization control, as we expected to image perforin inside of the cell; and granzyme B compared to lytic granule perforin (D48+ δG9+) as a positive co-localization control, as lytic granules contain both perforin and granzyme B.
Imaging flow cytometry reveals co-localization with recycling endosome compartments
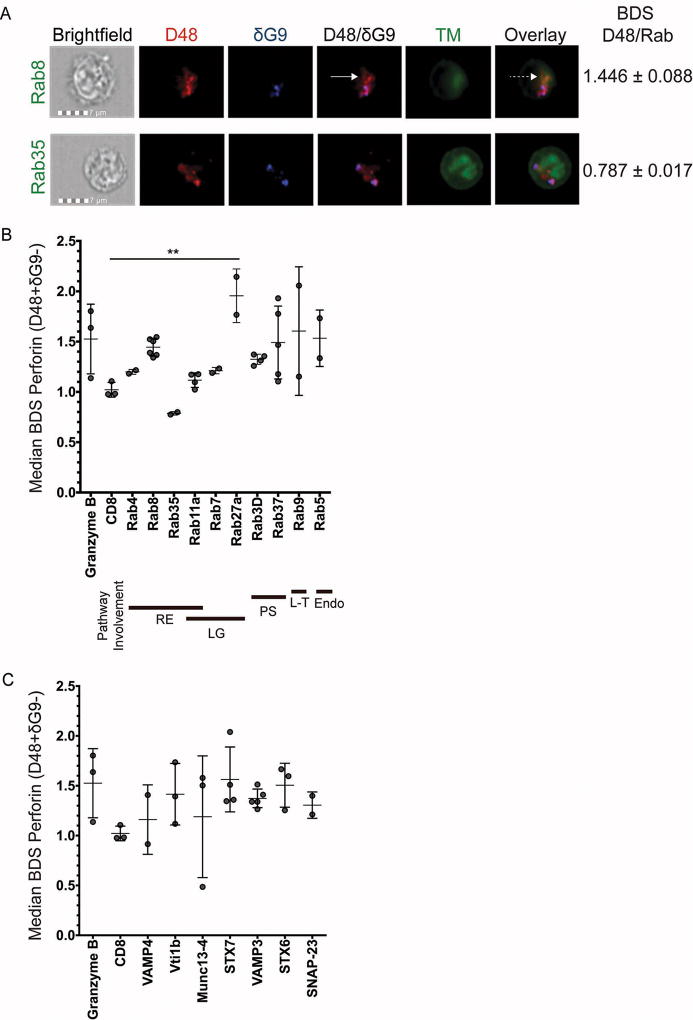
For our initial screening, we focused on a panel of rab-labeled vesicles implicated in protein secretion from immune cells: Rab4, rab8, rab11a, and rab35, rab3D and rab37. Also included as negative controls: rab5, since it is involved in endocytosis and therefore not expected to co-localize with secreted proteins[16]; rab7, as it is known to co-localize with late endosomes and lytic granules[17, 18]; and rab9, as it is involved in trafficking between late endosomes and the trans-golgi network (TGN)[19]. As displayed in Figure 1A, new perforin (D48+ δG9- appears red, but not blue) co-localized with the recycling endosome component rab8 (top row green; co-localization appears orange in the overlay; BDS = 1.446) but not with Rab35 (bottom row green; no co-localization in the overlay; BDS = 0.787). Representative images for all the markers we screened are illustrated in Figure S2. Figuress 1B and 1C depict the median BDS scores of new perforin (D48+ δG9-) with our panel of Rab and SNARE markers, respectively; these data are summarized in Table 1. New perforin appeared to co-localize with rab8 (BDS= 1.446), rab11a (BDS=1.117), and rab4 (BDS=1.199). New perforin also co-localized with rab37 (BDS=1.491). Surprisingly, rab5 also appeared to co-localize with new perforin (BDS=1.534). In addition, Rab9 (BDS=1.605) and rab27a (BDS=1.956) appeared to co-localize with new perforin, whereas Rab3D (BDS=1.325) and rab35 (BDS=0.787) did not co-localize appreciably.
Fig 1. High-throughput analysis of perforin co-localization by imaging flow cytometry.
Antigen-specific CTL from normal human donors were expanded ex vivo for 10–14 days, re-stimulated with their cognate antigen for 6 hours, and then stained with the indicated antibodies plus anti-CD8 and anti-CD56. An imaging flow cytometer was used to capture images at 60X magnification. Representative images showing the localization of perforin with trafficking markers (TM) are shown in (A). Solid white arrows point to new perforin (D48+ δG9-) and the dashed white arrow shows co-localization between new perforin and rab8. B–C) Bright detail similarity (BDS) values indicate the similarity of the TM image with the image of new perforin (masked D48+ δG9-). The plotted BDS scores represent the median score of all D48+ δG9- spots within the cells, with each symbol denoting one experiment with one human donor. Horizontal lines depict the mean of all donors. Error bars show the standard deviation of all donors combined. Granzyme B and CD8 represent positive and negative co-localization controls, respectively. 2–4 experiments were performed for each trafficking marker, and 10–1200 cells containing new perforin were analyzed from each donor. Bold lines underneath the×axis in (B) represent the pathway(s) each rab is associated with: RE=recycling endosome, LG=lytic granule-associated, PS=(general) protein secretion, L-T=Lysosome-to-TGN trafficking, Endo=endocytosis. **p<0.01, t test compared to negative control (CD8).
Table 1.
Bright Detail Similarity (BDS) scores.
| Trafficking marker | New perforin (D48+ δG9−) BDS (Median ± St. Dev) |
# of Donors |
N (cells analyzed per donor) |
|---|---|---|---|
| Granzyme B (positive control) | 1.526±0.3467 | 3 | 4, 5, 16 |
| CD8 (negative control) | 1.022±0.0730 | 3 | 48, 49, 72 |
| Rab3D | 1.325±0.0504 | 4 | 100, 119, 178, 301 |
| Rab4 | 1.199±0.0240 | 2 | 2, 151 |
| Rab5 | 1.534±0.2807 | 2 | 61, 83 |
| Rab7 | 1.211±0.0311 | 2 | 372, 1298 |
| Rab8 | 1.446±0.0888 | 6 | 157, 269, 318, 346, 394, 530 |
| Rab9 | 1.605±0.6392 | 2 | 367, 766 |
| Rab11a | 1.117±0.0725 | 4 | 61, 70, 136, 175 |
| Rab27a | 1.956±0.2666** | 2 | 102, 658 |
| Rab35 | 0.787±0.0173 | 2 | 87, 126 |
| Rab37 | 1.491±0.3621 | 5 | 16, 78, 153, 212, 236 |
| Munc13-4 | 1.189±0.3467 | 3 | 183, 274, 348 |
| SNAP-23 | 1.306±0.1322 | 2 | 9, 100 |
| Syntaxin 6 (STX6) | 1.505±0.2205 | 3 | 16, 37, 284 |
| Syntaxin 7 (STX7) | 1.564±0.3256 | 4 | 84, 182, 248, 411 |
| VAMP3 | 1.373±0.0933 | 5 | 45, 53, 108, 376, 423 |
| VAMP4 | 1.161±0.3484 | 2 | 88, 220 |
| Vti1b | 1.415±0.3091 | 3 | 32, 91, 648 |
p<0.01 t test vs negative control (CD8).
Values in Table 1 correspond to the graphs in Fig 1B–C.
Another family of proteins that regulate efficient intracellular protein transport is SNAREs. These proteins orchestrate the proper orientation and union of vesicles, and are expressed on unique compartments of the various pathways. Consistent with our hypothesis for the recycling endosome pathway, we tested for the SNAREs on vesicles of this pathway as new perforin carriers: Syntaxins 6 and 7 (STX6 and STX7), VAMP3, VAMP4, vti1b, and SNAP-23. Akin to the rabs, the SNAREs we studied initially have reported roles in mediating immune cell/T cell function: recycling pathways (vti1b, VAMP3, VAMP4, STX6, STX7); cytokine transport (vti1b, STX6, STX7, VAMP3, SNAP-23); granule secretion (STX7, SNAP-23, vti1b)[20–20–24]. In our study, the SNAREs VAMP4 (BDS=1.161) and vti1b (BDS=1.415) overlapped with new perforin, and STX6 (BDS=1.505) and STX7 (BDS=1.564) showed co-localization as well (Figure 1C, Table 1, Figure S2). VAMP3 (BDS=1.373) overlapped with granule perforin, but not new perforin. SNAP-23 did not co-localize with either type of perforin (BDS=1.306).
