Abstract
Increasing incidence of viral infections in crop plants adversely affects their growth and yield. Tomato (Solanum lycopersicum) is considered to be a favorite host for viruses with over 50 species of begomoviruses naturally infecting this crop. Tomato leaf curl virus (ToLCV) is among the most widespread and devastating begomoviruses affecting tomato production. microRNAs (miRs) have been established as key regulators of gene expression and plant development. The miR pathways are disturbed during infection by viruses. Thus, comprehension of regulatory miR networks is crucial in understanding the effect of viral pathogenicity. To identify key miRs involved in ToLCV infection, a high throughput approach involving next generation sequencing was employed. Healthy and infected leaf tissues of two tomato varieties, differing in their susceptibility to ToLCV infection were analyzed. NGS data analysis followed by computational predictions, led to identification of 91 known miRs, 15 novel homologs and 53 novel miRs covering two different varieties of tomato, susceptible (Pusa Ruby) and tolerant (LA1777) to ToLCV infection. The cleaved targets of these miRs were identified using online available degradome libraries from leaf, flower and fruit of tomato and showed their involvement in various biological pathways through KEGG Orthology. With detailed comparative profiling of expression pattern of these miRs, we could associate the specific miRs with the resistant and infected genotypes. This study depicted that in depth analysis of miR expression patterns and their functions will help in identification of molecules that can be used for manipulation of gene expression to increase crop production and developing resistance against diseases.
Electronic supplementary material
The online version of this article (10.1007/s12298-017-0482-3) contains supplementary material, which is available to authorized users.
Keywords: microRNA, Next generation sequencing, Degradome, Begomoviruses, Tomato leaf curl New Delhi virus
Introduction
Geminivirus are phytopathogens that profoundly affect diverse crop plants in tropical and subtropical regions of various countries (Moffat 1999; Boulton 2003; Mansoor et al. 2003). The Geminivirus represent an evolving class of viruses with several new strains that have broadened their host range and made them increasingly virulent (Seal et al. 2006). Tomato leaf curl virus (ToLCV) is a member of the Begomovirus genus, within the Geminivirus family, infecting wide spectrum of vegetable crops such as tomato, chilly etc. and causing acute loss in yields (Moffat 1999; Moriones and Navas-Castillo 2000).
Tomato (Solanum lycopersicum) is a very important crop plant, widely consumed as a rich source of antioxidants. It is placed among the top Solanacea vegetables and appears to be a favorite host for the Begomovirus. More than 50 species of Begomovirus have been identified as natural pathogens of this crop with most of the members being identified during the last two decades. The viruses clustered under the generic name ‘Tomato leaf curl virus’ and ‘Tomato yellow leaf curl virus’ cause the most devastating tomato diseases. These viruses induce characteristic leaf curl diseases (Briddon et al. 2000), which manifest as leaf size reduction, upward or downward curling of the leaf (Yang et al. 2011) crinkling of interveinal areas, marginal and interveinal chlorosis, development of enations, purple discoloration of the abaxial surface of leaves, shortening of internodes, development of small branches, reduction in fruit size and decline in fruit yields. The ToLCV is transmitted by the whitefly, Bemisia tabaci (Hunter et al. 1998). It has a circular, single-stranded monopartite or bipartite (DNA-A and DNA-B) genome (Azzam et al. 1994; Pascal et al. 1994; Rigden et al. 1994; Ingham et al. 1995).
MicroRNAs (miRs) act as key regulatory molecules in various biologically significant processes. These are small non-coding RNAs, 21–24 nucleotides in length. They are processed from stem-loop regions of precursor miRs that are derived from long primary transcripts by the action of Dicer-like (DCL) enzyme. The mature miRs are loaded into cytoplasmic silencing complexes (RISC), where they generally direct cleavage or repression of the complementary mRNAs (Bartel 2004; Jones-Rhoades and Bartel 2004; Naqvi et al. 2009). The role of miRs in leaf morphogenesis, flower development, as well as root and shoot development, has already been confirmed in different model plants (Achard et al. 2004; Chen 2004; Kidner and Martienssen 2004; Mallory et al. 2004; Guo et al. 2005; Naqvi et al. 2009; Zhai et al. 2011). During the past few years there has been an exponential increase in number of miRs, identified and functionally annotated from various plant species, in public databases. The NGS technology along with the computational approaches has been extensively exploited as rapid, precise and affordable technique to identify miRs (Tripathi et al. 2015). This has been successful in plants, where miRs and their target mRNAs often have near perfect complementarity (Rhoades et al. 2002; Bonnet et al. 2004; Jones-Rhoades and Bartel 2004). In tomato, 30 miRs were first reported in the miRBase release 12 in 2008 and this number has increased to 110 in the current miRBase release 21 (Kozomara and Griffiths-Jones 2014). However, the increase is less in comparison to that seen in other model plant species, indicating the vast undetected repertoire of tomato miRs.
The role of miRs in leaf development is well established (Kidner and Martienssen 2004; Mallory et al. 2004) as exemplified by miR164 that regulates CUC2 and determines leaf patterning by controlling leaf margins (Nikovics et al. 2006). Similarly, miR159 and miR319 in Arabidopsis have been reported to play crucial roles in retaining leaf phenotype by targeting the members of MYB and TCP transcription factors, respectively (Palatnik et al. 2003). Since the involvement of miRs in biotic responses and leaf patterning is now well recognized, we desired to explore the molecular principles behind the ToLCV mediated leaf deformations. An earlier work validated 9 novel miRs having high complementarity to ToLCV ORFs, from healthy and ToLCV New Delhi strain (ToLCV-ND) infected tomato leaves (Naqvi et al. 2009; Wang and Luan 2015; Zhao et al. 2015). It is thus evident that the leaf curling induced by ToLCV can be employed as a model, to investigate the impacts of miR mediated regulations of biological actions. This necessitates the identification of the global picture of miR expression patterns and studies on the effect of their deregulations, in response to virus infection. In this report, we describe the miR expression status in the tomato leaves during response to infection with ToLCV-ND. To achieve this, three NGS libraries were constructed from leaf tissues of ToLCV susceptible (Pusa Ruby) and tolerant (LA1777) tomato varieties. A computational pipeline developed in the lab was used to predict novel miRs. The comparative analysis was performed to gain understanding on the differential response to ToLCV-ND infection.
Materials and methods
Plant material and growth conditions
Tomato cv Pusa Ruby seeds were obtained from IARI, New Delhi while tomato accession LA1777 were collected from Dr. Manoj Prasad, NIPGR, New Delhi. Seeds were soaked overnight and then kept in moist in germinating sheets for germination at 30 °C. As they reached seedling stage they were transferred to vermiculite for further growth in green house under 28 °C, 14 h light/10 h dark and 70% humidity condition. These were used as healthy control plant for experiment. For ToLCV-ND infection, tomato cv Pusa Ruby plants were agroinfiltrated with Agrobacterium tumefaciens EHA105 harbouring plasmid constructs, containing dimers of ToLCNDV ‘A’ [HQ264185.1] and ToLCNDV ‘B’ [HQ2641856.1] genome. The detailed procedure has been discussed and described in earlier reports (Naqvi et al. 2010; Pradhan et al. 2015).
RNA isolation
Trizol reagent was used for total RNA isolation (following the manufacturer’s protocol, Invitrogen) from the leaf samples obtained from ToLCV susceptible tomato cv Pusa Ruby (PR) and ToLCV resistant tomato cv LA1777 (LA). The small RNA enrichment was done with 50% PEG and 0.1% NaCl precipitation followed by ethanol purification (Pradhan et al. 2015).
Small RNA library sequencing and analysis
Small RNA library preparation
Three small RNA libraries were prepared using Healthy Pusa Ruby (PRHL), ToLCV-ND infected Pusa Ruby (PRIL) and Healthy LA1777 (LA) tissues. RNA bands falling in range of 15–30 nt were gel purified and ligated with 3′ and 5′ adapters. First strand cDNA synthesis was carried out using manufacturer’s instructions and then amplified using adaptor specific primers. The amplified small RNA samples were sequenced using GAII platform (Illumina, USA).
Pre-processing of sequencing data
The data was obtained as sequences of length 36 bases, along with the base quality scores and read counts. Each sequence included a part of the 3′ sequencing adapter depending upon the length of the small RNA. Only those reads were retained which contained the exact match to the first seven nucleotides of the adapter sequence. The obtained raw sequence reads were trimmed to remove the 3′ adapter bases and sequences having length in between 18 and 24 nucleotides were collected. These were clustered based on sequence identity such that all identical reads in each small RNA library were grouped into single sequence tags containing information of the unique sequence with their abundances in each library.
Identification of known miRs
The small RNA sequences of all three libraries were matched to the miR sequences in miRBase 21.0 to identify known miRs using Bowtie (Langmead et al. 2009). Only those sequences showing 100% match to known mature miRs were selected and the digital expression status of each sequence was calculated as transcript per million (TPM). The miR expression value was normalized as
To observe the differential expression pattern of the miRs, reads with ≥ 5 tags in atleast one library were selected and a comparison across the libraries was performed, by calculating fold-change from the normalized expression (Peng et al. 2011).
Prediction of novel miRs
Novel miRs were predicted using two independent approaches and all the predictions were further validated by searching their corresponding miR* sequences within the NGS data. The first one involved miRDeep-P (Yang and Li 2011), it is a modified package of miRDeep, which is being used specifically for the identification and prediction of known and novel plant miRs from deep sequencing data. It consists of core algorithms developed in Perl (a programming language) that filters the putative miRs based on the probabilistic model of miR biogenesis and assigns them an appropriate score. The second pipeline was developed in-house and the design included the stepwise execution for alignment of all the unique tags to the genome ITAG2.3 release (retrieved from Solgenomics Tomato Genome page) with zero mismatches using Bowtie 0.12.7. The reads aligning to the genome were further matched to known miRs (mirBase Rel. 21) and other small non-coding RNAs, rRNA, snRNA, snoRNA and tRNAs, Gp-II Intron, SRPs (Signal recognition particle) ITAG_infernal 2.3 (retrieved from Sol Genomics Network). The matching reads were excluded from further analysis. For the remaining reads, the region flanking (250 bases upstream and downstream) to the aligned sequence was retrieved from genomic sequences and this sequence was considered as candidate precursor sequence. All the candidate precursors were folded using RNAfold (Lorenz et al. 2011). Structures containing the sequenced region within the branch of the folded precursor were removed. The structures with free energy at most −20 kcal/mol were further screened for classification as putative miRs on the basis of plant miR criteria (Meyers et al. 2008). On the basis of alignment positions, the small RNA sequences showing overlap and a common stretch of 18–20 nt were clustered and the most abundant sequence in a cluster is considered as putative miR. The putative miRs with their star sequence, which were common between both the predictions, were selected as novel miRs. TPMs and fold changes were calculated as for all identified miRs.
