Abstract
Maize is an important crop for billions of people as food, feed, and industrial raw material. It is a prime driver of the global agricultural economy as well as the livelihoods of millions of farmers. Genetic interventions, such as breeding, hybridization and transgenesis have led to increased productivity of this crop in the last 100 years. The technique of genome editing is the latest advancement in genetics. Genome editing can be used for targeted deletions, additions, and corrections in the genome, all aimed at genetic enhancement of crops. The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR associated protein 9 (CRISPR/Cas9) system is a recent genome editing technique that is considered simple, precise, robust and the most revolutionary. This review summarizes the current state of the art and predicts future directions in the use of the CRISPR/Cas9 tool in maize crop improvement.
Keywords: CRISPR, Cas9, Gene editing, Genome modification, Maize
Introduction
Maize (Zea mays L.) is the most produced grain crop globally. Its myriad end uses and the ease of cultivation over varied environmental and soil conditions has made it a desirable crop across the world. In addition to human consumption, it is used as feed for livestock, raw materials for chemical and food industries and as biofuel (Pegoraro et al. 2011). To further improve its agronomical traits, scientists have continuously worked to modify its genome through genetic techniques. Traditionally, maize genes were modified or edited via irradiation (such as gamma or fast neutron) and chemical mutagens (such as, ethyl methanesulfonate). These techniques could introduce mutations in the plant genome during exposure and deoxyribonucleic acid (DNA) repair processes. Mutation breeding is generally not precise. It can lead to both positive and negative outcomes with no control over regions of the genome to be modified. Transposon tagging is another frequently used technique in maize genetics, whereby specific transposons are used to cause mutations and thus permit gene discovery (Walbot 2000). This technique is both time consuming and can be expensive (Brutnell 2002). It also leads to random mutations and is cumbersome to perform for large screens (Feng et al. 2016).
The limitations of random mutagenesis stimulated research on targeted genome modification techniques. Such techniques have evolved during the last decade, and they are considered to have increased the fidelity of gene editing by approximately a thousand fold (Puchta et al. 1993; Svitashev et al. 2016). The basic principle of targeted genome editing requires designing of nucleases to cause a double stranded break (DSB) in the DNA at the target site. Most commonly, the broken DNA site leads to mutation either by endogenous repair mechanisms (Non homologous end—joining, NHEJ) or by using an externally added homologous DNA repair template (Homologous—directed repair, HDR). The Zinc Finger Nucleases (ZFNs) and Transcription Activator-Like Effector Nucleases (TALENs) were the first generation targeted genome editing techniques (Joung and Sander 2013). Although successful to some extent, these techniques have some disadvantages. Engineering of ZFNs and TALENs is difficult and minimally a pair of ZFNs or TALENs is required, because both the up-stream and the down-stream regions of any specific locus must be targeted (Beumer et al. 2013). Multiplexing to edit several targets would require many ZFNs or TALENs. Each ZFN or TALEN protein must be genetically engineered to tailor it to generate DSBs at the desired location (Kim and Kim 2014).