Newly synthesized perforin localizes with rab8, rab4, rab37, rab11a, VAMP4, and vti1b
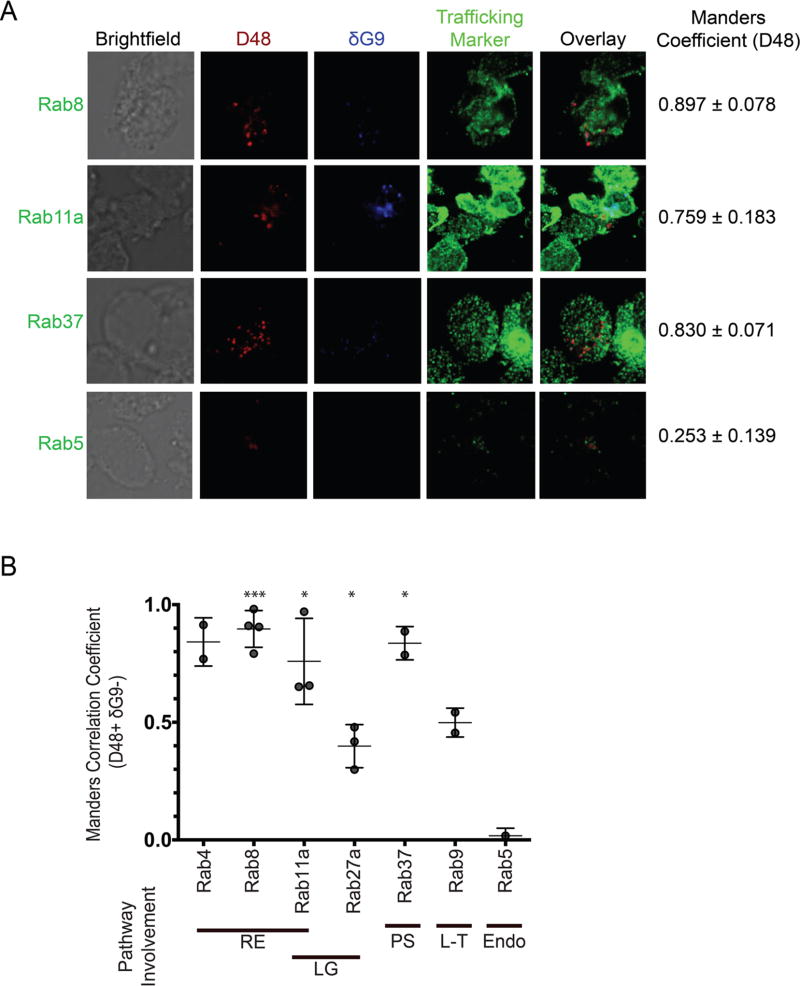
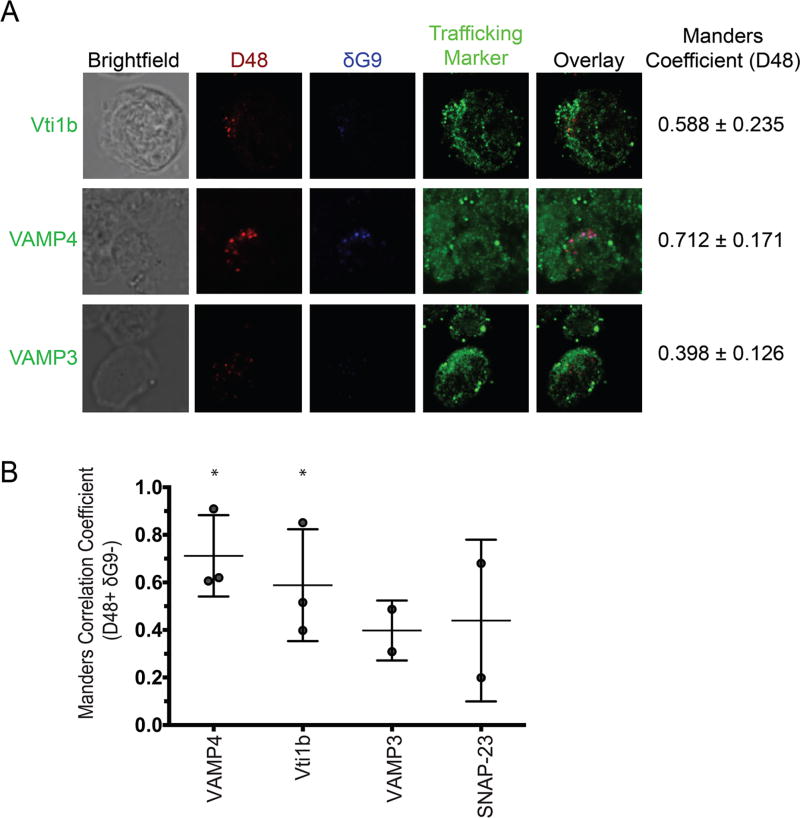
IFC analysis identified the recycling endosome-associated proteins rab8, rab4, and rab11a, rab37, and the SNAREs vti1b and VAMP4, as being co-localized with new perforin. However, IFC is limited in resolution and may not necessarily distinguish between markers that are co-localized truly from those that are only spatially close. Furthermore, the dynamic range of the BDS scale is quite narrow, which complicates the resolution of biologically relevant scores. Consequently, when the BDS scores of each rab/SNARE were compared to the negative control (CD8), only one comparison (rab27a vs. CD8) achieved statistical significance (p<0.01). Thus, we sought to confirm our putative trafficking markers by high-resolution confocal microscopy. We relied on the Manders M1 coefficient to quantify the degree of co-localization between D48+ δG9- perforin, again using a mask to specifically analyze D48+ δG9- only, and our array of candidate trafficking markers (Table 2). M1 values range between 0 and 1: 1 represents perfect co-localization whereas 0 represents none[25]. As such, the M1 values we obtained were statistically compared to 0. This analysis confirmed the co-localization of the recycling endosome components rab8 (M1=0.8970, p=0.0002), rab11a (M1=0.7590, p=0.0188), rab4 (M1=0.8415, p=0.0547), and rab37 (M1=0.8300, p=0.0380) with new perforin (Figure 2A–B, Table 2, Figure S3). Our negative controls Rab5 (M1=0.0175, p=0.2430), rab9 (M1=0.4985, p=0.0554), and rab27a (M1=0.3987, p=0.0171) – involved in lytic granule secretion -- did not show high degrees of overlap with new perforin, validating our hypothesis of an auxiliary transport pathway for new perforin. Additionally, VAMP4 showed a high degree of co-localization with new perforin (M1=0.7120, p=0.0187), and vti1b showed also showed co-localization with new perforin, but at a lower degree (M1=0.5160, p=0.0493) (Figure 3A–B, Table 2, Figure S3). Consistent with IFC analysis, VAMP3 (M1= 0.4395, p=0.1401) and SNAP-23 (M1=0.3980, p=0.3188) did not co-localize with new perforin (Figure 3, Figure S4). These results refined our interpretation of the IFC analyses, and affirmed the recycling endosome pathway as a real mode for the intracellular transport of new perforin protein.
Table 2.
Manders (M1) scores.
| Trafficking marker |
Manders (M1) (Median) |
# of Donors |
N (cells analyzed per donor) |
|---|---|---|---|
| Rab4 | 0.8415 | 2 | 8, 19 |
| Rab5 | 0.0175 | 1 | 6 |
| Rab8 | 0.8970 *** | 4 | 4, 13, 14, 17 |
| Rab9 | 0.4985 | 2 | 4, 8 |
| Rab11a | 0.7590 * | 3 | 5, 5, 12 |
| Rab27a | 0.3987 * | 3 | 2, 14, 24 |
| Rab37 | 0.8300 * | 2 | 15, 17 |
| SNAP-23 | 0.4395 | 2 | 2, 10 |
| VAMP3 | 0.3980 | 2 | 9, 21 |
| VAMP4 | 0.7120 * | 3 | 6, 6, 15 |
| Vti1b | 0.588 * | 3 | 2, 9, 18 |
Fig 2. Newly synthesized (D48+ dG9-) perforin co-localizes with rab4, rab8, rab11a, and rab37.