The conservation analysis and search for novel homologs (NH) was performed against all miRBase mature miRs using Bowtie and allowing up to three mismatches. The physical maps of all identified novel miRs and precursors of known miRs were plotted with the help of MapChart 2.3 (Voorrips 2002) by converting the base pair (bp) positions to million base pairs (Mbps). The miRs localized within a distance of ≤ 2 kb were clustered in a group.
Validation of microRNA expression in infected tissue
Total RNA was isolated from leaf tissues using TRIzol reagent, following the manufacturer’s protocol. End point stem loop RT-PCR was performed to validate the expression of selected miRs in infected tissues by following protocol as described earlier (Sharma et al. 2015). Briefly, 500 ng total RNA was used to synthesize cDNA using miR-specific stem-loop primer with Superscript reverse transcriptase III (Invitrogen) as per manufacturer’s specifications. A pulsed RT reaction was performed in a thermal cycler as follows: 30 min at 16 °C, 60 cycles at 30 °C for 30 s, 42 °C for 30 s and 50 °C for 1 s. RT enzyme was inactivated by incubating the reaction at 85 °C for 5 min. 1 μl of direct cDNA was used for PCR using specific forward primer and reverse primers.
Target prediction and validation using degradome sequencing
Potential gene targets for known and novel tomato miRs were searched using degradome libraries of leaf (GSM553688), fruit (GSM553690), and flower (GSM553689) tissues which were downloaded from GEO (Gene Expression Ominibus; http://www.ncbi.nlm.nih.gov/geo/) at NCBI (Barrett et al. 2009). The degradome sequence tags were converted to FASTA format and used as input to CleaveLand 4.3 (Addo-Quaye et al. 2009) to identify cleaved targets with not more than 0.05 p value. All these targets were searched against PlantTFDB (Zhang et al. 2011) to identify the miRs involved in transcription regulatory activities. Transcripts having ≤ 4.5 alignment score were considered as potential target transcripts.
To completely examine the function of the miRs and their targets, the regulator pathway annotation was performed based on visualization of the pathways collected in the KEGG database by searching KEGG Orthology (KO) of each cleaved transcript retrieved with the help of UniProt Database (Huntley et al. 2015) (http://www.uniprot.org/). The target expression was retrieved from tomato electronic Fluorescent Protein (eFP) Browser (http://bar.utoronto.ca/efp_tomato/cgi-bin/efpWeb.cgi). The number of targets transcripts for each miR and number of miRs targeting a transcript were also calculated.
Results and discussion
Composition of small RNA sequencing libraries
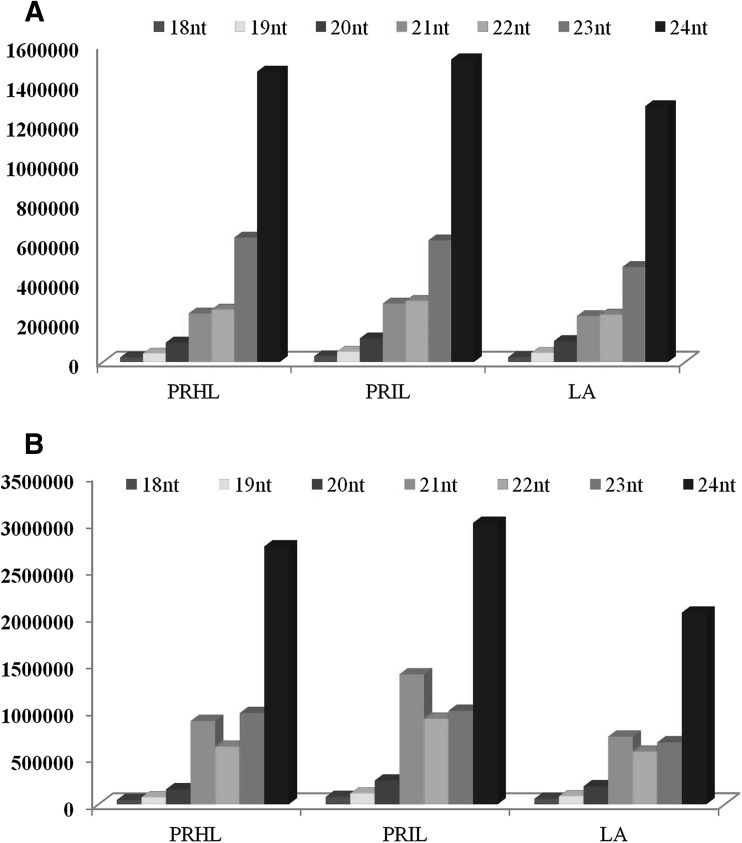
The sequencing of tomato small RNA libraries generated around 5.0–7.0 million reads containing approximately 2.5–3.0 million unique small RNAs of length ranging from 18 to 24 nt. The Healthy Pusa Ruby (PRHL) and ToLCV-ND infected Pusa Ruby (PRIL) libraries have been submitted to the NCBI with accession number GSM1288580 and GSM1288581, respectively (Saraf et al. 2015). Approximately 75% of the unique reads in all three libraries showed alignment with the genome (Table 1). The sequences were analyzed for size distribution on the basis of total abundance and number of unique sequence tags (Fig. 1). In general there was an increased accumulation of the 24 nt sequences in the three libraries, with these sequences constituting 56–68% of unique small RNAs. It was also seen that the 24 nt small RNAs exhibited high sequence diversity. This indicated a dominant role for the 24 nt long siRNAs and miRs which are known to act at the transcriptional level by modulating the methylation patterns on the genome (Chinnusamy and Zhu 2009). The 21 nt size class, which is characteristic for canonical miRs, accounted for 36–46% of the total small RNA pool (Fig. 1), with the predicted miRs constituting 10–16% of the population. There was an increased accumulation of the miRs in PRIL library. These observations were consistent with DCL cleavage products and previous reports from other species such as Arabidopsis thaliana, Medicago truncatula, Cucumis sativa, Zea mays and Populus trichocarpa (Fahlgren et al. 2007; Morin et al. 2008; Moxon et al. 2008; Szittya et al. 2008; Jiao et al. 2011; Martinez et al. 2011; Puzey et al. 2012; Nosaka et al. 2013).
Table 1.
Description of analyzed small RNA libraries
| PRHL | PRIL | LA | |
|---|---|---|---|
| Total no. of reads | 6,114,153 | 7,414,409 | 4,961,769 |
| No. of adaptor trimmed reads (18–24nt) | 5,507,145 | 6,747,228 | 4,325,308 |
| No. of unique reads (18–24nt) after adapter trimming | 2,768,250 | 2,945,834 | 2,419,188 |
| No. unique reads aligned to genome | 2,071,445 (74.83%) | 2,224,335 (75.51%) | 1,814,504 (75.00%) |
| Reads aligned to other sRNAs | 9244 | 10,789 | 9159 |
| No. of known mirBase 19 | 35 | 34 | 37 |
| No. of known mirBase 21 | 83 | 81 | 84 |
| Novel miRs predicted miRDeepP* | 93 | 99 | 88 |
| Novel miR with their star* present in libraries | 63 | 68 | 60 |
| Total Novel miR predicted In-house pipeline* | 403 | 388 | 364 |
| Common Novel miRs* | 59 | 61 | 57 |
* Predicted by using miRBaseRel 19 and from the prediction 9 miRs are now reported in Rel 21
Fig. 1.
Length distribution of small RNA based on a number of unique reads b relative abundance. Pusa Ruby (PRHL), ToLCNDV infected Pusa Ruby (PRIL), ToLCNDV resistant Tomato LA1777 variety (LA)
Prediction of novel putative miRs
Analysis of the small RNA libraries using miRDeep-P predicted 199 novel putative miRs. However the miR* sequences could be predicted, from at least one of the three libraries, for only 90 putative miRs (this included 9 known miRs from miRBase release 21). In an alternative strategy, as described in the materials and methods, 682 novel putative miRs were predicted using the in-house developed pipeline. A comparison revealed that 68 unique putative miRs with 84 precursors were common between both prediction algorithms among these 8 miRs viz. Sly-5, Sly-13, Sly-39, Sly-44, Sly-63, NH–Sly-12(827), NH–Sly-30(166) and NH-Sly-56(408) have been reported as novel miRs in an earlier report (Zhao et al. 2015). This finding provided the conviction that the parameters used for predicting the novel miRs are accurate. In this manuscript, the known miRs are prefixed with “sly-miR” while the novel putative miRs are prefixed with “Sly”, and novel homologs are prefixed by “NH-Sly” followed by the name of the miR family in brackets, for easy understanding.
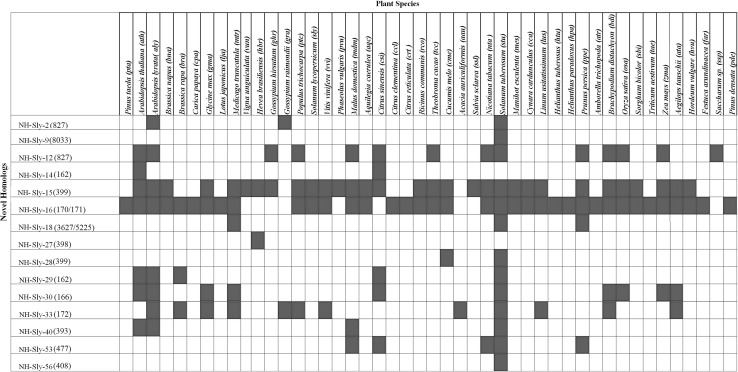
Conservation analysis was performed for the 68 newly predicted tomato miRs by searching for their homologs across the plant species (Fig. 2). It was found that 15 tomato miR sequences were conserved as they showed alignment with 46 different plant species, mainly dicots and these were considered to be the newly identified family members of known miRs or novel homologs (NH). The remaining 53 sequences that were present at 65 different genomic locations did not show homologs in other plants. The detailed description of all these novel miRs, their precursor, the miR*, information on their genomic locations and family details of novel homolog are listed in Supplementary File 1.