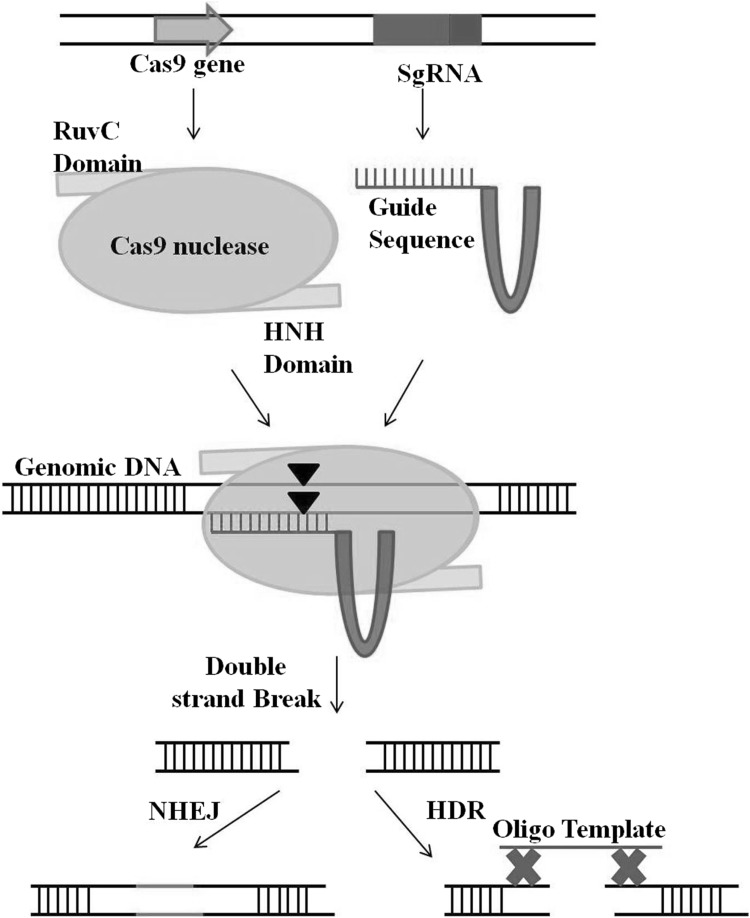
The CRISPR/Cas9 system is a newly introduced technique (Jinek et al. 2012; Gilbert et al. 2013; O’Connell et al. 2014). It’s rather simple design, flexibility of operation, and low cost has revolutionised the field of genome editing (Doudna and Charpentier 2014; Murugan et al. 2017). The system consists of the same Cas9 nuclease protein in all applications and a single guide RNA (sgRNA)—containing a 20 base pair (bp) target site sequence and a hairpin structure (Fig. 1). The Cas9 protein induces a DSB in the 20 bp targeted genomic locus specified by the sgRNA: the DSB is introduced adjacent to the NGG sequence (Protospacer Adjacent Motif, PAM; N means any nucleotide). Interestingly, just, 20 bp is sufficient to provide allele specificity in the single copy regions of a genome like maize. The small size makes construction of sgRNA easy (Newman and Ausubel 2016; Shan et al. 2014). The delivery of the CRISPR/Cas9 system (protein plus sgRNA) inside the cell can be transient or involve stable maize transformation. Multiplexing is also possible via this technique (Shan et al. 2014). Unlike other nucleases, the ability of CRISPR/Cas9 to target methylated DNA makes it a more versatile technique for plant genome editing (Bortesi and Fischer 2015). The targeting efficiency of the CRISPR/Cas9 system is also remarkably better compared to either TALENs or ZFNs (Reis et al. 2014).
Fig. 1.
Representative model depicting CRISPR/Cas9 system for genome modification. The Cas9 protein contains two catalytic nuclease domains: RuvC and HNH. It generates a double stranded break (DSB) at target sites with complementarity to single guide RNA (sgRNA) which can later be edited via Non homologous end—joining (NHEJ) or Homologous—directed repair (HDR)
The CRISPR/Cas9 system has no species limitation. It has been utilized in a wide range of families and genera within the plant kingdom including sorghum (Sun et al. 2015), wheat (Zhang et al. 2016), rice (Mazumdar et al. 2016), Aradidopsis thaliana (Jiang et al. 2014), tobacco (Gao et al. 2015), tomato (Brooks et al. 2014), potato (Wang et al. 2015) and orange (Jia and Wang 2014). In this review, we focus on the current status of maize genome editing using the CRISPR/Cas9 technique.
Factors affecting CRISPR/Cas9 activity
The prerequisite for genome modification by CRISPR/Cas9, is the successful delivery of the Cas9 protein and the sgRNA to the nucleus via an effective vector. The Cas9 gene and the sgRNA gene could be assembled in one plasmid or on separate plasmids (Jiang et al. 2013; Li et al. 2013; Brooks et al. 2014). In most cases reported thus far, the Cas9 gene is driven by commonly used promoters in plant transformation, like ubiquitin and Cauliflower mosaic virus 35S (CaMV 35S) promoters. In basic plant biology studies, where studying functions of genes is the main interest, the Cas9 gene is often attached to both a molecular tag to allow detection/purification of the protein, and a Nuclear Localization Signal (NLS) to facilitate Cas9 protein entry inside the nucleus (Mazumdar et al. 2016). The pCAMBIA-based sgRNA module vectors work efficiently in many plant species (Xing et al. 2014). In these module vectors, two or more sgRNA expression cassettes are assembled using Golden Gate or Gibson Assembly methods. The vector worked successfully in maize with a high mutation efficiency (60–95%) reported in transgenic lines. A large number of biallelic mutations thus generated could also be transmitted to next generation with high efficiency (Xing et al. 2014; Char et al. 2017).