Antigen-specific CTL from normal human donors were expanded ex vivo for 10–14 days, re-stimulated with their cognate antigen for 6 hours, and then stained with the indicated antibodies and imaged on a confocal microscope with a 100X oil objective. A) Representative images showing perforin localization with rab proteins. Manders values shown in (A) represent the mean values of all the experiments combined. B) Manders correlation coefficients between perforin and the indicated trafficking marker were measured after selecting new perforin (D48+ δG9-) spots only. Each symbol represents the mean score of 1 donor, and the horizontal lines represent the mean of all donors. Errors bars represent the standard deviation of all the donors combined. The shading of the graph shows the pathway associations of each rab. 2–6 experiments with separate human donors were performed for each trafficking marker, and 2–20 cells were analyzed from each donor. Bold lines underneath the×axis in (B) represent the pathway(s) each rab is associated with: RE=recycling endosome, LG=lytic granule-associated, PS=(general) protein secretion, L-T=Lysosome-to-TGN trafficking, Endo=endocytosis. *p<0.05, ***p<0.001 t test compared to a theoretical mean of 0 (no co-localization).
Fig 3. Newly synthesized (D48+ dG9-) perforin co-localizes with vti1b and VAMP4.
Antigen-specific CTL from normal human donors were expanded ex vivo for 10–14 days, re-stimulated with their cognate antigen for 6 hours, and then stained with the indicated antibodies and imaged on a confocal microscope with a 100X oil objective. A) Representative images showing perforin localization with SNARE proteins. Manders values shown in (A) represent the mean values of all the experiments combined. B) Manders correlation coefficients between perforin and the indicated trafficking marker were measured after selecting new perforin (D48+ δG9-) spots only. Each symbol represents the mean score of 1 donor, and the horizontal lines represent the mean of all donors. Errors bars represent the standard deviation of all the donors combined. 2–3 experiments with separate human donors were performed for each trafficking marker, and 5–20 cells were analyzed from each donor. *p<0.05, t test compared to a theoretical mean of 0 (no co-localization).
Granzyme B-independent trafficking of newly synthesized perforin
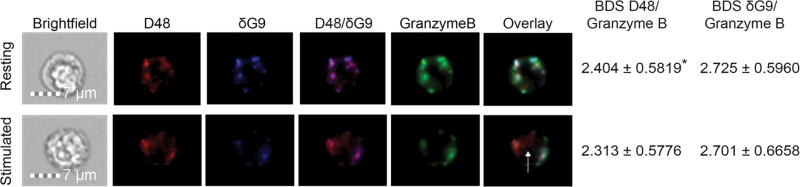
In addition to perforin, lytic granules carry an assortment of serine proteases, one of which is granzyme B (GrzB)[26, 27]. Given its independence from lytic granules, we investigated whether new perforin trafficked with or without of granzyme B. Granule perforin (D48+ δG9+) largely co-localized with granzyme B, as expected, (BDS D48/GrzB=2.404, δG9/GrzB=2.725 in resting CD8+ cells) whereas some new (D48+ δG9-) perforin co-localized with granzyme B and some did not (Figure 4; BDS D48/GrzB=2.313, δG9/GrzB=2.701 in stimulated CD8+ cells). The co-localization of granzyme B and D48+ perforin significantly decreased (p=0.0471) upon stimulation, whereas co-localization between perforin δG9 and granzyme B did not decrease significantly (p=0.6385), consistent with the production of new perforin upon stimulation and incomplete co-localization with granzyme B. These results demonstrate that newly synthesized perforin does not traffic reliably with granzyme B, consistent with its independence from the lytic granule pathway.
Fig 4. Newly synthesized perforin may traffic with or independently of granzyme B.
Antigen-specific CTL from normal human donors were expanded ex vivo for 10–14 days, re-stimulated with their cognate antigen for 6 hours, and then stained with the indicated antibodies plus anti-CD8. Images were captured by imaging flow cytometry at 60X magnification. Representative images are shown. The white arrow indicates new perforin trafficking independently of granzyme B. BDS values indicate the similarity of the granzyme B image with the image of perforin D48 (BDS D48) and perforin δG9 (BDS δG9). BDS scores given represent the median score of all CD8+/CD56-/granzymeB+/perforin D48+ cells in the pooled experiments. The data represent 3 experiments with 5–20 cells analyzed per donor. *p<0.05, t test (resting vs. stimulated).
Ablation of recycling endosomes increases perforin retention in rab8+ vesicles and decreases perforin intensity
Our microscopy results support our hypothesis that the intracellular transport of new perforin protein involves the recycling endosome pathway. To validate our hypothesis formally, we devised a unique recycling endosome ablation assay we adapted from existing literature[28] to suit our experimental setting. Our objective was to inhibit recycling endosome function through enzymatic inactivation while leaving the functions of early and late endosomes intact[29–31]. Since recycling endosomes are involved in the shuttling of TCR components to and from the immunological synapse[32], we tested our improvised system initially by attempting to inhibit the internalization of CD3 upon stimulation. Normally, CD3 is internalized from the cell surface of a cell into recycling endosomes, as is the iron-binding protein transferrin (Tfn). To ablate recycling endosome function, we added Tfn conjugated to horseradish peroxidase (Tfn-HRP) to stimulated CTL, which becomes internalized into the recycling endosomes, and then added HRP’s substrate 3-3’-diaminobenzidine (DAB) and hydrogen peroxide. This process results in a build-up of a solid substrate in the recycling endosome, thereby halting further progression of proteins in the recycling pathways. As illustrated in Figure S5a, HRP-labeled transferrin was successfully internalized into CD8+ T cells; and upon enzymatic ablation decrease in CD3 internalization was observed (Figure S5b–c; Internalization score=0.482 control, 0.254 ablated, ****p<0.0001). Thus, we successfully created a recycling endosome ablation assay for primary, human CD8 T cells; which to our knowledge is unprecedented.
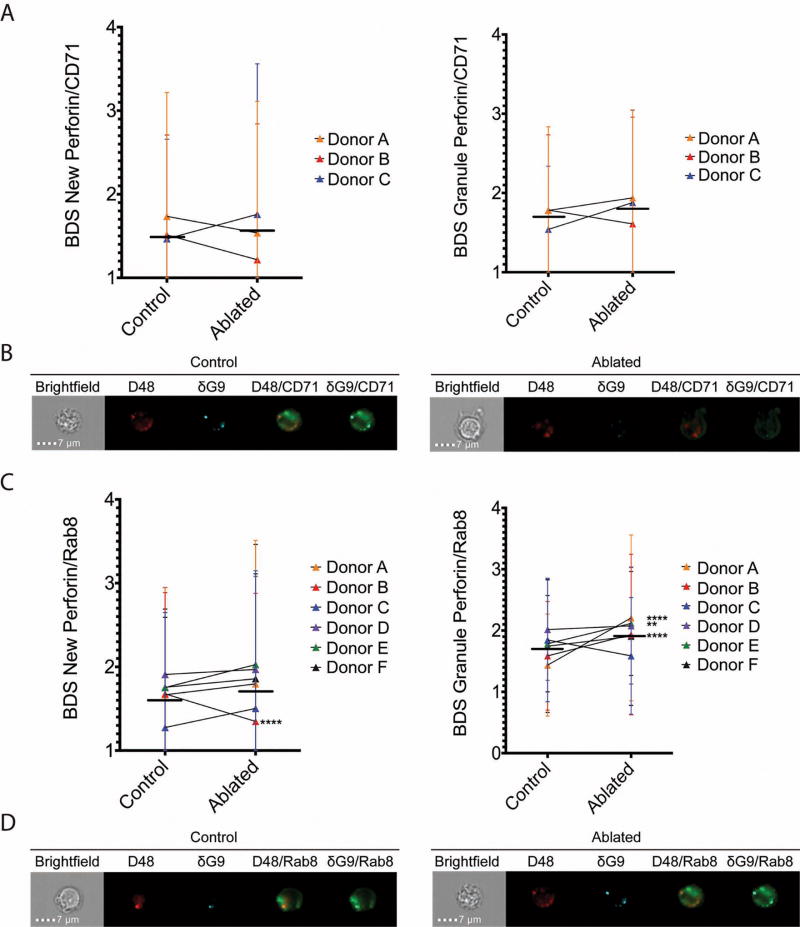
Initially, we used the transferrin receptor (CD71) as a surrogate marker of recycling endosomes, as it is a marker commonly used to identify these compartments. If recycling endosomes truly transport new perforin, we hypothesized our ablation procedure would result in an increase in co-localization between new perforin (D48+δG9-) and CD71 as new perforin would be prevented from exiting the recycling vesicles. We analyzed stimulated CTLs from six different human donor subjects (Donors A-F). In Donors A, B, and C, ablation did not result in a statistically significant change in the median co-localization score between new perforin and CD71 or granule (D48+ δG9+) perforin and CD71 (Figure 5A, Tables 3 and 4).