Fig. 2.
Schematic representation of predicted miRs showing homology with 46 plant species. Red and white boxes denote presence or absence of sequence homology, NH (Novel Homologs)
Among the conserved putative miRs, NH-Sly-15(399) and NH-Sly-16(170/171) were conserved across the 43 plant species and these were identified as novel members of the miR399 and miR170/miR171 family, respectively. The homolog of NH-Sly-27(398) was present only in Hevea brasiliensis while NH-Sly-9(8033) and NH-Sly-56(408) showed alignments to only Solanum tuberosum. The putative miR NH-Sly-14(162) was identified as a new member of miR162a-5p family whereas NH-Sly-12(827) and NH-Sly-2(827) were homologous to miR827. NH-Sly-27(398) and NH-Sly-28(399) showed similarity with Hbr-miR398 and miR399 family respectively, while NH-Sly-29(162) was homologous to miR162 family and NH-Sly-30(166) showed similarity to miR166 family. NH-Sly-33(172) was identified as a homologue of miR172 family but unlike miR172 it was predicted to target Alpha-L-arabinofuranosidase/beta-D-xylosidase. New members of miR393, miR477 and miR408 family were also identified and named as NH-Sly-40(393), NH-Sly-53(477) and NH-Sly-56(408) respectively.
Secondary structure analysis of the precursor sequences showed that they were capable of folding into stem-loops with complementarity between the predicted miR and miR* duplex (Supplementary File 2). The number of predicted as well as known miRs among all three libraries is shown in Table 1 and their digital distribution is provided as Supplementary File 3. The structures of few selected miRs predicted from the PRHL and LA tissues are shown in Fig. 3. To confirm the accuracy of our predictions the novel putative miR sequences were aligned against EST (Expressed Sequence Tags) database of NCBI by allowing up to 3 mismatches. It was observed that sequences of 4 predicted miRs viz. NH-Sly-9(8033), NH-Sly-15(399), Sly-34 and Sly-58 aligned to the ESTs (Table 2).
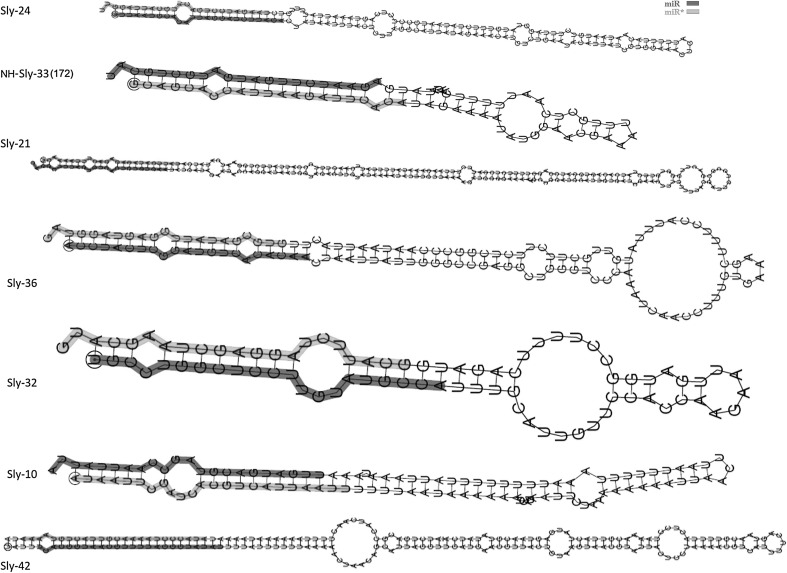
Fig. 3.
Secondary structures of predicted miRs uniquely expressed in LA and PRHL tissues. Predicted miRs, Sly-21, Sly-24 and NH-Sly-33(172) were unique to ToLCV tolerant LA variety, predicted miRs Sly-36, Sly-32, Sly-10, and Sly-42 were unique to PRHL
Table 2.
Predicted miRs showing alignment with ESTs database of S. lycopersicum
| Novel miRNAseq showing hits in ESTdb (S. Lycopersicum) | Aligned EST |
|---|---|
| Sly-34 : CATCAATGATGCAGGAGCTGA | BI933366.1 |
| NH-Sly-9: TTCCAAAGCTGCAGAAATGAG | AW096339.2, BP908468.1, BP911251.1 |
| NH-Sly-15: TGCCAAAGGAGAATTGCCCTG | DB684524.1 |
| Sly-58: ACCTATGTTGGTCGGACTCTC | BG630760.1, AI781029.1, |AI780730.1 |
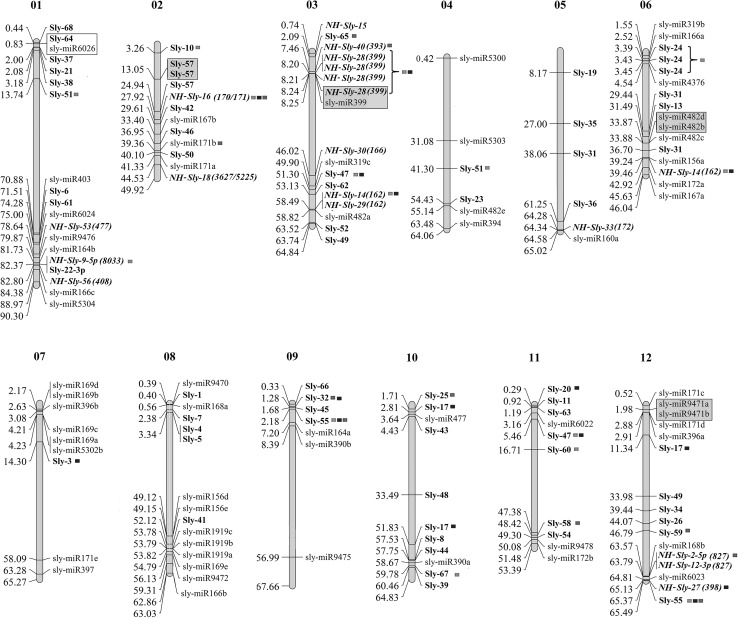
Genomic localization of predicted novel miRs and homologs
The genomic distribution of all the known and novel putative miRs and their physical location on the twelve chromosomes of S. lycopersicum is plotted (Fig. 4). The chromosome wise distribution and frequency analysis showed a higher frequency (12 miRs) of predicted miRs on chromosome 1 whereas only one novel miR, Sly-3, was present on chromosome 7.
Fig. 4.
Physical map showing distribution of novel miRs on the tomato chromosomes. The numbers in the left indicates the chromosomal location (in Mbps) and the respective miR IDs are provided in the right. Predicted miRs are distinguished in bold font and the conserved molecules among these are depicted in Bold and Italics fonts and prefixed with NH. The miRs with positional difference of less than 2 kb are clustered together and are shown in open boxes. Clustered miRs belonging to same family are shown in grey boxes. miRs whose targets are identified in degradome are marked with green (leaf), blue (flower) and red (fruit)
The miRs localized within a distance of ≤ 2 kb were clustered together. The distribution map revealed that putative miR, Sly-64, was clustered with a known miR sly-miR6026 on chromosome 1 with a positional difference of only 827 bp. Whereas NH-Sly-28(399) and sly-miR-399 were clustered on Chromosome 3 with a positional difference of 1271 bp. This is similar to the reports on the presence of pre-miR399 cluster on chromosome 3, from other plants species such as M. truncatula, Hordeum vulagre and Oryza sativa (Cui et al. 2009; Li et al. 2012; Naqvi et al. 2010). Physical mapping also revealed that two family members sly-miR482b and sly-miR482d were clustered on chromosome 6 (Li et al. 2012), over a narrow region of just 322 bp. The other clusters were spaced over a bit larger distances like Sly-1 and a sly-miR9470 were clustered with a difference of 1791 bp on chromosome 8. On chromosome 2, two members of putative Sly-57 were clustered together with a positional difference of 1741 bp. Similarly, sly-miR9471a & b were clustered on chromosome 12 over a distance of 1611 bp. These findings may possibly aid in understanding, the outer attributes of the real pre-miRs and offer an insight into structure of primary transcripts and the transcription of clustered miRs as polycistronic transcripts.
It was observed that NH-Sly-9(8033) was located on the 5′ arm while Sly-22 was on the 3′ arms of same pre-miR while NH-Sly-2(827) and NH-Sly-12(827) were present on 5′ and 3′ arms, respectively on same precursor. Though these miRs were originating from different arms of the same precursor, they do not represent the complementary miR* sequences. This indicates the presence of several miR-like miRs that have been earlier reported in other plant species (Zhang et al. 2010). The co-transcription and processing of these miRs was evident from their similar expression patterns. Both the miRs did not express in PRHL and exhibited low expression levels in PRIL but their expression was high in LA tissues. We could also detect the iso-miRs as exemplified by the putative miRs NH-Sly-14(162) and NH-Sly-29(162) that share the same precursor on chromosome 3 but differ in a single base, Thymidine, at the first position. NH-Sly-29(162) shows higher expression in PRHL and its levels were down regulated by eight fold during ToLCV infection. In LA tissues, the miR shows a 12-fold down regulation. In contrast the expression of NH-Sly-14(162) is up regulated in PRIL and LA tissues.