The mutation efficiency of CRISPR/Cas9 depends on many factors, like, gene specificity of the target sites; the position of the PAM sequence; the nature of the sgRNA sequence; the promoters of Cas9 gene and sgRNA; the target tissues; and the transformation technique employed. Genes as targets should be carefully selected as some genes are crucially important for cell growth and gene knock-out of such genes could be lethal to the plant. In crops, genes are usually present in multiple copies due to rearrangements, polyploidization or duplication. Therefore, non specificity of sgRNA could lead to biallelic mutation or mosaicism. For example, the acetolactate synthase (Als) gene family comprises of Als1 and Als2 at chromosomes 4 and 5 respectively, in maize. In a study, the non-gene specific sgRNA led to biallelic mutation in both the Als genes leading to non stable event recovery. The desired mutation was achieved after designing Als2 specific sgRNA based on nucleotide polymorphism between Als1 and Als2 (Svitashev et al. 2015). Furthermore, the GC content of the targeted sites affects the stability of DNA-sgRNA hybrid. High GC content allows for stable DNA-RNA hybrid, but, more stable DNA-sgRNA hybrid also increases the chances of off-targets. GC content up to 35% in the target region has shown good Cas9 enzyme activity with minimal off- targeting (Peng et al. 2016). The PAM sequences are usually NGG, but, NAG could also be used with weakened binding efficiency of the nuclease with the genomic DNA (Xie et al. 2014). The binding efficiency of NRG is only one-fifth of NGG sequence. The binding efficiency of nuclease differs with each base of the PAM sequence. The first nucleotide in PAM is least conserved but the G at second position improves the binding efficiency by 90% and therefore, the stability of NRG is relatively less than NGG (Zhang et al. 2015).
The sgRNA sequence is comprised of the “seed” and the “non seed” region. Initial studies revealed that the seed sequence is at 10–12 base pair adjacent to the PAM sequence. Wu et al. (2014) observed that true seed sequence is limited to one to five base pairs proximal to PAM. The seed sequence determines the specificity of Cas9–sgRNA binding (Mali et al. 2013). Purines are more favoured at maximum position of sgRNA. It was also observed that G at distal region of the PAM can assist in better loading of nuclease (Wang et al. 2014). The sgRNA activity also depends on its length. The efficiency of 19 nucleotide long sgRNA is better than a truncated 17–18 nucleotide sgRNA and a longer 22–23 nucleotide sgRNA. The sequence features in and around the target sequence could also influence action of sgRNA (Peng et al. 2016).
Table 1 lists the promoters driving Cas9 gene and sgRNA reported by different groups for maize genome editing. In general, CaMV 35S promoter has been used extensively for Cas9 gene expression in dicots. For maize and other monocots, ubiquitin gene promoter has been more commonly used for driving expression of the Cas9 gene. The sgRNA is generally expressed using plant RNA polymerase III promoters such as U6 and U3. These promoters have a defined transcription start nucleotide, that is “G” for U6 and “A” for U3 (Belhaj et al. 2013). The start nucleotide is not stringent in terms of preference of type of RNA polymerase III but can affect the targeted gene mutagenesis efficiency to some extent (Zhu et al. 2016).
Table 1.
List of some maize genes edited via CRISPR/Cas9 technology
| Gene name | Promoter driving Cas9 expression | Promoter driving sgRNA expression | Tissue type for maize transformation | References |
|---|---|---|---|---|
| Inositol phosphate kinases, IpK | 35S | U3 | Protoplast | Liang et al. (2014) |
| High affinity K+ transporter, Hkt1 | Ubiquitin | U3 | Immature embryo | Xing et al. (2014) |
| Acetolactate synthase, Als2 | Ubiquitin | U6 | Immature embryo | Svitashev et al. (2015) |
| Liguleless, lg11 | Ubiquitin | U6 | Immature embryo | Svitashev et al. (2015) |
| Male fertility gene, Ms26 | Ubiquitin | U6 | Immature embryo | Svitashev et al. (2015) |
| Male fertility gene, Ms45 | Ubiquitin | U6 | Immature embryo | Svitashev et al. (2015) |
| MADS-box transcription factor 47 | Ubiquitin | U6 | Immature embryo | Qi et al. (2016) |
| Ribosomal protein, Rpl | Ubiquitin | U6 | Immature embryo | Qi et al. (2016) |
| IspH protein, Zmzb7 | 35S | U3 | Protoplast | Feng et al. (2016) |
| Phytoene synthase1, Psy1 | Ubiquitin | U6 | Immature embryo | Zhu et al. (2016) |
| Argonaute protein, Ago18 | Ubiquitin | U6 | Immature embryo | Char et al. (2017) |
| Dihydroflavonol 4-reductase (dfr) | Ubiquitin | U6 | Immature embryo | Char et al. (2017) |
| Anthocyaninless 1(a1) and homolog (a4) | Ubiquitin | U6 | Immature embryo | Char et al. (2017) |
| Auxin regulated gene involved in organ size, ArgoS8 | Ubiquitin | U6 | Immature embryo | Shi et al. (2017) |
Transformation techniques suitable for CRISPR/Cas9 based genome editing