Fig 5. Ablation of recycling endosomes increases perforin localization with rab8.
CD8+ T cells were isolated by negative selection and ablated of their recycling endosome function after stimulation with anti-CD3 and anti-CD28/CD49d for 3 hours. 3–6 separate donors (Donors A–F) were used, with 60–1300 cells analyzed per donor. Median BDS (similarity) scores of new perforin and granule perforin with CD71 (A–B) and rab8 (C–D) are shown, including representative images (B and D). Horizontal lines represent the mean of all donors combined, and error bars show the standard deviation of each donor. **p<0.01, ****p<0.0001, 2 way ANOVA with Holm-Sidak multiple comparisons test between control and ablated cells.
Table 3.
Results of recycling endosome ablation – new perforin.
| Control | Ablated | |||||
|---|---|---|---|---|---|---|
| Donor | N (cells) |
BDS New Perforin/ Rab8 |
BDS New Perforin/ CD71 |
BDS New Perforin/ Rab8 |
BDS New Perforin/ CD71 |
N (cells) |
| A | 1306 | 1.666 | 1.467 | 1.797 | 1.759 | 219 |
| B | 1630 | 1.677 | 1.513 | 1.348 **** | 1.216 | 605 |
| C | 74 | 1.275 | 1.736 | 1.275 | 1.537 | 64 |
| D | 467 | 1.909 | N/A | 1.969 | N/A | 337 |
| E | 528 | 1.753 | N/A | 2.027 | N/A | 131 |
| F | 569 | 1.756 | N/A | 1.858 | N/A | 115 |
Values in Table 3 correspond to Fig 5.
p<0.0001 vs. control.
Table 4.
Results of recycling endosome ablation – granule perforin.
| Control | Ablated | |||||
|---|---|---|---|---|---|---|
| Donor | N (cells) |
BDS Granule Perforin/ Rab8 |
BDS Granule Perforin/ CD71 |
BDS Granule Perforin/ Rab8 |
BDS Granule Perforin/ CD71 |
N cells) |
| A | 1306 | 1.436 | 1.540 | 2.206 **** | 1.879 | 219 |
| B | 1630 | 1.587 | 1.784 | 1.934 **** | 1.610 | 605 |
| C | 74 | 1.845 | 1.779 | 1.586 | 1.938 | 64 |
| D | 467 | 2.016 | N/A | 2.077 | N/A | 337 |
| E | 528 | 1.785 | N/A | 2.115 | N/A | 131 |
| F | 569 | 1.745 | N/A | 1.906 ** | N/A | 115 |
Given that rab8 is thought to regulate Golgi-to-recycling endosome traffic, we hypothesized that ablation of recycling function would result in the accumulation of new perforin in rab8+ vesicles, preventing its release from the cell (Figure 5B, Tables 3 and 4)[29, 31, 33]. Co-localization of rab8 and new perforin increased in 5 of the 6 donors, although in all but one case the change did not achieve statistical significance. Co-localization of granule perforin and rab8 also increased in 5 donors and decreased in 1 donor.
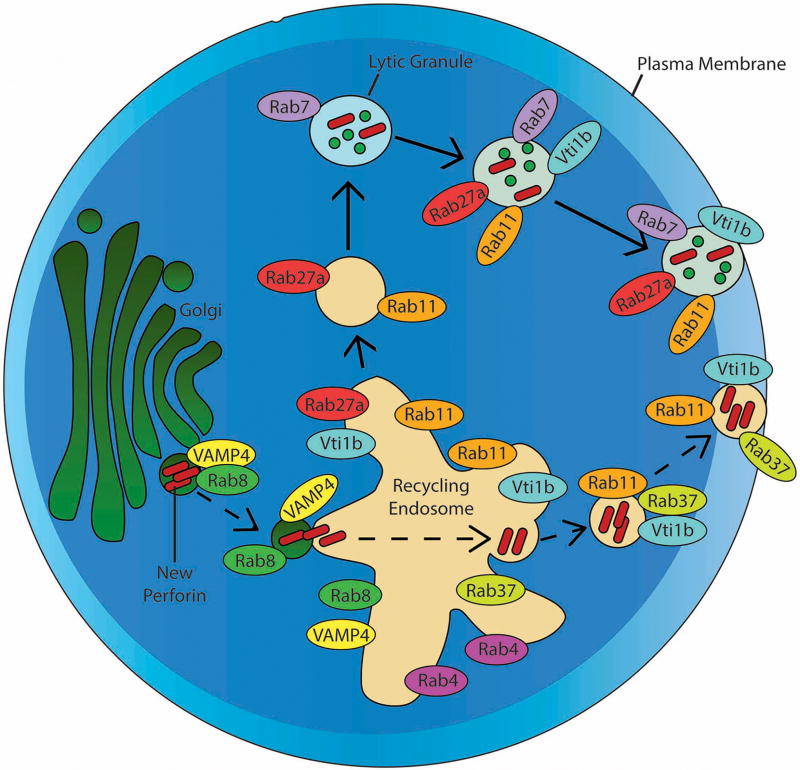
Overall, our recycling endosome loss-of-function experiments suggest rab8+ vesicles serve as an important sorting center for freshly translated perforin protein (Figure 6), from which it may proceed to the default lytic granule pathway or divert to the recycling endosome pathway for express delivery to the immunologic synapse.
Fig 6. Model for trafficking of newly synthesized perforin in human CTL.
Newly synthesized (D48+ δG9-) perforin traffics from the golgi apparatus to the recycling endosome (RE), a process which is mediated by rab8 and likely VAMP4. The perforin becomes localized with rab11a, rab4, rab37, and vti1b in the RE and is secreted from the RE, which is located in close proximity to the immunological synapse upon stimulation of the CTL[32]. Also shown are the known mechanisms of lytic granule (D48+ δG9+ perforin) trafficking to the immunological synapse, which involves the fusion of a recycling endosome-derived rab11a+ rab27a+ vesicle with rab7+ late endosomes.
DISCUSSION
The objective of these studies was to address the mechanism by which new perforin protein arrives at an immunologic synapse independent of secretory lysosomes. While some (if not most) new perforin protein likely replenishes the lytic granule compartment in the cytoplasm, it has been demonstrated that antigen-specific human CTLs direct an appreciable amount to the IS for immediate effect[6]. In order to sustain a cytotoxic response, we hypothesized that human CTLs usurp an auxiliary pathway for the rapid deployment of new perforin protein to the site of engagement. Thus, the question at hand: If not via lytic granules, how does new perforin transit from the Golgi to the synapse? Our data herein illustrate that new perforin (D48+ δG9-) co-localizes with rab8, rab4, rab11a, and rab37. Therefore, we conclude an auxiliary pathway to promote immediate and directed cytotoxicity does exist, constituted by recycling endosomes.