Differential expression profiling
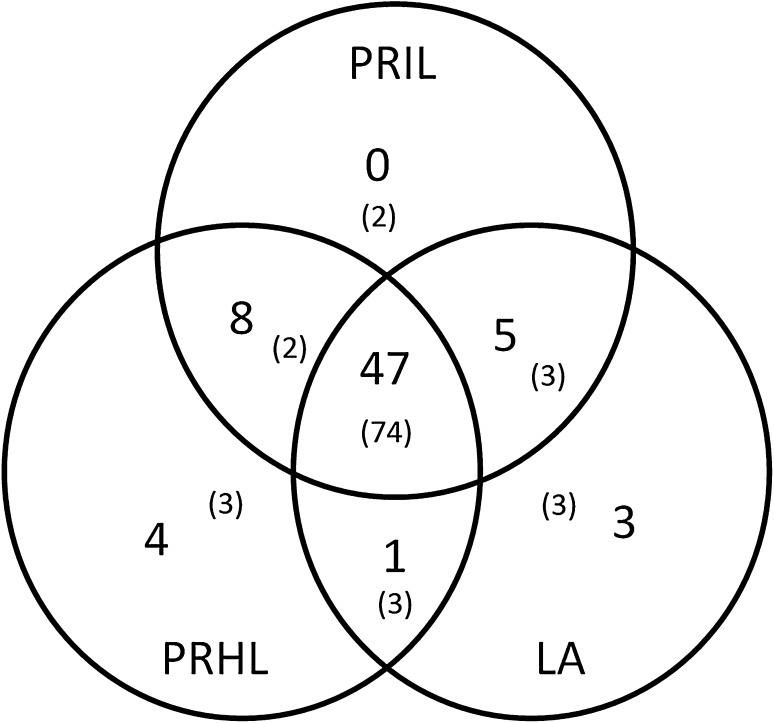
The comparative analysis of miR distribution in PRHL, PRIL and LA showed that 37 predicted, 10 novel homologs and 74 known miRs were common to all three libraries. It was observed that 10 predicted, 2 novel homologs and 7 known miRs, were present in Pusa Ruby libraries but absent from the LA library (Fig. 5). Among this pool 6 predicted miRs, 2 novel homologs and 2 known miRs were present in both PRHL and PRIL libraries but expression of 4 predicted miRs and 3 known miRs (Fig. 5) was restricted to PRHL indicating their down regulation upon viral infection. While sly-miR319c-3p and sly-miR169e-5p selectively expressed in the PRIL library indicating that the up regulation correlated with the virus infection and/or symptoms. The expression of three predicted miRs, Sly-21, Sly-24 and NH-Sly-33(172) and three known miRs were specific to ToLCV tolerant LA variety (Fig. 5). Moreover 3 predicted, 2 novel homologs and 3 known miRs were commonly expressed in both PRIL and LA tissues.
Fig. 5.
Venn diagram showing the distribution of predicted miRs in PRHL, PRIL and LA leaf tissues. The values in brackets represent the known miRs
In the common pool a lot of variation was observed (Table 3). The expression levels of miRs such as miR166, miR159/319, miR164 and miR160 having a role in regulating leaf development were altered under viral infection. This probably caused the leaf deformities characteristically associated with the ToLCV infection (Naqvi et al. 2010). On the basis of this differential expression analysis, two putative miRs (Sly-37 and Sly-45) and 2 known miRs (sly-miR397 and sly-miR482a) were selected for experimental validation in root and shoot tissues of infected plants by stem-loop RT-PCR analysis (Supplementary File 4). The digital expression profiles were verified by the experimental analysis thereby confirming our computational analysis.
Table 3.
List of differentially expressed miRs
| miR-ID | PRHL | PRIL | LA | PRIL/PRHL (fold change) | PRHL/LA (fold change) |
|---|---|---|---|---|---|
| NH-Sly-18(3627/5225) | 39.74385 | 395.0416 | 120.9246 | ↑9.939690488 | ↓3.042599137 |
| NH-Sly-27(398) | 2.943989 | 399.7621 | 484.1015 | ↑135.7892711 | ↓164.4372703 |
| NH-Sly-29(162) | 19.79015 | 2.292833 | 2.01541 | ↓8.631309423 | ↑9.819414869 |
| NH-Sly-30(166) | 1.471995 | 0.809235 | 0.201541 | ↓1.818994961 | ↑7.30369701 |
| NH-Sly-40(393) | 52.17403 | 24.27705 | 7.053936 | ↓2.149108862 | ↑7.396442372 |
| Sly-45 | 32.05677 | 0.674363 | 0.201541 | ↓47.53640169 | ↑159.0582905 |
| NH-Sly-56(408) | 4.415984 | 51.92592 | 31.4404 | ↑11.75863009 | ↓7.119681978 |
| Sly-1 | 7.032863 | 10.7898 | 19.75102 | ↑1.534197584 | ↓2.808389761 |
| NH-Sly-14(162) | 9.649742 | 6.339008 | 3.426197 | ↓1.522279471 | ↑2.816458326 |
| NH-Sly-15(399) | 2.126214 | 0.404618 | 1.813869 | ↓5.254874338 | ↑1.172198286 |
| NH-Sly-28(399) | 25.51457 | 7.013371 | 10.07705 | ↓3.637989922 | ↑2.531948297 |
| Sly-34 | 0.163555 | 0.674363 | 0.604623 | ↑4.123156002 | ↓3.69675795 |
| Sly-37 | 0.163555 | 0.53949 | 2.620033 | ↑3.298524804 | ↓16.01928445 |
| Sly-44 | 0.490665 | 0.809235 | 1.007705 | ↑1.649262402 | ↓2.053754416 |
| Sly-5 | 0.98133 | 3.641558 | 8.263182 | ↑3.710840404 | ↓8.420393106 |
| Sly-61 | 1.63555 | 0.53949 | 0.403082 | ↓3.031658268 | ↑4.05760945 |
| Sly-8 | 1.799104 | 2.832323 | 7.457018 | ↑1.57429593 | ↓4.144849824 |
| sly-miR156d-3p | 1.30844 | 0.944108 | 0.403082 | ↓2.3422219 | ↑3.246088 |
| sly-miR166c-5p | 23.2248 | 3.23694 | 1.410787 | ↓7.17492457 | ↑16.4623 |
| sly-miR167b-3p | 4.906649 | 0.674363 | 0.403082 | ↓7.275979845 | ↑12.17283 |
| sly-miR168a-3p | 21.91636 | 5.799518 | 5.240067 | ↓3.778997284 | ↑4.182459 |
| sly-miR168a,b-5p | 4764.356 | 4826.548 | 1034.712 | ↑1.013053627 | ↑4.604525 |
| sly-miR168b-3p | 34.8372 | 27.51399 | 13.50325 | ↓1.266163159 | ↑2.579913 |
| sly-miR169e-3p | 223.5796 | 156.7219 | 27.40958 | ↓1.426601327 | ↑8.156988 |
| sly-miR171c | 9.486187 | 4.450793 | 0.806164 | ↓2.131347631 | ↑11.76707 |
| sly-miR171e | 2.453324 | 25.49091 | 12.69708 | ↑10.39035314 | ↓5.175461 |
| sly-miR1919a,b | 10.79463 | 10.52006 | 4.635444 | ↓1.026099722 | ↑2.328715 |
| sly-miR1919c-3p | 10.79463 | 10.52006 | 4.635444 | ↓1.026099722 | ↑2.328715 |
| sly-miR390a-3p | 7.196418 | 2.967195 | 1.813869 | ↓2.425326615 | ↑3.96744 |
| sly-miR390b-3p | 0.32711 | 0.674363 | 6.650854 | ↑2.061578003 | ↓20.33216873 |
| sly-miR390b-5p | 75.56239 | 129.6125 | 307.5516 | ↑1.715304295 | ↓4.070167845 |
| sly-miR394-3p | 20.60792 | 7.552861 | 2.620033 | ↓2.728492442 | ↑7.86552 |
| sly-miR396a-3p | 79.32415 | 33.04377 | 2.01541 | ↓2.400578384 | ↑39.35881 |
| sly-miR396a-5p | 59.69756 | 88.61124 | 416.5853 | ↑1.484336162 | ↓6.978263638 |
| sly-miR396b | 30.91189 | 77.68657 | 66.10546 | ↑2.513161756 | ↓2.138512536 |
| sly-miR399 | 10.95818 | 0.404618 | 6.852395 | ↓27.08281387 | ↑1.599175 |
| sly-miR4376 | 3584.47 | 5082.266 | 698.3396 | ↑1.41785695 | ↑5.132847 |
| sly-miR482a | 7.196418 | 17.6683 | 33.45581 | ↑2.455151985 | ↓4.64895318 |
| sly-miR482d-5p | 9.813297 | 3.77643 | 1.007705 | ↓2.598564231 | ↑9.738263 |
| sly-miR482e-5p | 92.40855 | 31.02068 | 14.30941 | ↓2.978933777 | ↑6.457885 |
| sly-miR5300 | 532.6985 | 371.4389 | 172.5191 | ↓1.434148291 | ↑3.087765 |
| sly-miR6022 | 1081.916 | 3460.559 | 3047.502 | ↑3.198546842 | ↓2.816763768 |
| sly-miR6023 | 16.68261 | 44.6428 | 69.93473 | ↑2.676009094 | ↓4.192075192 |
| sly-miR6024 | 20.44437 | 42.08023 | 172.9222 | ↑2.058279478 | ↓8.458182191 |
| sly-miR9471b-5p | 3.925319 | 0.944108 | 2.418492 | ↓4.157702769 | ↑1.623044 |
| sly-miR9472-5p | 3.271099 | 80.11427 | 116.2892 | ↑24.49154668 | ↓35.55048896 |
| sly-miR9476-5p | 7.359973 | 6.878498 | 2.01541 | ↓1.069997036 | ↑3.651849 |
| sly-miR9478-3p | 0.32711 | 1.213853 | 1.007705 | ↑3.710840405 | ↓3.080631626 |
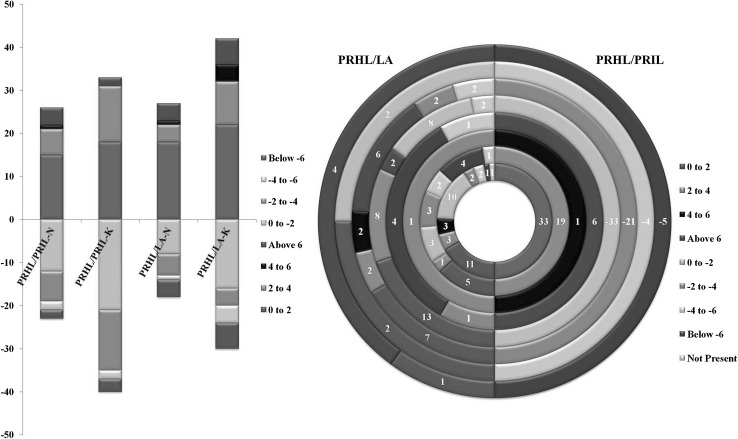
The differential expression analysis (of reads with ≥ 5 tags in at least one library) revealed that within the Pusa Ruby tissues ~ 40% predicted and 46% of known miRs were more than twofold deregulated upon ToLCV infection (PRIL/PRHL) (Fig. 6a). All the miRs which were deregulated upon infection in pusa ruby leaf (PRIL/PRHL) were selected to see their expression patterns in resistant LA leaf tissues (PRHL/LA). The cutoff was decided for the deregulations considering that miRs, which were not significantly deregulated in PRIL may show higher level of deregulations of expression patterns in the LA tissues. The fold change comparisons revealed several interesting changes in expression patterns, with some of the ToLCV deregulated miRs, exhibiting a contrasting behavior in the LA tissues (Fig. 6b). It was observed that 33 miRs were up regulated by zero to twofold in PRIL, 19 miRs showed a two to fourfold up regulation and 6 miRs were more than sixfold up regulated in PRIL (Fig. 6b). Among the 36 miRs that were up regulated by zero to twofold in PRIL (Fig. 6b), 11 miRs exhibited no change in the expression patterns in LA tissues and 1 miR (Sly-46) was not expressed in LA tissues. 6 miRs were also up regulated in LA tissues and within this pool 3 miRs (sly-miR168a-5p, sly-miR168b-5p and sly-miR4376) showed a more than fourfold up regulation. It was also seen that 15 miRs were down regulated in LA tissues, with 5 miRs (Sly-8, Sly-1, Sly-44, sly-miR390b-5p and sly-miR396a-5p) exhibiting more than fourfold down regulation. Among the miRs showing a two to fourfold up regulation in PRIL, sly-miR156d-5p had similar levels of expression in LA while 12 miRs showed a down regulation in the expression levels in LA. Similarly 5 miRs (NH-Sly-18(3627/5225), NH-Sly-56(408), NH-Sly-27(398), sly-miR9472-5p and sly-miR171e exhibiting more than sixfold up regulation in PRIL were down regulated, while 1 miR was not expressed in the LA tissues.