Transformation alters the genetic constitution of cell through introduction of foreign DNA inside them. There are various techniques to accomplish transformation. Plant cells are surrounded by a thick cell wall. Therefore, it is difficult to use transfection or an electroporation mode of transformation, the two most common methods for mammalian cells. Plant transformation primarily relies on Agrobacterium-mediated or biolistic delivery of CRISPR/Cas9 reagents present in the DNA vectors (Svitashev et al. 2016). There have been rapid advances in maize cell transformation efficiency over last several years (Yadava et al. 2017). Protoplast transformation for CRISPR/Cas9 has also been attempted in maize (Table 1). Protoplasts are cell wall free plant cells, and after transformation some of them can reform cell walls and later regenerate into a plant. Protoplast regeneration in maize is rare. It has been observed that in majority of crop plants, it fails to give rise to a fertile plant or is limited to particular genotypes (Gasser and Fraley 1992; Svitashev et al. 2016). Nevertheless, protoplast transient assays can confirm the expression of the engineered Cas9 and sgRNA cassettes quickly. Maize transformation takes approximately 3–4 months from DNA delivery to plantlet regeneration, and a further 7–8 months to seed setting. Therefore, transient assay validation of vectors is very useful. Second, some sgRNAs do not generate desired particular mutation due to unknown reasons (Char et al. 2017). Therefore, a mutation test of a particular Cas9–sgRNA via protoplast transient assay before transformation of immature embryos could save time and resources by validating sgRNA expression and nuclease activity (Zhu et al. 2016; Feng et al. 2016). Agrobacterium strains harbouring different Cas9–sgRNA constructs, co-infecting immature embryos simultaneously could be used as an alternative strategy for multiplex gene editing. Also, T-DNA Agrobacterium vectors can be built with one Cas9 gene and several sgRNA genes. Char et al. (2017) observed that the mutation frequency of immature embryos co-infected by a cocktail of A. tumefaecians were similar to single strain infection. Mutations at each target sites by separate Cas9–sgRNA cassette, were independent of each other to generate transgenic maize lines. This strategy was found to be cost effective and a time saver method.
Examples and special considerations in CRISPR/Cas9-based genome editing in maize
In crop species, genes and genomes are often present in multiple copies, because of local duplications, and/or polyploidization. In some cases, all copies of a gene have to be edited to get the desired mutant phenotype. CRISPR/Cas9 technique could be used to generate homozygous nulliplex mutants or a large diversity of allele combinations in a polyploid plant species through mutations in the different homeologous genes. Currently, the focus in research using CRISPR/Cas9 is on gene knock-out i.e. generating a null mutation. Loss of gene function can be easily phenotypically identified or can be proven by polymerase chain reaction and related molecular experiments (Wu et al. 2014). Table 1 highlights the list of genes edited in maize using CRISPR/Cas9 technology to date. The published reports of maize genes targeted via CRISPR/Cas9 system are fewer than rice and sorghum but private and many public labs have successfully made mutants and are conducting research on the mutants. The era of publishing examples of mutation efficiency of CRISPR/Cas9 can be considered to be over, while the products of this technology are on the anvil.
Liang et al. (2014) provided an early example of editing maize gene using CRISPR technology and they also performed a comparative study of the editing efficiency with TALENs. The CRISPR/Cas9 system induced targeted mutations in the phytic acid biosynthesis gene-ZmIPK in the maize protoplasts with an efficiency of 13.1% as compared to 9.1% obtained with TALENs. The CRISPR technology also showed 10–20-folds higher mutation frequency in maize, as compared to homing endonucleases (Svitashev et al. 2015). Recently, Char et al. (2017) designed an easy public sector system—‘ISU Maize CRISPR’, consisting of E. coli cloning vector and Agrobacterium binary vector for efficient site specific mutagenesis in maize. The vector could be used to insert up to four sgRNA for single or multiplex mutagenesis (Char et al. 2017). This example from maize is a major development towards progress in CRISPR/Cas9 mediated multiplex gene editing in crops. Qi et al. (2016) demonstrated yet another novel multiplex gene editing strategy based on the tRNA-processing system. Maize glycine-tRNA was used to construct multiple tRNA–sgRNA units for the simultaneous production of multiple sgRNAs under the control of one maize U6 promoter. They observed that editing efficiency of tRNA–sgRNA strategy was higher than simplex editing system. They could successfully achieve mutagenesis of multiple genomic loci or deletion of short chromosomal fragments—a major development in gene editing in plants, demonstrated first in maize. As compared to some other plants, the maize genome has large proportion of heterochromatic regions. It was observed that the CRISPR/Cas9 system works independent of the chromatin state. The mutation frequency in the heterochromatic region of maize was found comparable to the euchromatic region (Feng et al. 2016).