Given that rab8 vesicles regulate traffic from the Golgi apparatus to recycling endosomes[29], we hypothesized that rab8 is important for new perforin transport. Indeed, we observed a tenable degree of co-localization between the two. Furthermore, ablation of recycling endosome function increased the co-localization between new perforin and rab8-labeled compartments. A methodological precedent for the ablation of recycling endosomes in human CD8 T cells does not exist, however we devised an experimental design adapted from similar endeavors[28]. While our data in this regard are imperfect, we believe they represent the best possible proof-of-concept premise given the restrictions of our context: difficulty in manipulating human primary CD8 T cells, induction of a variable response in a constrained time frame, and lack of reagents to specifically block recycling endosome function. By rendering the recycling endosome pathway inert, new perforin protein originally destined for it was trapped in the rab8 compartment; therefore the latter must be new perforin’s conduit between the Golgi and a recycling endosome pathway. It is possible that rab8, or the recycling endosome itself, serves as a “decision point” for new perforin: enter the default secretory pathway in order to replenish a CTL’s supply of lytic granules, or traffic via a recycling endosome pathway to the plasma membrane for immediate use (Figure 6). However, the determinants of new perforin’s fate at this stage are not known. The strength and/or persistence of T cell receptor (TCR) signaling could be key; as robust and constant antigenic stimulation would communicate an urgent need for new perforin at the IS to sustain a cytotoxic response. A direct relationship between the strength of TCR stimulation and the degree of polyfunctional response subsequently has been shown[34]. New perforin’s fate may also be influenced by post-translational modification of the protein; Brennan et al. proved the type of N-linked glycosylation on perforin denotes its final intracellular destination[8]. Model systems of secretory pathways have identified “signal patch” recognition motifs -- such as an amino- or carboxy-terminal peptide -- in the tertiary structure of the protein or a specific glycosyl modification. Might new perforin possess such a feature(s) to direct its fate? Future work in our laboratory will determine the molecular signals that dictate the fate of new perforin.
The accumulation of new perforin at the rab8 stage is not conducive to perforin stability; after ablation treatment there was a decrease in the overall intensity of perforin protein inside the cells. This result may signify degradation of perforin. Studies of rab8 deficiency in murine epithelial cells report the degradation of apical proteins by lysosomes[35]. An alternative explanation is that new perforin may have been diverted to another pathway, perhaps the constitutive pathway, through which it may have been discharged non-specifically, as first postulated previously[7]. We consider this outcome unlikely, since the viability of the total cell population under scrutiny did not change appreciably after treatment (data not shown). We presume a rab8-labeled compartment is only a transient carrier, and cannot accommodate perforin stably. Due to its cytotoxic nature, perforin is a protein that requires exceptionally special conditions for its storage. As such, it is logical that once a CTL synthesizes new perforin protein it must transport it to a safe destination rapidly: either the IS for immediate use or secretory granules for storage.
We believe in times of need, when a CTL must deploy new perforin to the IS for immediate use, recycling endosomes serve as an auxiliary pathway. Previous studies characterized recycling endosomes as sorting centers, from which cargo can be targeted to other organelles, including lysosomes, or the plasma membrane[14, 28, 29, 33]. Rab11a is involved in regulating the flow of traffic through the recycling endosome[36, 37]. Rab4 and rab37 are involved in “fast” recycling pathways, which shuttle proteins to the plasma membrane within minutes, while rab11a is associated with “slow” recycling; a process which takes hours rather than minutes to traffic proteins to the synapse[38, 39]. Human CD8 T cells synthesize new perforin protein as early as 2 hours after stimulation, but it may take up to 4–6 hours for perforin to appear at the synapse[5, 6]. Since new perforin co-localizes with components of both the fast (rab4 and rab37) and slow (rab11a) pathways, our next endeavor will be to delineate which recycling pathway new perforin favors under distinct conditions. The co-localization of perforin with recycling markers could result from fusion of a recycling endosome-derived vesicle with another vesicle already containing perforin, similar to what has been previously demonstrated during lytic granule maturation[13].
New perforin co-localized with the SNAREs vti1b and VAMP4, which have been associated with cytokine secretion from CD4+ T cells and macrophages and golgi-to-endosome trafficking, respectively. Vti1b has also been shown to regulate docking of lytic granules in CTL[40]. The lytic granule docking process is also mediated by munc13-4, rab27a, munc18-2, and syntaxin11[14]. The lack of co-localization between new perforin and rab27a or munc13-4 suggests new perforin-containing vesicles may utilize a docking mechanism at the immunological synapse separate than that used by lytic granules. Dysfunction of components involved in lytic granule secretion results in severe disease due to dysregulation of the immune system. Mutations in rab27a cause Griscelli syndrome type 2, which is characterized by inhibited CTL killing as well as hypopigmentation of the skin and hair[41]. Familial lymphohistiocytosis (FHL), a disease characterized by impaired CTL and NK cell killing, fever, and hepatosplenomegaly, is caused by mutations in the genes encoding perforin (FHL type 2, FHL2)[2], munc13-4 (FHL3)[42], syntaxin11 (FHL4)[43] and MUNC18-2 (FHL5)[44]. The MUNC18-2 mutation, however, can be bypassed by addition of IL-2[44]. It is possible that IL-2 induces the activity of the new perforin trafficking pathway described here, which would provide a secondary means of cytotoxicity for FHL5 patients.
Our data show that new perforin trafficked both with and independently of granzyme B. This suggests some new perforin traffics separately from granzyme B for a period of time. In this scenario, new perforin: 1) could be secreted kinetically and spatially apart from all granzymes, 2) could be secreted in parallel to lytic granules as a means of supplementing the traditional delivery of lytic granules, or 3) could arrive at the synapse on its own but then unite with granzymes via a unique fusion event. The leading paradigm for the delivery of cytotoxic protein mediators is that perforin and granzyme B are exocytosed by the CTL and then internalized by the target cell via endocytosis[45]. It will be interesting to determine if new perforin abides by this model or operates distinctively. While perforin is generally thought of as acting in tandem with granzyme B, it is interesting to speculate on a granzyme or granulysin-independent function of perforin. It is possible that new perforin may be secreted in high enough quantities to result in target death by cell membrane lysis rather than apoptosis, or that it may be directed towards non-cellular targets for an unknown function.
Here we utilized a relatively new technology, IFC, to examine co-localization between new perforin and numerous trafficking vesicles. This technique is advantageous in that it allows for hundreds or thousands of images to be captured, which is not practical by traditional confocal microscopy, and thus allows for imaging of relatively small populations of cells without upstream selection. However, imaging flow cytometers are not yet capable of the resolution afforded by confocal microscopes. It is not possible to distinguish individual trafficking vesicles, and some co-localization seen between two proteins by IFC may be because they are spatially close, but not co-localized actually. The range of BDS scores obtained in these experiments was relatively small, making it difficult to determine whether co-localization was present based on this value alone. BDS scores may theoretically encompass an infinite range of values, with a score of 0 indicating no colocalization between the probes of interest and higher numbers indicating greater degrees of colocalization. However, the BDS scores of cells which show true colocalization will vary based on the probes measured, as these scores depend upon the total area of each probe. In these studies, all BDS scores are generally below 2.5—this is due to the fact that perforin encompasses a very small area of the cell, whereas the rabs and SNAREs are more ubiquitous and highly expressed in most cells. When studying protein trafficking, IFC is useful for enumerating a short list of potential trafficking mediators involved, but it is crucial to follow up these studies with high-resolution confocal microscopy. The use of BDS values is more appropriate when studying change in response to a treatment relative to a control group, as we did with our recycling endosome ablation experiments. Still, it is possible that the limitations inherent to the BDS scores are responsible for the statistical insignificance we obtained in our ablation assays, despite clear trends to that effect. In addition, the number of perforin+ cells generally decreases after ablation relative to the controls, possibly due to a secretion defect related to recycling endosome dysfunction. This further limits the amount of analyzable cells.
Perforin upregulation may be a result of exposure to certain pathogens, or a consequence of frequent stimulation of antigen-specific CTL[46, 47]. Therefore, rapid upregulation of perforin is a means by which CTL sustain their cytotoxic function, especially when pathogen burden is high. This may also explain, in part, how CTL are able to execute serial killing, and how patients with certain types of FHL are able to maintain some residual cytotoxic activity despite deficiencies in lytic granule exocytosis[44, 48]. Here, we postulate an auxiliary, lytic granule-independent trafficking pathway an activated CTL may usurp to deploy new perforin expressly to sustain host defense in the face of a life-threatening challenge.