Fig. 6.
Differential Expression Profiling a Fold change distribution of known and predicted miRNAs deregulated in PRIL, PRHL and LA tissues. b Comparative distribution of all deregulated miRs of PRHL/PRIL in PRHL/LA. PRHL/PRIL-N (Deregulated predicted novel miRs in Pusa Ruby infected samples as compared to PRHL) PRHL/PRIL-K (Deregulated known miRs in Pusa Ruby infected samples as compared to PRHL), PRHL/LA-N (Deregulated predicted novel miRs in LA samples as compared to PRHL) and PRHL/LA-K (Deregulated known miRs in LA samples as compared to PRHL)
In a similar manner the expression pattern of miRs showing down regulation upon ToLCV infection was studied in LA tissues. It was observed that 33 miRs exhibited up to twofold down regulation in PRIL. Among these 2 miRs (NH-Sly-30(166) and sly-miR169e-3p) were up regulated by more than sixfold and 8 miRs (NH-Sly-14(162), sly-miR1919a, sly-miR1919b, sly-miR1919c-3p, sly-miR168b-3p, sly-miR5300, sly-miR156d-3p and sly-miR9476-5p) were up regulated to two to fourfold in LA. It was seen that 21 miRs were down regulated by two to fourfold in PRIL and within this group 6 miRs (NH-Sly-40(393), sly-miR482e-5p, sly-miR394-3p, sly-miR482d-5p, sly-miR171c and sly-miR396a-3p) were up regulated by more than sixfold while 4 miRs (NH-Sly-28(399), sly-miR390a-3p, sly-miR168a-3p, Sly-61) were up regulated by two to sixfold in LA and expression of 2 miRs (sly-miR477-3p and Sly-43) was not seen in LA. Likewise 4 miRs exhibited four to sixfold down regulation in PRIL, among these 2 miRs (NH-Sly-15(399), sly-miR9471b-5p) were up regulated to the range of 0–2 fold and 2 miRs were down regulated to the same level in LA. 5 miRs, which were down regulated by more than sixfold in PRIL showed up regulation (NH-Sly-29(162), sly-miR399, sly-miR166c-5p, sly-miR167b-3p and Sly-45) in the LA tissues, among these sly-miR399 was up regulated by zero to twofold while the remaining showed more than sixfold up-regulation. This indicated that the miR profiles were significantly influenced upon viral infection (Table 3).
On comparing PRHL and LA leaves it was seen that 47.22% known miRs and 42.22% predicted miRs exhibited a twofold or more deregulation, with a majority being up regulated in the LA tissues. The total percentage of up regulated miRs is higher in PRHL/LA (56.8%) in contrast with PRIL/PRHL (Fig. 6a). The comparative analysis also showed that miRs Sly-47 and NH-Sly-9(8033) that were highly up regulated in PRIL tissues were absent in LA. The predicted miR Sly-34 which showed more than fourfold up regulation in PRIL was ~ fourfold down regulated in LA. The differences observed in LA could be attributed to the varietal differences.
Target identification
The global miR profiling data evidently suggested that a vast set of miRs was differentially altered in ToLCV-ND infected leaf tissues of tomato cv Pusa Ruby. A set of miRs was also identified, which showed differential expression between the susceptible and tolerant varieties. Identification of the genes targeted by miRs is essential to understand their regulatory action on biological functions. Thus, the transcribed regions of the tomato genome were used to find sequences complementary to the miR candidates. Using CleaveLand around 135 unique targets with 168 miR:target associations were identified from three publically available tomato degradome libraries. This includes 101 miR:target interactions involving 71 unique transcripts, showing an alignment score of ≤ 4.5 in three PARE libraries. The distribution of targets showed 83, 70 and 84 cleaved transcripts in tomato leaf, fruit and flower degradome libraries, respectively. The complete list included 78 unique transcripts with 21 sequences emerging as common among all three tissues.
To understand the functional association of all identified cleaved transcripts with various biological pathways, KEGG Orthology (KO) search for each target was conducted. The transcripts having KO annotations are listed in Table 4 and the details of all these targets and their associated libraries are listed in Supplementary File 5. T-plots, a representation of miR directed cleavage, of the targets were identified using degradome sequencing. It shows the distribution of the degraded sequence tags along the whole transcript sequence (mRNA) on the basis of alignment. Black zigzag lines represent the detected cleavage and red colored dot on the target transcript indicates the cleavage site detected in the degradome library. The detailed target plots (t-plots) are available in Supplementary File 6. In this analysis, the cleaved targets of the predicted miRs were found to be associated with various crucial pathways indicating that the deregulation or absence of these miRs can severely affect the functioning of these biological pathways. This is well exemplified by Sly-32, which targets Mannosyl-oligosaccharide 1 2-alpha-mannosidase,”(MAN1), associated with N-Glycan biosynthesis and protein processing in endoplasmic reticulum during fruit developmental. Sly-32 also inhibits 40s ribosomal protein S6 (RPS6) that is abundantly expressed in mature fruit and roots.
Table 4.
Degradome validated cleaved targets and their KEGG orthology
| miR-ID | Transcript | Description | Degradome Tissue | KOID | KO-DISC | Pathways |
|---|---|---|---|---|---|---|
| Sly-32 | Solyc08g074240.2.1 | 40S ribosomal protein S6 | Flower | K02991 | Small subunit ribosomal protein S6e | Ribosome, HIF-1 signaling pathway, mTOR signaling pathway, PI3 K-Akt signaling pathway. |
| Sly-20 | Solyc10g017620.2.1 | 5-dehydro-2-deoxygluconokinase 1 | Flower | K00847 | Fructokinase | Fructose and mannose metabolism, starch and sucrose metabolism, amino sugar and nucleotide sugar metabolism |
| NH-Sly-33(172) | Solyc01g104950.2.1 | Alpha-l-arabinofuranosidase/beta-d-xylosidase | Fruit | K15920 | Beta-D-xylosidase 4 | Starch and sucrose metabolism, amino sugar and nucleotide sugar metabolism |
| sly-miR172b | Solyc04g049800.2.1 | AP2-like ethylene-responsive transcription factor | Leaf, flower, fruit | K09284 | AP2-like factor, euAP2 lineage | |
| sly-miR172a | Solyc10g084340.1.1 | AP2-like ethylene-responsive transcription factor | Flower | K09284 | AP2-like factor, euAP2 lineage | |
| sly-miR172b | Solyc06g075510.2.1 | AP2-like ethylene-responsive transcription factor | Leaf | K09284 | AP2-like factor, euAP2 lineage | |
| sly-miR172b | Solyc10g084340.1.1 | AP2-like ethylene-responsive transcription factor | Leaf | K09284 | AP2-like factor, euAP2 lineage | |
| sly-miR172a | Solyc02g064960.2.1 | AP2-like ethylene-responsive transcription factor | Flower | K09284 | AP2-like factor, euAP2 lineage | |
| sly-miR172a | Solyc03g044300.2.1 | AP2-like ethylene-responsive transcription factor | Flower, fruit | K09284 | AP2-like factor, euAP2 lineage | |
| sly-miR172a | Solyc09g007260.2.1 | AP2-like ethylene-responsive transcription factor | Flower | K09285 | AP2-like factor, euAP2 lineage | |
| sly-miR172b | Solyc11g072600.1.1 | AP2-like ethylene-responsive transcription factor | Leaf, flower, Fruit | K09284 | AP2-like factor, euAP2 lineage | |
| sly-miR172b | Solyc09g007260.2.1 | AP2-like ethylene-responsive transcription factor | Leaf | K09284 | AP2-like factor, euAP2 lineage | |
| sly-miR169e-5p | Solyc10g054440.1.1 | Arginine decarboxylase | Flower | K01583 | Arginine decarboxylase | Arginine and proline metabolism |
| sly-miR168a-5p | Solyc06g072300.2.1 | ARGONAUTE 1 | Leaf, flower | K11593 | Eukaryotic translation initiation factor 2C | |
| Sly-59 | Solyc02g070100.2.1 | ATP-dependent RNA helicase | Fruit | K14810 | ATP-dependent RNA helicase DDX56/DBP9 | |
| sly-miR9471b-3p | Solyc01g080250.2.1 | BTB/POZ domain containing protein expressed | Fruit | K10523 | Speckle-type POZ protein | |
| sly-miR5302a | Solyc02g064680.2.1 | Calcium-transporting ATPase 1 | Flower | K01537 | Ca2 + -transporting ATPase | |
| sly-miR166a | Solyc03g120910.2.1 | Class III homeodomain-leucine zipper | Fruit | K09338 | Homeobox-leucine zipper protein | |
| sly-miR166b | Solyc03g120910.2.1 | Class III homeodomain-leucine zipper | Leaf | K09338 | Homeobox-leucine zipper protein | |
| sly-miR166c-3p | Solyc03g120910.2.1 | Class III homeodomain-leucine zipper | Flower | K09338 | Homeobox-leucine zipper protein | |
| sly-miR166c-3p | Solyc12g044410.1.1 | Class III homeodomain-leucine zipper | Leaf, flower, fruit | K09338 | Homeobox-leucine zipper protein | |
| sly-miR166a | Solyc02g024070.2.1 | Class III homeodomain-leucine zipper | Fruit | K09338 | Homeobox-leucine zipper protein | |
| sly-miR166a | Solyc11g069470.1.1 | Class III homeodomain-leucine zipper | Leaf | K09338 | Homeobox-leucine zipper protein | |