To date, reports of modifying genome architecture in vivo through Cas9–sgRNA plus donor templates for high fidelity homology directed repair have been few. The repair template could be either single stranded DNA (ssDNA) or the double stranded DNA (dsDNA) vector. Svitashev et al. (2015) observed that both—the ssDNA and dsDNA vector repair template, edited the Als2 gene and yielded chlorsulfuron resistant maize plants. In the case of gene editing via insertion of a donor template, biolistic transformation is preferable to Agrobacterium-mediated transformation. The possible explanation could be that, the multiple copies of DNA molecules are inserted via bombardment compared to the low copy number of T-DNA insertion. The mRNA expression level of the ARGOS8 gene was changed by replacing the promoter of ARGOS8 with the constitutively expressed Gos2 promoter or via insertion of Gos2 promoter at 5′-UTR of ARGOS8 in the maize inbred line—PH184C. The replacement of the promoter required two sgRNA for DSB as either ends of ARGOS8 promoter and insertion required one sgRNA. Both the events led to ARGOS8 variants with comparable frequency of approximately 1%, but the replacement of promoter also witnessed along with DNA fragment deletion due to NHEJ (Shi et al. 2017).
As described previously, the expression of the Cas9–sgRNA could be either transient or stable. The stable integration is insertion of foreign gene into plant genome whereas transiently transformed plant cell expresses the foreign gene without its integration into the genome. The stable integration of Cas9–sgRNA can lead to somatic mutations in T0 and T1 plants or later generation and consequently may result in chimeric plants as seen in many dicots. However, in maize transformation, the mutations appear to occur in individual embryo cells that then regenerate—thus minimizing chimeras, in contrast to many dicots where chimeras are common. Svitashev observed segregation distortion in progenies of only two T0 plants (Svitashev et al. 2015). Sometimes, such mutations do not follow Mendelian segregation ratios because they are present in only the ear or the tassel. This problem could be overcome in the following ways: (1) Genetic segregation and selection of null segregants with the desired mutant alleles but carrying no CRISPR transgene (Char et al. 2017); and (2) the stable pre-integration of a Cas9 followed by transient expression of the desired sgRNA. The sgRNA can be expressed transiently, if it is delivered in the form of RNA instead of a DNA expression cassette. But the mutation frequency of sgRNA delivered as RNA was approximately 100-fold lower compared to sgRNA delivered as DNA. Svitashev et al. discussed that the possible reason could be failure to meet required simultaneous coincident function of Cas9 and sgRNA as Cas9 was delivered as DNA vector and sgRNA as RNA (Svitashev et al. 2015). The strategy of stable pre-integration of Cas9 could also be used if sequential mutation in every generation is required and sgRNA can still interact with the target DNA.
The Cas9–sgRNA transformed maize plants should be handled by standard genetic technique in which all ears are covered to prevent controlled crosses, as maize is highly cross-pollinated crop. Plants can be detassled to prevent pollen flow or bagged. An inadvertent hybridization of Cas9–sgRNA pollen with any non-target maize plant can result in “gene drive,” in which the gene modified by sgRNA may be preferentially inherited through sexual reproduction and alter the entire population (Pennisi 2015).
Most CRISPR/Cas9 mutants are biallelic. Thus, re-editing of the target gene is also possible if the sgRNA can still interact. The development and characterization of the transgenic line bearing the active Cas9 protein requires time and resources, however, once available any sgRNA can be used. Transgene free technology to edit the genome has always been a desirable goal. Recently, Cas9 and sgRNA in the form of ribonucleoprotein (RNP) complexes were delivered to maize cells via particle bombardment. This can mitigate many effects of off-site targeting and plant mosaicism (Svitashev et al. 2016).
Mitigating off-tragets
Off-site targeting is non-specific cleavage of untargeted sequences. This is major concern in the CRISPR/Cas9 based genome editing as it causes unknown collateral damage to the genome, such as genomic instability and disruption of functionality of otherwise normal genes. The off-targets can be evaluated and predicted by various web based tools such as CROP-IT (Singh et al. 2015), CCTop (Stemmer et al. 2015) and CRISPOR (Haeussler et al. 2016). The improvement in offsite targeting specificity in CRISPR/Cas9 system will provide accurate and precise genotype–phenotype correlations. The wild-type Cas9 enzyme has two nuclease domains: RuvC and HNH. If one of the domains is inactivated then it functions as nickase. The pairing of nCas9 (nickase) with two sgRNAs, each cleaving single strand DNA, reduced the off targets by 50–1500 times without compromising cleavage efficiency (Shen et al. 2014; Zhang et al. 2015). The concept of nCas9 is described in greater detail in the following section. Recently, a new hyper-accurate Cas9 variant (HypaCas9) was designed that exhibited high genome-wide specificity without compromising on-target activity (Chen et al. 2017).