MATERIALS AND METHODS
Cells
Normal human peripheral blood mononuclear cells (PBMCs) were isolated from 1) source leukocyte preparations received from Gulf Coast Regional Blood Center, Houston, TX or 2) blood specimens collected from healthy adult donors after informed consent was obtained in accordance with protocol #H33095, reviewed and approved by the Internal Review Board of Baylor College of Medicine and Texas Children’s Hospital. PBMCs were isolated by density gradient centrifugation (Ficoll-Paque; GE Healthcare Life Sciences, Pittsburgh, PA) and rested overnight at 37°C in complete medium—RPMI (Life Technologies, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Flowery Branch, GA), 1% penicillin-streptomycin (Life Technologies), 1% HEPES (Life Technologies), 1% sodium pyruvate (Corning, Manassas, VA), 1% L-glutamine (Life Technologies), and 1% MEM non-essential amino acid solution (Life Technologies).
Peptide pools and stimuli
PBMCs from normal donors were stimulated with one of the following peptide sets: Cytomegalovirus, Epstein barr virus, and Influenza (CEF) peptide pool (NIH AIDS Reagent Program); CMV pp65 peptide pool (NIH AIDS Reagent Program); Influenza non-structural protein (BEI Resources); Influenza matrix protein (BEI Resources, Manassas, VA), or staphylococcal enterotoxin B (SEB; Millipore, Darmstadt, Germany). All peptide stimulations were performed at a final concentration of 2 μM; SEB was used at a final concentration of 1 µg/mL.
Antibodies
For imaging flow cytometry experiments: anti-CD8 APC-Cy7 (BD Biosciences, San Jose, CA or Tonbo Biosciences, San Diego, CA) and anti-CD56 PE-CF594 (BD Biosciences) were used for surface staining; all intracellular stains included anti-perforin D48 PE or BV421 (Biolegend, San Diego, CA) and anti-perforin δG9 FITC (Biolegend) and one of the following unconjugated antibodies: anti-CD71 (Cell Signaling Technology, Danvers, MA), anti-granzyme B (BD Biosciences), anti-rab3D (Abcam, Cambridge, MA), anti-rab4 (Pierce Antibodies, Waltham, MA), anti-rab5 (Cell Signaling Technology), anti-rab7 (Santa Cruz Biotechnology, Dallas, TX), anti-rab8 (Cell Signaling Technology), anti-rab11a (Cell Signaling Technology), anti-rab27a (Abcam), anti-rab35 (Abcam), anti-rab37 (Abcam), anti-syntaxin6 (Cell Signaling Technology), anti-syntaxin7 (R&D systems, Minneapolis, MN), anti-vti1b (Abcam), anti-VAMP3 (Abcam), anti-VAMP4 (Abcam), or anti-SNAP23 (Abcam). The unconjugated antibodies were detected using goat anti-rabbit AF647, chicken anti-goat AF647, or donkey anti-sheep AF647 secondary antibodies (Invitrogen, Carlsbad, CA). For confocal microscopy, perforin D48 FITC (Abcam), perforin δG9 AF647 (Biolegend), and goat anti-rabbit or donkey anti-sheep AF568 secondary antibodies (Invitrogen) were used, along with the selected rab or SNARE antibody.
Stimulation and staining procedures
For imaging flow cytometry experiments, cells were washed with PBS (Life Technologies) and resuspended in complete medium at 1x106 cells/mL in the presence of co-stimulatory antibodies anti-CD28 and anti CD49d (1 μg/mL final concentration; BD Biosciences) and the appropriate peptide pool, SEB, or anti-CD3 (Tonbo Biosciences); the cells were incubated for 4 or 6 hours at 37°C with 5% CO2. Extracellular stains were then added to the cells and incubated for 22 minutes, followed by a wash with PBS. The cells were permeabilized using the Cytofix/Cytoperm kit (BD Biosciences) according to the manufacturer’s instructions, followed by two washes with Perm/Wash buffer (BD Biosciences) or Flow Cytometry Perm buffer (Tonbo Biosciences). A cocktail of intracellular antibodies was then added to the cells for 45 minutes. When unlabeled primary antibodies were used, an appropriate secondary antibody was diluted in 1:100 human AB serum (Atlanta Biologicals) in DI water, and then added to the cells for a separate incubation period of 22 minutes. Both the intracellular and secondary antibody steps were followed with a wash in Perm/Wash or Flow Cytometry Perm buffer, and the cells were fixed in 1% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) upon completion of staining. All incubations with dye and antibodies were done in the dark at room temperature.
For confocal microscopy experiments, CD8+ T lymphocytes were isolated by negative selection (StemCell Technologies, Vancouver, BC) from healthy donor expanded cells and stimulated for 4–6 hours with the appropriate peptide and anti-CD28/49d. The cells were then adhered to poly-l-lysine pre-coated slides for 20 minutes at 37°C, fixed and permeabilized (BD fixation/permeabilization kit) for 12 minutes, and stained with a cocktail of antibodies for 45 minutes. Secondary antibodies were used to detect rabs and SNAREs, and were previously diluted in 1:100 human AB serum to prevent non-specific binding. Secondary antibodies were applied to the slides for 25 minutes, separately from other intracellular antibodies. Slides were washed in between steps in either PBS (pre-permeabilization) or 1% BSA (Thermo Fisher, Waltham, MA) + 0.1% saponin (Thermo Fisher) (post-permeabilization). The 1% BDS + 0.1% saponin solution was also used to dilute the antibody cocktail.
Expansion of antigen-specific CTLs
The capacity of human CTLs to rapidly upregulate perforin is not ubiquitous. In order to study the intracellular transport of new perforin dependably, we required a steady source of robust perforin-upregulating, antigen-specific CTLs. Thus, we identified healthy human donors with new perforin responses to SEB, CEF, or influenza peptide pools (as described previously[46]) and expanded those CTL in complete medium supplemented with IL-2 (50 U/mL final concentration; Roche, Basal, Switzerland), the appropriate peptide pool, and CD28/49d (1 µg/mL final concentration) for 10–14 days. This approach allowed for the production of perforin-upregulating CTL, which commonly made up 20% or more of the total CTL population after expansion (data not shown).
Imaging flow cytometry and analysis
Perforin-containing CTLs were visualized using imaging flow cytometry. Between 1,000 and 5,000 CD8+ CD56- Perforin D48+ events were acquired on an Imagestream X MarkII (Amnis Corporation, Seattle, WA) for each specimen. The number of analyzable cells containing new perforin varied between donors, ranging from 10–1200 cells per donor for co-localization experiments. 2–6 donors were used per trafficking marker. The images were captured using a 60X lens. Compensation tubes for each antibody used were prepared using OneComp eBeads (eBioscience, San Diego, CA).
The localization of perforin with protein trafficking mediators was analyzed using the co-localization wizard in IDEAS software version 6.0 (Amnis). Spot masks were created in the IDEAS mask manager to select for perforin D48+ or δG9+ spots (spot to cell background ratio of 3, radius of 6 pixels) within the cell. Then, new Bright Detail Similarity (BDS) spot features were created to detect new perforin (D48 mask and excluding δG9 mask) and granule perforin (D48 mask plus δG9 mask). BDS scores are based on the Pearson’s correlation coefficient and represent the overlap between 2 probes of interest. The gating strategy used for these experiments is shown in Supplementary Figure S1.
Internalization of CD3 was measured using the Internalization Wizard in IDEAS. The internalization score represents the ratio of CD3 intensity inside the cell to the intensity of CD3 of the entire cell. The inside of the cell was identified using an erosion mask on the brightfield channel (channel 1), with an adaptive erosion coefficient of 50. Cells were gated on single, in-focus, CD3+ CD8+ cells.
Confocal microscopy and analysis
Cells were imaged on a laser-scanning microscope (Leica Microsystems, Wetzlar, Germany) SP8 equipped with a tunable white light laser and 100X oil objective. Emission was detected with HyD detectors, and excitation/emission settings were adjusted with fluorescence minus one (FMO) controls so that overlap between markers was minimized. Data were collected in LAS software (Leica) and exported into Volocity (PerkinElmer, Waltham, MA) for analysis. 5–20 new perforin-containing cells were analyzed per experiment with each human donor; and 2–4 donors were used per trafficking marker. Volocity software was used to identify the perforin, D48+ dG9+ granule or D48+ dG9- new perforin, present in each cell and calculate its overlap with protein trafficking markers. Data were thresholded at a standard deviation of 3 above the mean fluorescence for analysis. Manders correlation coefficients were obtained using the co-localization module in Volocity.