| sly-miR166b | Solyc02g024070.2.1 | Class III homeodomain-leucine zipper | Flower | K09338 | Homeobox-leucine zipper protein | |
| sly-miR166b | Solyc11g069470.1.1 | Class III homeodomain-leucine zipper | Flower, Fruit | K09338 | Homeobox-leucine zipper protein | |
| sly-miR166c-3p | Solyc02g024070.2.1 | Class III homeodomain-leucine zipper | Leaf | K09338 | Homeobox-leucine zipper protein | |
| NH-Sly-2 | Solyc09g090100.2.1 | Cryptochrome 2 | Fruit | K12119 | Cryptochrome 2 | Circadian rhythm—plant |
| sly-miR166c-5p | Solyc06g067920.2.1 | Cytochrome c1 | Leaf | K00413 | Ubiquinol-cytochrome c reductase cytochrome c1 subunit | Oxidative phosphorylation, two-component system, |
| Sly-24 | Solyc08g075010.2.1 | Glucose-repressible alcohol dehydrogenase transcriptional effector | Leaf | K12603 | CCR4-NOT transcription complex subunit 6 | RNA degradation |
| sly-miR396b | Solyc05g054050.2.1 | Glutamate decarboxylase | flower, fruit | K01580 | Glutamate decarboxylase | Alanine, aspartate and glutamate metabolism, beta-alanine metabolism, taurine and hypotaurine metabolism, butanoate metabolism, GABAergic synapse |
| sly-miR319c-5p | Solyc07g007550.2.1 | Heparanase | Flower | K07964 | Heparanase 1 | Glycosaminoglycan degradation, proteoglycans in cancer |
| Sly-32 | Solyc03g123900.2.1 | Mannosyl-oligosaccharide 1 2-alpha-mannosidase | Fruit | K01230 | Mannosyl-oligosaccharide alpha-1,2-mannosidase | N-Glycan biosynthesis, various types of N-glycan biosynthesis, protein processing in endoplasmic reticulum |
| sly-miR390b-5p | Solyc10g008500.2.1 | Mpv17 | Flower | K13348 | Protein Mpv17 | Peroxisome |
| Sly-65 | Solyc02g070240.2.1 | mRNA 3'-end-processing protein YTH1 | Fruit | K14404 | Cleavage and polyadenylation specificity factor subunit 4 | mRNA surveillance pathway, Influenza A |
| sly-miR159 | Solyc01g009070.2.1 | MYB transcription factor | Leaf, flower, fruit | K09422 | Myb proto-oncogene protein, plant | |
| sly-miR159 | Solyc06g073640.2.1 | MYB transcription factor | Leaf, flower | K09422 | Myb proto-oncogene protein, plant | |
| Sly-55 | Solyc01g067290.2.1 | NAD-dependent epimerase/dehydratase | Leaf | K19073 | Divinyl chlorophyllide a 8-vinyl-reductase | Porphyrin and chlorophyll metabolism |
| sly-miR171c | Solyc07g007870.2.1 | NADH flavin oxidoreductase/12-oxophytodienoate reductase | Fruit | K05894 | 12-oxophytodienoic acid reductase | alpha-Linolenic acid metabolism |
| sly-miR169b | Solyc01g087240.2.1 | Nuclear transcription factor Y subunit A-1 | Leaf, flower, fruit | K08064 | Nuclear transcription factor Y, alpha | |
| sly-miR169c | Solyc08g062210.2.1 | Nuclear transcription factor Y subunit A-3 | Leaf, flower, fruit | K08064 | Nuclear transcription factor Y, alpha | |
| Sly-10 | Solyc01g100860.2.1 | ADP-ribosylation factor | Leaf | K07937 | ADP-ribosylation factor 1 | Endocytosis, vibrio cholerae infection, legionellosis |
| sly-miR162 | Solyc10g005130.2.1 | Ribonuclease 3-like protein 3 | Leaf, flower, fruit | K11592 | Endoribonuclease Dicer [EC:3.1.26.-] | |
| sly-miR9470-3p | Solyc02g063150.2.1 | Ribulose bisphosphate carboxylase small chain | Leaf | K01602 | Ribulose-bisphosphate carboxylase small chain | Glyoxylate and dicarboxylate metabolism, carbon fixation in photosynthetic organisms, carbon metabolism |
| Sly-60 | Solyc09g007850.2.1 | RNA-binding protein | Leaf | K11294 | Nucleolin | Pathogenic Escherichia coli infection |
| sly-miR9478-5p | Solyc04g077020.2.1 | Tubulin alpha-3 chain | Leaf, Fruit | K07374 | Tubulin alpha | Phagosome, gap junction, pathogenic Escherichia coli infection |
| sly-miR1919b | Solyc03g111340.2.1 | Ubiquitin-like modifier-activating enzyme 5 | Leaf, flower, fruit | K12164 | Ubiquitin-like modifier-activating enzyme 5 |
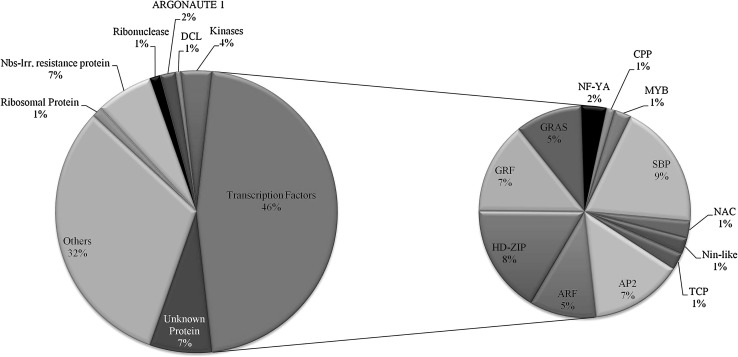
A majority of the predicted target transcripts include the transcription factors, such as AP2/ERF (APETALA2/ethylene responsive factor), ARF (Auxin response factors), CPP (Tesmin/TSO1-like CXC domain-containing protein), GRAS, GRF (Growth Regulating Factor), HD-ZIP (Homeo domain leucine zipper), MYB, NAC, NF-YA (Nuclear transcription factor Y), Nin-like, SBP (Squamosa promoter-binding protein) and TCP (TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor). Transcription factors primarily regulate a network of biological processes by controlling the expression of a large number of genes (Fig. 7). They are therefore, good targets for manipulation of metabolic pathways involved in plant defense and plant’s response against stresses. The AP2 transcription factor is an important pathogenesis related gene family constituting 7% of total target transcription factor identified in present study. They are reported to participate in response to both biotic and abiotic stresses in plants (Li et al. 2011; Licausi et al. 2013) by binding to the promoters of stress responsive genes. AP2/ERF also plays a vital role during TYLCV infection and its promoter binding ability differs in resistant and susceptible varieties (Huang et al. 2016). The SBP family comprises 9% of the miR targeted transcription factors. They are also shown to be involved in biotic and abiotic stress responses (Chen et al. 2002; Wang et al. 2009). Similarly, NACs are unique to plants and are one of the major stress responsive transcription factors (Nuruzzaman et al. 2013). The MYB transcription factors are involved in determining the structure of leaf and have been shown to cause leaf deformation in ToLCNDV infected tomato plants, due to the change in levels of miR159 (Naqvi et al. 2010; Hanley-Bowdoin et al., 2013; Carbonell and Carrington, 2015). The homeodomain-leucine zipper family of transcription factors plays an important role during geminivirus infection. These transcription factors are induced by viral infection and activate abnormal cell divisions which results in characteristic symptoms of infection (Park et al. 2011). It has been reported that expression of five class III HD-Zip transcription factor increases after TYLCV infection due to the corresponding decrease in the levels of miR166 (Wang et al. 2015). Moreover ~ 7% of cleaved transcripts belonged to NBS-LRR disease resistant proteins (Zhai et al. 2011). Their expression increases upon pathogen infection and the miRs viz. miR482b-c, miR6024 and miR6027-3p, which target them, are down regulated in the PRIL library. ERF transcription factors recognize and bind GCC box but the binding ability differed in resistant and susceptible varieties against TYLCV (Huang et al., 2016).
Fig. 7.
Functional distribution of cleaved targets identified in degradome libraries. ARF: Auxin Response Factor, GRF: Growth Regulating Factor, AP2: APETALA2/ethylene responsive factor, HD-ZIP homeo domain leucine zipper, NF-YA nuclear transcription factor Y, CPP tesmin/TSO1-like CXC domain-containing protein
Interestingly, this list included the AGO1 transcript, targeted by sly-miR403-3p and sly-miR168a,b-5p, and DCL1 transcript targeted by sly-miR162. Both AGO1 and DCL1 play key roles in miR biogenesis and function, respectively and thus can regulate global miR fluctuations (Naqvi et al. 2010; Hanley-Bowdoin et al. 2013; Carbonell and Carrington 2015). It was observed that these three miRs were up regulated in the PRIL libraries. The up regulation of sly-miR168 and sly-miR162 upon ToLCV infection has been also confirmed by microarray data analysis (Naqvi et al. 2010). This indicates the deviation of host miRs to suppress the host RNAi machinery. The other ToLCV up regulated miRs include miR NH-Sly-33(172) which targets the Alpha-L-arabinofuranosidase/beta-D-xylosidase mRNA, a defense related gene involved in cell wall reorganization (Chen et al. 2013); sly-miR160a, which targets ARF3, 16 and 17 as well as sly-miR167a and sly-miR167b-5p that target ARF8 and 8-1, respectively. The manipulation of ARFs due to miR deregulations results in phenotypic defects that are characteristic of viral infections (Alazem and Lin 2015). The up regulation of sly-miR319 suppresses TCP Transcription factor family that is involved in growth suppression (Naqvi et al. 2010). Ap2/ERF transcription factor is an important pathogenesis related gene family and its overexpression can mediate an increase in resistance against fungal, bacterial and viral attack. It also plays a vital role during TYLCV infection. The ToLCV down regulated miRs include putative miR, Sly-32 which targets the ribosomal protein encoding transcripts. This indicates up regulation of the corresponding ribosomal proteins. It has been shown that invading Geminiviruses require the ribosomal proteins for successful infection (Yang et al. 2009; Warner and McIntosh 2009).