Future prospects of CRISPR/Cas9 based gene editing in maize
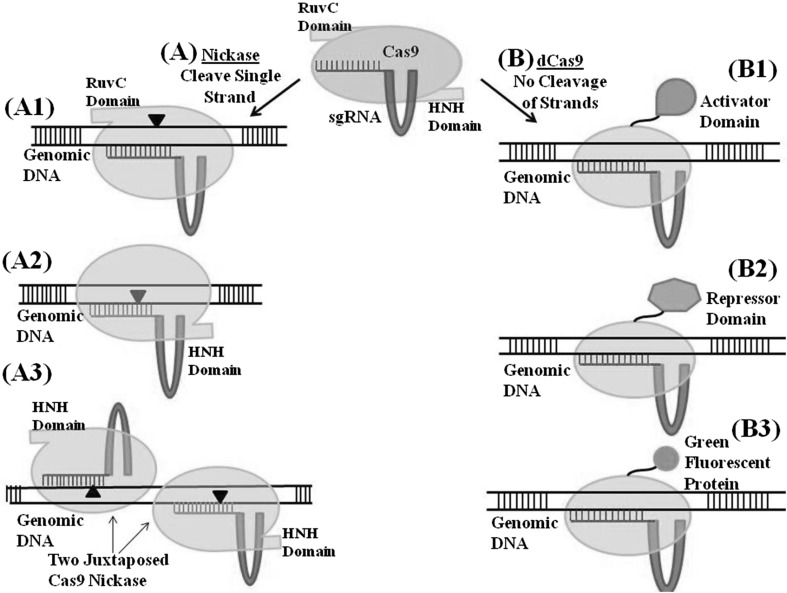
Researchers are continuously searching new innovative methods to broaden the scope and accuracy of mutagenesis by CRISPR/Cas9. The Cas9 has two catalytic nuclease domains—HNH and RuvC. The HNH domain cleaves the DNA stand complementary to sgRNA whereas the RuvC cleaves the non complementary strand. When both nuclease domains are active, Cas9 along with its sgRNA generates a DSB (Fig. 1). If one domain is mutated and loses its catalytic property, then Cas9 is a single strand nickase (nCas9), that is, the Cas9 will cleave only one strand but gene modification may still occur using either the NHEJ or HDR mechanism (Mali et al. 2013) (Fig. 2a). The Cas9 nickases could be used either singly or in pairs. The paired Cas9 nickase works analogously to a pair of ZFNs, creating a displaced DSB (Sander and Joung 2014). Recently, a pair of Cas9 nickases along with sgRNA was used in rice, and the accuracy was so high that even one nucleotide mismatch with the target sequence failed to induce mutation (Mikami et al. 2016). The fusion of nCas9 with cytidine deaminase enabled efficient and site specific C to T base editing in rice, wheat and maize (Zong et al. 2017). The paired sgRNA/Cas9 nickase could also be used to target gene insertions. But, in this case, the flexibility of the targeted gene replacement will be limited by the length between the double nicks. Recently, Zhao et al. (2016) performed gene replacement rather than gene insertion in Arabidopsis using dual-sgRNA/Cas9. The technique of duality created double DSBs and resulted in deletion of large intentional deletion length. In maize, they could successfully create deletion ranging from 1 to 300 kb. The genomic tool which can handle large sequences with versatile application such as deletion, knock-in and replacement in gene function studies, are the future of the gene editing technology for crop genetic improvement.
Fig. 2.
Representative model depicting the newly described alternative forms of the Cas9 protein. a Cas9 nickase created by mutation of either of RuvC or HNH nuclease domain; a1 Cas9 nickase created by mutation in the HNH domain cleaves non complementary DNA strand; a2 Cas9 nickase created by mutation in the RuvC nuclease domain, cleaves complementary DNA strand; a3 Paired nickase creates a displaced double stranded break. This strategy improves specificity. b The catalytically inactive or nuclease deficient or ‘dead’ Cas9 (dCas9) (that is mutations in both the RuvC and HNH domains) can specifically target genome based on sgRNA sequence, without cleaving DNA. The dCas9 can be fused to various effector domains such as transcriptional activator, repressor or GFP protein to perform other functions at the target site
The nuclease deficient Cas9 is termed a dead Cas9 (dCas9). It has lost its catalytic activity due to mutation in both the nickase domain. The dCas9 can specifically target genome based on sgRNA sequence, without DNA cleavage. The dCas9 can be fused with various transcriptional domains to work as an activator (CRISPRa) or as a gene repressor system (CRISPRi). The dCas9 can also be attached to various proteins for their programmable localization on DNA. For example, fusion of reporter genes with dCas9 for molecular visualization can be accomplished using this approach (Fig. 2b) (Shalem et al. 2015; Radzisheuskaya et al. 2016).