Recycling endosome ablation
The ablation of recycling endosomes was performed using a procedure modified after Ang et al[28]. CD8+ T cells from normal human donors, without antigen-specific expansion, were isolated by negative selection (Stem Cell Technologies). The cells were washed twice with PBS and starved of transferrin for 1 hour in dye-free RPMI 1640 medium (Life Technologies) at 37°C, 5% CO2. CTLs were then stimulated with anti-CD3 and anti-CD28/CD49d. HRP-labeled transferrin (Final concentration 0.01 mg/mL; Fitzgerald Industries, Acton, MA) was added with the stimulation. The cells were then incubated for an additional 4 hours at 37°C, after which the cells were washed with PBS and chased for 20 minutes in RPMI. A solution of 3-3’-diaminobenzidine (DAB, 0.1 mg/mL final concentration, Sigma Aldrich, St. Louis, MO) and hydrogen peroxide (0.025% final concentration, Fisher Scientific) was used to perform ablation: the cells were incubated on ice for 1 hour in the dark. A separate tube of cells given DAB without hydrogen peroxide was used as a control. Cells from 3 human donors were used; and 60–1300 new perforin-containing cells per donor were analyzed by imaging flow cytometry.
Statistical analysis and correlation analysis
All statistical analyses -- one-way ANOVA, unpaired multiple comparison t tests, and one sample t tests -- were performed in Prism version 6.02 (GraphPad Software, La Jolla, CA). Error bars in all figures represent the standard deviation. Comparisons are non-significant unless otherwise indicated.
Co-localization in the confocal microscopy images was calculated using the Manders correlation coefficient, M1. The scores range from 0 to 1, with 0 indicating no co-localization and 1 indicating perfect co-localization. M1 values were obtained using Volocity 6.3 software (Perkin Elmer) after selecting perforin D48+ δG9- spots. One sample t-tests were used to compare M1 values to a theoretical mean of 0, which would be indicative of no co-localization.
Figures
All figures were assembled using Illustrator CS6 software (Adobe Systems, San Jose, CA).
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Pinaki Banerjee and Dr. Emily Mace (Center for Human Immunobiology, Texas Children’s Hospital, Houston, TX) for providing technical advice.
Funding for these studies was provided by National Institutes of Health (NIH) T32 grant AI053831 (KEL) and R01 grant AI067946 (JSO). The funding institution had no role in collection, analysis, or interpretation of data.
Footnotes
Compliance with ethical standards
Normal human peripheral blood mononuclear cells (PBMCs) were isolated from 1) source leukocyte preparations received from Gulf Coast Regional Blood Center, Houston, TX or 2) blood specimens collected from healthy adult donors after informed consent was obtained in accordance with protocol #H33095, reviewed and approved by the Internal Review Board of Baylor College of Medicine and Texas Children’s Hospital. All procedures performed in studies involving human participants were in accordance with the 1964 Helsinki declaration and its later amendments. Informed consent was obtained from all individual participants included in the study.
Competing interests
The authors declare that they have no competing interests.
Author’s contributions
All authors contributed to the research design, KEL preformed experiments and analyzed data, KEL and GM wrote and edited the manuscript, and JSO edited the manuscript.
References
- 1.Kagi D, Ledermann B, Burki K, Seiler P, Odermatt B, Olsen KJ, et al. Cytotoxicity mediated by T cells and natural killer cells is greatly impaired in perforin-deficient mice. Nature. 1994;369(6475):31–7. doi: 10.1038/369031a0. [DOI] [PubMed] [Google Scholar]
- 2.Stepp SE, Dufourcq-Lagelouse R, Le Deist F, Bhawan S, Certain S, Mathew PA, et al. Perforin gene defects in familial hemophagocytic lymphohistiocytosis. Science. 1999;286(5446):1957–9. doi: 10.1126/science.286.5446.1957. [DOI] [PubMed] [Google Scholar]
- 3.Sanderson CJ, Glauert AM. The mechanism of T cell mediated cytotoxicity. V. Morphological studies by electron microscopy. Proc R Soc Lond B Biol Sci. 1977;198(1132):315–23. doi: 10.1098/rspb.1977.0100. [DOI] [PubMed] [Google Scholar]
- 4.Rothstein TL, Mage M, Jones G, McHugh LL. Cytotoxic T lymphocyte sequential killing of immobilized allogeneic tumor target cells measured by time-lapse microcinematography. J Immunol. 1978;121(5):1652–6. [PubMed] [Google Scholar]
- 5.Hersperger AR, Makedonas G, Betts MR. Flow cytometric detection of perforin upregulation in human CD8 T cells. Cytometry A. 2008;73(11):1050–7. doi: 10.1002/cyto.a.20596. [DOI] [PubMed] [Google Scholar]
- 6.Makedonas G, Banerjee PP, Pandey R, Hersperger AR, Sanborn KB, Hardy GA, et al. Rapid up-regulation and granule-independent transport of perforin to the immunological synapse define a novel mechanism of antigen-specific CD8+ T cell cytotoxic activity. J Immunol. 2009;182(9):5560–9. doi: 10.4049/jimmunol.0803945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isaaz S, Baetz K, Olsen K, Podack E, Griffiths GM. Serial killing by cytotoxic T lymphocytes: T cell receptor triggers degranulation, re-filling of the lytic granules and secretion of lytic proteins via a non-granule pathway. Eur J Immunol. 1995;25(4):1071–9. doi: 10.1002/eji.1830250432. [DOI] [PubMed] [Google Scholar]
- 8.Brennan AJ, Chia J, Browne KA, Ciccone A, Ellis S, Lopez JA, et al. Protection from endogenous perforin: glycans and the C terminus regulate exocytic trafficking in cytotoxic lymphocytes. Immunity. 34(6):879–92. doi: 10.1016/j.immuni.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz SL, Cao C, Pylypenko O, Rak A, Wandinger-Ness A. Rab GTPases at a glance. J Cell Sci. 2007;120(Pt 22):3905–10. doi: 10.1242/jcs.015909. [DOI] [PubMed] [Google Scholar]
- 10.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2(2):107–17. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 11.Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7(9):631–43. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 12.Huse M, Lillemeier BF, Kuhns MS, Chen DS, Davis MM. T cells use two directionally distinct pathways for cytokine secretion. Nat Immunol. 2006;7(3):247–55. doi: 10.1038/ni1304. [DOI] [PubMed] [Google Scholar]
- 13.Menager MM, Menasche G, Romao M, Knapnougel P, Ho CH, Garfa M, et al. Secretory cytotoxic granule maturation and exocytosis require the effector protein hMunc13-4. Nat Immunol. 2007;8(3):257–67. doi: 10.1038/ni1431. [DOI] [PubMed] [Google Scholar]
- 14.van der Sluijs P, Zibouche M, van Kerkhof P. Late steps in secretory lysosome exocytosis in cytotoxic lymphocytes. Front Immunol. 4:359. doi: 10.3389/fimmu.2013.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stow JL, Low PC, Offenhauser C, Sangermani D. Cytokine secretion in macrophages and other cells: pathways and mediators. Immunobiology. 2009;214(7):601–12. doi: 10.1016/j.imbio.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 16.Bucci C, Parton RG, Mather IH, Stunnenberg H, Simons K, Hoflack B, et al. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70(5):715–28. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- 17.Vitelli R, Santillo M, Lattero D, Chiariello M, Bifulco M, Bruni CB, et al. Role of the small GTPase Rab7 in the late endocytic pathway. J Biol Chem. 1997;272(7):4391–7. doi: 10.1074/jbc.272.7.4391. [DOI] [PubMed] [Google Scholar]