Conclusion
Plants respond to the viral invasion by modulating their transcriptome to activate enzymes/proteins involved in evading the attack. This also involves activating the RNAi defense pathways, indicating an important role for both siRNAs and miRs. The miRs are also key regulators of various aspects of plant development and the literature is flooded with reports on the knowledge of the abundance and function of miRs. Here, we report 15 novel homologs and 53 novel miRs from tomato that may have important roles in modulating the plants response to ToLCV. These include miRs originating from 5′ and 3′ arms of the pre-miR, with many of them clustered together on the genome indicating their potential processing from polycistronic transcripts. The length of these newly identified miRs varied from 18 to 24 nt. The structural analysis suggested that their putative precursor sequences could fold into stable stem-loop structures containing the miR/miR* sequence in the folded arm.
We observed that 4 miRs were specifically expressed in the healthy leaves and not in the infected tissue of Pusa Ruby and 2 known miRs sly-miR319c-3p, sly-miR169e-5p were specifically induced under ToLCV infection. The differential regulation of miR expression between tomato varieties was also observed with three miRs (Sly-21, Sly-24 and NH-Sly-33(172)) showing expression only in the ToLCV resistant LA variety. Only one miR was common between PRIL and the LA tissues. The comparative analysis of miR distribution showed that ~ 45% miRs were more than twofold deregulated upon ToLCV infection within the Pusa Ruby tissues and a similar pattern was seen in the LA tissues.
Our analysis also identified 135 target transcripts for these miRs. Most of these included transcription factors that were regulated in response to external cues and hormonal signals. Some of these may thus play critical roles in the establishment/evasion of viral infection. The changes in the expression levels of the miRs, disturbs the cellular transcriptome to down regulate transcripts that may prove to be helpful to plants in coping against various biotic and abiotic stress and up regulating transcripts that the virus may utilize for their own benefits. The siRNA mediated plant defense responses against viral infections are well studied, but the roles of host miRs in plant viral immunity/sensitivity have not been well investigated. The functional studies of these miRs are required to understand their role in ToLCV-ND infection. Some of these can serve as ideal candidates for early identification of virus besides providing useful tools for manipulating tomato plants for conferring viral resistance.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary File 1: Lists all the predicted novel miRs, their precursors, genomic loci, associated family and isoforms
Supplementary File 2: Secondary structure of all predicted miRs and their isoforms. miR is highlighted with blue and miR* is highlighted with red color and the circle at the last base represents 5′ end of the precursor
Supplementary File 3: Expression values represented as TPM (Transcript Per Million) for both known and predicted miRs
Supplementary File 4: Validation of miRs by stem-loop RT-PCR in tissues infiltrated with agrobacterium clone. Lane 1: Low range DNA ladder (25 bp), 7 s: shoot tissue 7 days post infiltration, 7 r: root tissue 7 days post infiltration, 28 s: shoot tissue 28 days post infiltration, 28r: root tissue 28 days post infiltration
Supplementary File 5: List of all the degradome validated targets
Supplementary File 6: Target-plots (t-plots) and cleavage sites of all the degradome validated targets. Red dot indicates signatures consistent with miR-directed cleavage site. The abundance of each signature is plotted as a function of its position in the transcript
Acknowledgements
This research was supported by grants from Department of Biotechnology, New Delhi, India. The authors acknowledge the efforts of Dr. Afsar Raza Naqvi in preparing the tomato sRNA libraries.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s12298-017-0482-3) contains supplementary material, which is available to authorized users.
Contributor Information
Anita Tripathi, Email: anita@icgeb.res.in.
Kavita Goswami, Email: kavita@icgeb.res.in.
Manish Tiwari, Email: manishbiotechie@gmail.com.
Sunil K. Mukherjee, Email: sunilm@icgeb.res.in
Neeti Sanan-Mishra, Phone: +91-11-2674 1358/61, Email: neeti@icgeb.res.in.
References
- Achard P, Herr A, Baulcombe DC, Harberd NP. Modulation of floral development by a gibberellin-regulated microRNA. Development. 2004;131(14):3357–3365. doi: 10.1242/dev.01206. [DOI] [PubMed] [Google Scholar]
- Addo-Quaye C, Miller W, Axtell MJ. CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics. 2009;25(1):130–131. doi: 10.1093/bioinformatics/btn604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alazem M, Lin NS. Roles of plant hormones in the regulation of host–virus interactions. Mol Plant Pathol. 2015;16(5):529–540. doi: 10.1111/mpp.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzam O, Frazer J, De La Rosa D, Beaver JS, Ahlquist P, Maxwell DP. Whitefly transmission and efficient ssDNA accumulation of bean golden mosaic geminivirus require functional coat protein. Virology. 1994;204(1):289–296. doi: 10.1006/viro.1994.1533. [DOI] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37(suppl 1):D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bonnet E, Wuyts J, Rouzé P, Van de Peer Y. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics. 2004;20(17):2911–2917. doi: 10.1093/bioinformatics/bth374. [DOI] [PubMed] [Google Scholar]
- Boulton MI. Geminiviruses: major threats to world agriculture. Ann Appl Biol. 2003;142(2):143–143. doi: 10.1111/j.1744-7348.2003.tb00239.x. [DOI] [Google Scholar]
- Briddon RW, Mansoor S, Bedford ID, Pinner MS, Markham PG. Clones of cotton leaf curl geminivirus induce symptoms atypical of cotton leaf curl disease. Virus Genes. 2000;20(1):19–26. doi: 10.1023/A:1008151921937. [DOI] [PubMed] [Google Scholar]
- Carbonell A, Carrington JC. Antiviral roles of plant ARGONAUTES. Curr Opin Plant Biol. 2015;27:111–117. doi: 10.1016/j.pbi.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303(5666):2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang H-S, Eulgem T, Mauch F, Luan S, Zou G, Whitham SA. Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell. 2002;14(3):559–574. doi: 10.1105/tpc.010410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Lv Y, Zhao T, Li N, Yang Y, Yu W, He X, Liu T, Zhang B. Comparative transcriptome profiling of a resistant vs. susceptible tomato (Solanum lycopersicum) cultivar in response to infection by tomato yellow leaf curl virus. PLoS ONE. 2013;8(11):e80816. doi: 10.1371/journal.pone.0080816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J-K. RNA-directed DNA methylation and demethylation in plants. Sci China, Ser C Life Sci. 2009;52(4):331–343. doi: 10.1007/s11427-009-0052-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Xu SM, Mu DS, Yang ZM. Genomic analysis of rice microRNA promoters and clusters. Gene. 2009;431(1–2):61–66. doi: 10.1016/j.gene.2008.11.016. [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, Carrington JC. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE. 2007;2(2):e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H-S, Xie Q, Fei J-F, Chua N-H. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell Online. 2005;17(5):1376–1386. doi: 10.1105/tpc.105.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S. Geminiviruses: masters at redirecting and reprogramming plant processes. Nat Rev Microbiol. 2013;11(11):777–788. doi: 10.1038/nrmicro3117. [DOI] [PubMed] [Google Scholar]
- Huang Y, Zhang B-L, Sun S, Xing G-M, Wang F, Li M-Y, Tian Y-S, Xiong A-S. AP2/ERF transcription factors involved in response to Tomato yellow leaf curly virus in Tomato. Plant Genome. 2016 doi: 10.3835/plantgenome2015.09.0082. [DOI] [PubMed] [Google Scholar]
- Hunter W, Hiebert E, Webb S, Tsai J, Polston J. Location of geminiviruses in the whitefly Bemisia tabaci (Homoptera: Aleyrodidae) Plant Dis. 1998;82(10):1147–1151. doi: 10.1094/PDIS.1998.82.10.1147. [DOI] [PubMed] [Google Scholar]
- Huntley RP, Sawford T, Mutowo-Meullenet P, Shypitsyna A, Bonilla C, Martin MJ, O’Donovan C. The GOA database: gene Ontology annotation updates for 2015. Nucleic Acids Res. 2015;43:d1057–1063. doi: 10.1093/nar/gku1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham DJ, Pascal E, Lazarowitz SG. Both bipartite geminivirus movement proteins define viral host range, but only BL1 determines viral pathogenicity. Virology. 1995;207(1):191–204. doi: 10.1006/viro.1995.1066. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Song W, Zhang M, Lai J. Identification of novel maize miRNAs by measuring the precision of precursor processing. BMC Plant Biol. 2011;11:141. doi: 10.1186/1471-2229-11-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14(6):787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature. 2004;428(6978):81–84. doi: 10.1038/nature02366. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C-W, Su R-C, Cheng C-P, You S-J, Hsieh T-H, Chao T-C, Chan M-T. Tomato RAV transcription factor is a pivotal modulator involved in the AP2/EREBP-mediated defense pathway. Plant Physiol. 2011;156(1):213–227. doi: 10.1104/pp.111.174268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J, Sun H, Kumar P, Baker B. MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci USA. 2012;109(5):1790–1795. doi: 10.1073/pnas.1118282109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F, Ohme-Takagi M, Perata P. APETALA2/ethylene responsive factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 2013;199(3):639–649. doi: 10.1111/nph.12291. [DOI] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Zu Siederdissen CH, Tafer h, Flamm C, Stadler PF, Hofacker IL. ViennaRNA package 2.0. Algorithms. Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J. 2004;23(16):3356–3364. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansoor S, Briddon RW, Zafar Y, Stanley J. Geminivirus disease complexes: an emerging threat. Trends in Plant Science. 2003;8(3):128–134. doi: 10.1016/S1360-1385(03)00007-4. [DOI] [PubMed] [Google Scholar]