Maize breeders could be benefitted greatly by the use of CRISPR/Cas9 technology. The conventional breeding depends on natural variation. Breeders perform extensive back crossing for introgressing a desired trait into an elite background. Genome editing can accelerate plant breeding by performing precise and predictable modifications directly on alleles in an elite background. The modifications introduced in the genome via CRISPR/Cas9 technology is indistinguishable from those introduced via conventional breeding or chemical or random mutagenesis. Therefore, the crop variety generated through this technology has been classified as non-genetically modified (non-GM) in some countries once the transgenic Cas9–sgRNA or any other foreign genetic element has been segregated from the stock (Belhaj et al. 2013). Increasingly, there has been a greater preference of using biolistic-based transformation techniques in genome editing applications, instead of Agrobacterium-mediated transformation, as the former does not involve use of any plant pathogen, which may have regulatory issues. It is expected that a variety of high amylopectin maize may be the first CRISPR edited maize to be grown commercially (Waltz 2016). The recent study on creating ARGOS8 gene variants in maize using CRISPR/Cas9 is a major demonstration of potential of this technology in future plant breeding. These alleleic variants increased grain yield by 314 kg per heactare under drought stress conditions in field trials (Shi et al. 2017).
The improvements in maize agronomic and quality traits are promising applications. Since the era of hybrid maize, natural allelic variations in a large number of genes, each with small effects, have been utilized to improve yield and stress tolerance. However, such alleles are present at relatively low frequencies in most elite breeding populations. As genomics leads to greater understanding of genetic variation, in future it may be possible to routinely design alleles through genome editing. The designed alleles, even if show small effects, can be extensively used and pyramided as there would be no regulatory costs of using these alleles. This is unlike transgenics, where regulatory costs of using every single transgene have to be thoroughly considered against the benefits it could offer. Maize is an industrial crop with a highly developed seed industry. In the past, as compared to other crops, new genetic technologies have been more widely and intensely applied to this crop. The rapid advances in genome editing in maize can once again harbinger a new technological era, which can be a potential trendsetter for all other crops.
Acknowledgements
The maize genome editing work in the laboratories of PY, KK and TK is funded by National Agricultural Science Fund competitive Grant NASF/GTR-5004/2015-16/204. PY is supported by a Fulbright- Nehru Grant (Award No. 2200/FNPDR/2016). PY is thankful to Prof. Virgina Walbot, Department of Biology, Stanford University for critical reading and editing of the manuscript.
Author contributions
IS and PY conceived the idea of review. AA wrote the primary draft, which was further augmented, edited and improved by PY. PKA, AP, KK and TK provided specific comments and improved the draft. All the authors read and approved it for publication.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Astha Agarwal and Pranjal Yadava have contributed equally to this work.
References
- Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V. Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods. 2013;9:39. doi: 10.1186/1746-4811-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer KJ, Trautman JK, Christian M, Dahlem TJ, Lake CM, Hawley RS, Grunwald DJ, Voytas DF, Carroll D. Comparing zinc finger nucleases and transcription activator-like effector nucleases for gene targeting in Drosophila. G3 Genes Genomes Genet. 2013;3(10):1717–1725. doi: 10.1534/g3.113.007260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortesi L, Fischer R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv. 2015;33:41–52. doi: 10.1016/j.biotechadv.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Brooks C, Nekrasov V, Lippman ZB, Van Eck J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 2014;166:1292–1297. doi: 10.1104/pp.114.247577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutnell TP. Transposon tagging in maize. Funct Integr Genomics. 2002;2:4–12. doi: 10.1007/s10142-001-0044-0. [DOI] [PubMed] [Google Scholar]
- Char SN, Neelakandan AK, Nahampun H, Frame B, Main M, Spalding MH, Becraft PW, Meyers BC, Walbot V, Wang K, Yang B. An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnol J. 2017;15:257–268. doi: 10.1111/pbi.12611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, Sternberg SH, Joung JK, Yildiz A, Doudna JA. Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature. 2017;550(7676):407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR–Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Feng C, Yuan J, Wang R, Liu Y, Birchler JA, Han F. Efficient targeted genome modification in maize using CRISPR/Cas9 system. J Genet Genomics. 2016;43:37–43. doi: 10.1016/j.jgg.2015.10.002. [DOI] [PubMed] [Google Scholar]