- 18.Daniele T, Hackmann Y, Ritter AT, Wenham M, Booth S, Bossi G, et al. A role for Rab7 in the movement of secretory granules in cytotoxic T lymphocytes. Traffic. 2011;12(7):902–11. doi: 10.1111/j.1600-0854.2011.01194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lombardi D, Soldati T, Riederer MA, Goda Y, Zerial M, Pfeffer SR. Rab9 functions in transport between late endosomes and the trans Golgi network. EMBO J. 1993;12(2):677–82. doi: 10.1002/j.1460-2075.1993.tb05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stow JL, Manderson AP, Murray RZ. SNAREing immunity: the role of SNAREs in the immune system. Nat Rev Immunol. 2006;6(12):919–29. doi: 10.1038/nri1980. [DOI] [PubMed] [Google Scholar]
- 21.Murray RZ, Wylie FG, Khromykh T, Hume DA, Stow JL. Syntaxin 6 and Vti1b form a novel SNARE complex, which is up-regulated in activated macrophages to facilitate exocytosis of tumor necrosis Factor-alpha. J Biol Chem. 2005;280(11):10478–83. doi: 10.1074/jbc.M414420200. [DOI] [PubMed] [Google Scholar]
- 22.Pattu V, Qu B, Marshall M, Becherer U, Junker C, Matti U, et al. Syntaxin7 is required for lytic granule release from cytotoxic T lymphocytes. Traffic. 12(7):890–901. doi: 10.1111/j.1600-0854.2011.01193.x. [DOI] [PubMed] [Google Scholar]
- 23.Pattu V, Qu B, Schwarz EC, Strauss B, Weins L, Bhat SS, et al. SNARE protein expression and localization in human cytotoxic T lymphocytes. Eur J Immunol. 42(2):470–5. doi: 10.1002/eji.201141915. [DOI] [PubMed] [Google Scholar]
- 24.Hepp R, Puri N, Hohenstein AC, Crawford GL, Whiteheart SW, Roche PA. Phosphorylation of SNAP-23 regulates exocytosis from mast cells. J Biol Chem. 2005;280(8):6610–20. doi: 10.1074/jbc.M412126200. [DOI] [PubMed] [Google Scholar]
- 25.Manders EM, Stap J, Brakenhoff GJ, van Driel R, Aten JA. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J Cell Sci. 1992;103(Pt 3):857–62. doi: 10.1242/jcs.103.3.857. [DOI] [PubMed] [Google Scholar]
- 26.Peters PJ, Borst J, Oorschot V, Fukuda M, Krahenbuhl O, Tschopp J, et al. Cytotoxic T lymphocyte granules are secretory lysosomes, containing both perforin and granzymes. J Exp Med. 1991;173(5):1099–109. doi: 10.1084/jem.173.5.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voskoboinik I, Whisstock JC, Trapani JA. Perforin and granzymes: function, dysfunction and human pathology. Nat Rev Immunol. 2015;15(6):388–400. doi: 10.1038/nri3839. [DOI] [PubMed] [Google Scholar]
- 28.Ang AL, Taguchi T, Francis S, Folsch H, Murrells LJ, Pypaert M, et al. Recycling endosomes can serve as intermediates during transport from the Golgi to the plasma membrane of MDCK cells. J Cell Biol. 2004;167(3):531–43. doi: 10.1083/jcb.200408165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henry L, Sheff DR. Rab8 regulates basolateral secretory, but not recycling, traffic at the recycling endosome. Mol Biol Cell. 2008;19(5):2059–68. doi: 10.1091/mbc.E07-09-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKenzie JE, Raisley B, Zhou X, Naslavsky N, Taguchi T, Caplan S, et al. Retromer guides STxB and CD8-M6PR from early to recycling endosomes, EHD1 guides STxB from recycling endosome to Golgi. Traffic. 2012;13(8):1140–59. doi: 10.1111/j.1600-0854.2012.01374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lock JG, Stow JL. Rab11 in recycling endosomes regulates the sorting and basolateral transport of E-cadherin. Mol Biol Cell. 2005;16(4):1744–55. doi: 10.1091/mbc.E04-10-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das V, Nal B, Dujeancourt A, Thoulouze MI, Galli T, Roux P, et al. Activation-induced polarized recycling targets T cell antigen receptors to the immunological synapse; involvement of SNARE complexes. Immunity. 2004;20(5):577–88. doi: 10.1016/s1074-7613(04)00106-2. [DOI] [PubMed] [Google Scholar]
- 33.Huber LA, Pimplikar S, Parton RG, Virta H, Zerial M, Simons K. Rab8, a small GTPase involved in vesicular traffic between the TGN and the basolateral plasma membrane. J Cell Biol. 1993;123(1):35–45. doi: 10.1083/jcb.123.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Almeida JR, Sauce D, Price DA, Papagno L, Shin SY, Moris A, et al. Antigen sensitivity is a major determinant of CD8+ T-cell polyfunctionality and HIV-suppressive activity. Blood. 2009;113(25):6351–60. doi: 10.1182/blood-2009-02-206557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato T, Mushiake S, Kato Y, Sato K, Sato M, Takeda N, et al. The Rab8 GTPase regulates apical protein localization in intestinal cells. Nature. 2007;448(7151):366–9. doi: 10.1038/nature05929. [DOI] [PubMed] [Google Scholar]
- 36.Ullrich O, Reinsch S, Urbe S, Zerial M, Parton RG. Rab11 regulates recycling through the pericentriolar recycling endosome. J Cell Biol. 1996;135(4):913–24. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonnichsen B, De Renzis S, Nielsen E, Rietdorf J, Zerial M. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol. 2000;149(4):901–14. doi: 10.1083/jcb.149.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Sluijs P, Hull M, Webster P, Male P, Goud B, Mellman I. The small GTP-binding protein rab4 controls an early sorting event on the endocytic pathway. Cell. 1992;70(5):729–40. doi: 10.1016/0092-8674(92)90307-x. [DOI] [PubMed] [Google Scholar]
- 39.Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol. 2009;10(9):597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qu B, Pattu V, Junker C, Schwarz EC, Bhat SS, Kummerow C, et al. Docking of lytic granules at the immunological synapse in human CTL requires Vti1b-dependent pairing with CD3 endosomes. J Immunol. 186(12):6894–904. doi: 10.4049/jimmunol.1003471. [DOI] [PubMed] [Google Scholar]
- 41.Menasche G, Pastural E, Feldmann J, Certain S, Ersoy F, Dupuis S, et al. Mutations in RAB27A cause Griscelli syndrome associated with haemophagocytic syndrome. Nat Genet. 2000;25(2):173–6. doi: 10.1038/76024. [DOI] [PubMed] [Google Scholar]
- 42.Feldmann J, Callebaut I, Raposo G, Certain S, Bacq D, Dumont C, et al. Munc13-4 is essential for cytolytic granules fusion and is mutated in a form of familial hemophagocytic lymphohistiocytosis (FHL3) Cell. 2003;115(4):461–73. doi: 10.1016/s0092-8674(03)00855-9. [DOI] [PubMed] [Google Scholar]
- 43.zur Stadt U, Schmidt S, Kasper B, Beutel K, Diler AS, Henter JI, et al. Linkage of familial hemophagocytic lymphohistiocytosis (FHL) type-4 to chromosome 6q24 and identification of mutations in syntaxin 11. Hum Mol Genet. 2005;14(6):827–34. doi: 10.1093/hmg/ddi076. [DOI] [PubMed] [Google Scholar]
- 44.zur Stadt U, Rohr J, Seifert W, Koch F, Grieve S, Pagel J, et al. Familial hemophagocytic lymphohistiocytosis type 5 (FHL-5) is caused by mutations in Munc18-2 and impaired binding to syntaxin 11. Am J Hum Genet. 2009;85(4):482–92. doi: 10.1016/j.ajhg.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thiery J, Keefe D, Boulant S, Boucrot E, Walch M, Martinvalet D, et al. Perforin pores in the endosomal membrane trigger the release of endocytosed granzyme B into the cytosol of target cells. Nat Immunol. 2011;12(8):770–7. doi: 10.1038/ni.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Makedonas G, Hutnick N, Haney D, Amick AC, Gardner J, Cosma G, et al. Perforin and IL-2 upregulation define qualitative differences among highly functional virus-specific human CD8 T cells. PLoS Pathog. 6(3):e1000798. doi: 10.1371/journal.ppat.1000798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hersperger AR, Pereyra F, Nason M, Demers K, Sheth P, Shin LY, et al. Perforin expression directly ex vivo by HIV-specific CD8 T-cells is a correlate of HIV elite control. PLoS Pathog. 6(5):e1000917. doi: 10.1371/journal.ppat.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feldmann J, Le Deist F, Ouachee-Chardin M, Certain S, Alexander S, Quartier P, et al. Functional consequences of perforin gene mutations in 22 patients with familial haemophagocytic lymphohistiocytosis. Br J Haematol. 2002;117(4):965–72. doi: 10.1046/j.1365-2141.2002.03534.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.