- Martinez G, Forment J, Llave C, Pallas V, Gomez G. High-throughput sequencing, characterization and detection of new and conserved cucumber miRNAs. PLoS ONE. 2011;6(5):e19523. doi: 10.1371/journal.pone.0019523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ, Griffiths-Jones S, Jacobsen SE, Mallory AC, Martienssen RA, Poethig RS, Qi Y, Vaucheret H, Voinnet O, Watanabe Y, Weigel D, Zhu JK. Criteria for annotation of plant MicroRNAs. Plant Cell. 2008;20(12):3186–3190. doi: 10.1105/tpc.108.064311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat AS. Geminiviruses emerge as serious crop threat. Science. 1999;286(5446):1835–1835. doi: 10.1126/science.286.5446.1835. [DOI] [Google Scholar]
- Morin RD, Aksay G, Dolgosheina E, Ebhardt HA, Magrini V, Mardis ER, Sahinalp SC, Unrau PJ. Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res. 2008;18(4):571–584. doi: 10.1101/gr.6897308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriones E, Navas-Castillo J. Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Res. 2000;71(1–2):123–134. doi: 10.1016/S0168-1702(00)00193-3. [DOI] [PubMed] [Google Scholar]
- Moxon S, Jing R, Szittya G, Schwach F, Rusholme Pilcher RL, Moulton V, Dalmay T. Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 2008;18(10):1602–1609. doi: 10.1101/gr.080127.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi AR, Islam MN, Choudhury NR, Haq QM. The fascinating world of RNA interference. Int J Biol Sci. 2009;5(2):97–117. doi: 10.7150/ijbs.5.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi AR, Haq Q, Mukherjee SK. MicroRNA profiling of Tomato leaf curl New Delhi virus (ToLCNDV) infected tomato leaves indicates that deregulation of mir159/319 and mir172 might be linked with leaf curl disease. Virol J. 2010;7(1):281. doi: 10.1186/1743-422X-7-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi AR, Choudhury NR, Mukherjee SK, Haq QMR. In silico analysis reveals that several tomato microRNA/microRNA* sequences exhibit propensity to bind to tomato leaf curl virus (ToLCV) associated genomes and most of their encoded open reading frames (ORFs) Plant Physiol Biochem. 2011;49(1):13–17. doi: 10.1016/j.plaphy.2010.09.013. [DOI] [PubMed] [Google Scholar]
- Nikovics K, Blein T, Peaucelle A, Ishida T, Morin H, Aida M, Laufs P. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell Online. 2006;18(11):2929–2945. doi: 10.1105/tpc.106.045617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosaka M, Ishiwata A, Shimizu-Sato S, Ono A, Ishimoto K, Noda Y, Sato Y. The copy number of rice CACTA DNA transposons carrying MIR820 does not correlate with MIR820 expression. Plant Signal Behav. 2013;8(8):e25169. doi: 10.4161/psb.25169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuruzzaman M, Sharoni AM, Kikuchi S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front Microbiol. 2013;4:248. doi: 10.3389/fmicb.2013.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. Control of leaf morphogenesis by microRNAs. Nature. 2003;425(6955):257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- Park J, Lee H-J, Cheon C-I, Kim S-H, Hur Y-S, Auh C-K, Im K-H, Yun D-J, Lee S, Davis KR. The Arabidopsis thaliana homeobox gene ATHB12 is involved in symptom development caused by geminivirus infection. PLoS ONE. 2011;6(5):e20054. doi: 10.1371/journal.pone.0020054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal E, Sanderfoot AA, Ward BM, Medville R, Turgeon R, Lazarowitz SG. The geminivirus BR1 movement protein binds single-stranded DNA and localizes to the cell nucleus. Plant Cell. 1994;6(7):995–1006. doi: 10.1105/tpc.6.7.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T, Lv Q, Zhang J, Li J, Du Y, Zhao Q. Differential expression of the microRNAs in superior and inferior spikelets in rice (Oryza sativa) J Exp Bot. 2011;62(14):4943–4954. doi: 10.1093/jxb/err205. [DOI] [PubMed] [Google Scholar]
- Pradhan B, Naqvi AR, Saraf S, Mukherjee SK, Dey N. Prediction and characterization of Tomato leaf curl New Delhi virus (ToLCNDV) responsive novel microRNAs in Solanum lycopersicum. Virus Res. 2015;195:183–195. doi: 10.1016/j.virusres.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Puzey JR, Karger A, Axtell M, Kramer EM. Deep annotation of Populus trichocarpa microRNAs from diverse tissue sets. PLoS ONE. 2012;7(3):e33034. doi: 10.1371/journal.pone.0033034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110(4):513–520. doi: 10.1016/S0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Rigden JE, Krake LR, Rezaian MA, Dry IB. ORF C4 of tomato leaf curl geminivirus is a determinant of symptom severity. Virology. 1994;204(2):847–850. doi: 10.1006/viro.1994.1606. [DOI] [PubMed] [Google Scholar]
- Saraf S, Sanan-Mishra N, Gursanscky NR, Carroll BJ, Gupta D, Mukherjee SK. 3′ and 5′ microRNA-end post-biogenesis modifications in plant transcriptomes: evidences from small RNA next generation sequencing data analysis. Biochem Biophys Res Commun. 2015;467(4):892–899. doi: 10.1016/j.bbrc.2015.10.048. [DOI] [PubMed] [Google Scholar]
- Seal S, VandenBosch F, Jeger M. Factors influencing begomovirus evolution and their increasing global significance: implications for sustainable control. Crit Rev Plant Sci. 2006;25(1):23–46. doi: 10.1080/07352680500365257. [DOI] [Google Scholar]
- Sharma N, Tripathi A, Sanan-Mishra N. Profiling the expression domains of a rice-specific microRNA under stress. Front Plant Sci. 2015;6:333. doi: 10.3389/fpls.2015.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szittya G, Moxon S, Santos DM, Jing R, Fevereiro MP, Moulton V, Dalmay T. High-throughput sequencing of Medicago truncatula short RNAs identifies eight new miRNA families. BMC Genom. 2008;9:593. doi: 10.1186/1471-2164-9-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A, Goswami K, Sanan-Mishra N. Role of bioinformatics in establishing microRNAs as modulators of abiotic stress responses: the new revolution. Front Physiol. 2015 doi: 10.3389/fphys.2015.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips R. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93(1):77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Wang W, Luan Y. The advance of tomato disease-related microRNAs. Plant Cell Rep. 2015;34(7):1089–1097. doi: 10.1007/s00299-015-1782-0. [DOI] [PubMed] [Google Scholar]
- Wang Y, Hu Z, Yang Y, Chen X, Chen G. Function annotation of an SBP-box gene in Arabidopsis based on analysis of co-expression networks and promoters. Int J Mol Sci. 2009;10(1):116–132. doi: 10.3390/ijms10010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yu W, Yang Y, Li X, Chen T, Liu T, Ma N, Yang X, Liu R, Zhang B. Genomewide analysis of tomato long non-coding RNAs and identification as endogenous target mimic for microRNA in response to TYLCV infection. Sci Rep. 2015;18(5):16946. doi: 10.1038/srep16946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34(1):3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Li L. miRDeep-P: a computational tool for analyzing the microRNA transcriptome in plants. Bioinformatics. 2011;27(18):2614–2615. doi: 10.1093/bioinformatics/btr430. [DOI] [PubMed] [Google Scholar]
- Yang C, Zhang C, Dittman JD, Whitham SA. Differential requirement of ribosomal protein S6 by plant RNA viruses with different translation initiation strategies. Virology. 2009;390(2):163–173. doi: 10.1016/j.virol.2009.05.018. [DOI] [PubMed] [Google Scholar]
- Yang X, Guo W, Ma X, An Q, Zhou X. Molecular characterization of Tomato leaf curl China virus, infecting tomato plants in China, and functional analyses of its associated betasatellite. Appl Environ Microbiol. 2011;77(9):3092–3101. doi: 10.1128/AEM.00017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Jeong D-H, De Paoli E, Park S, Rosen BD, Li Y, González AJ, Yan Z, Kitto SL, Grusak MA. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011;25(23):2540–2553. doi: 10.1101/gad.177527.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Gao S, Zhou X, Xia J, Chellappan P, Zhou X, Zhang X, Jin H. Multiple distinct small RNAs originate from the same microRNA precursors. Genome Biol. 2010;11(8):R81. doi: 10.1186/gb-2010-11-8-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Jin J, Tang L, Zhao Y, Gu X, Gao G, Luo J. PlantTFDB 2.0: Update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 2011;39(suppl 1):D1114–D1117. doi: 10.1093/nar/gkq1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Li Z, Fan J, Hu C, Yang R, Qi X, Chen H, Zhao F, Wang S. Identification of jasmonic acid-associated microRNAs and characterization of the regulatory roles of the miR319/TCP4 module under root-knot nematode stress in tomato. J Exp Bot. 2015;66(15):4653–4667. doi: 10.1093/jxb/erv238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1: Lists all the predicted novel miRs, their precursors, genomic loci, associated family and isoforms
Supplementary File 2: Secondary structure of all predicted miRs and their isoforms. miR is highlighted with blue and miR* is highlighted with red color and the circle at the last base represents 5′ end of the precursor
Supplementary File 3: Expression values represented as TPM (Transcript Per Million) for both known and predicted miRs
Supplementary File 4: Validation of miRs by stem-loop RT-PCR in tissues infiltrated with agrobacterium clone. Lane 1: Low range DNA ladder (25 bp), 7 s: shoot tissue 7 days post infiltration, 7 r: root tissue 7 days post infiltration, 28 s: shoot tissue 28 days post infiltration, 28r: root tissue 28 days post infiltration
Supplementary File 5: List of all the degradome validated targets
Supplementary File 6: Target-plots (t-plots) and cleavage sites of all the degradome validated targets. Red dot indicates signatures consistent with miR-directed cleavage site. The abundance of each signature is plotted as a function of its position in the transcript