- Gao J, Wang G, Ma S, Xie X, Wu X, Zhang X, Wu Y, Zhao P, Xia Q. CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol Biol. 2015;87:99–110. doi: 10.1007/s11103-014-0263-0. [DOI] [PubMed] [Google Scholar]
- Gasser CS, Fraley RT. Transgenic crops. Sci Am. 1992;266:62–69. doi: 10.1038/scientificamerican0692-62. [DOI] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianné J, Renaud JB, Maunoury SS, Shkumatava A, Teboul L, Joly JS. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Wang N. Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS ONE. 2014;9:e93806. doi: 10.1371/journal.pone.0093806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41:e188. doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Yang B, Weeks DP. Efficient CRISPR/Cas9-mediated gene editing in Arabidopsis thaliana and inheritance of modified genes in the T2 and T3 generations. PLoS ONE. 2014;9:e99225. doi: 10.1371/journal.pone.0099225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14(1):49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol. 2013;31:688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Zhang K, Chen K, Gao C. Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J Genet Genomics. 2014;41:63–68. doi: 10.1016/j.jgg.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar S, Quick WP, Bandyopadhyay A. CRISPR–Cas9 mediated genome editing in rice, advancements and future possibilities. Indian J Plant Physiol. 2016;21:437–445. doi: 10.1007/s40502-016-0252-1. [DOI] [Google Scholar]
- Mikami M, Toki S, Endo M. Precision targeted mutagenesis via Cas9 paired nickases in rice. Plant Cell Physiol. 2016;57:1058–1068. doi: 10.1093/pcp/pcw049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan K, Babu K, Sundaresan R, Rajan R, Sashital DG. The Revolution continues: newly discovered systems expand the CRISPR-Cas toolkit. Mol Cell. 2017;68(1):15–25. doi: 10.1016/j.molcel.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M, Ausubel FM. Introduction to gene editing and manipulation using CRISPR/Cas9 technology. Curr Protoc Mol Biol. 2016;115:1–6. doi: 10.1002/cpmb.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516(7530):263–266. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegoraro C, Mertz LM, da Maia LC, Rombaldi CV, de Oliveira AC. Importance of heat shock proteins in maize. J Crop Sci Biotechnol. 2011;14:85–95. doi: 10.1007/s12892-010-0119-3. [DOI] [Google Scholar]
- Peng R, Lin G, Li J. Potential pitfalls of CRISPR/Cas9-mediated genome editing. FEBS J. 2016;283:1218–1231. doi: 10.1111/febs.13586. [DOI] [PubMed] [Google Scholar]
- Pennisi E (2015) US researchers call for greater oversight of powerful genetic technology. Science Insider. http://news.sciencemag.org/biology/2014/07/us-researchers-call-greater-oversight-powerful-genetic-technology. Accessed 3 May 2015
- Puchta H, Dujon B, Hohn B. Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucl Acids Res. 1993;21:5034–5040. doi: 10.1093/nar/21.22.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Zhu T, Tian Z, Li C, Zhang W, Song R. High-efficiency CRISPR/Cas9 multiplex gene editing using the glycine tRNA-processing system-based strategy in maize. BMC Biotechnol. 2016;16:58. doi: 10.1186/s12896-016-0289-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzisheuskaya A, Shlyueva D, Müller I, Helin K. Optimizing sgRNA position markedly improves the efficiency of CRISPR/dCas9-mediated transcriptional repression. Nucl Acids Res. 2016;44:e141. doi: 10.1093/nar/gkw583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis A, Hornblower B, Robb B, Tzertzinis G. CRISPR/Cas9 and targeted genome editing: a new era in molecular biology. NEB Expr. 2014;1:3–6. [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR–Cas9. Nat Rev Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Li J, Gao C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc. 2014;9:2395–2410. doi: 10.1038/nprot.2014.157. [DOI] [PubMed] [Google Scholar]
- Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, Wang L, Hodgking A, Iyer V, Huang X, Skarnes WC. Efficient genome modification by CRISPR–Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11:399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- Shi J, Gao H, Wang H, Lafitte HR, Archibald RL, Yang M, Hakimi SM, Mo H, Habben JE. ARGOS8 variants generated by CRISPR–Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol J. 2017;15:207–216. doi: 10.1111/pbi.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kuscu C, Quinlan A, Qi Y, Adli M. Cas9-chromatin binding information enables more accurate CRISPR off-target prediction. Nucleic Acids Res. 2015;43:e118. doi: 10.1093/nar/gkv575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer M, Thumberger T, del Sol Keyer M, Wittbrodt J, Mateo JL. CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS ONE. 2015;10:e0124633. doi: 10.1371/journal.pone.0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Hu Z, Chen R, Jiang Q, Song G, Zhang H, Xi Y. Targeted mutagenesis in soybean using the CRISPR–Cas9 system. Sci Rep. 2015;5:10342. doi: 10.1038/srep10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitashev S, Young JK, Schwartz C, Gao H, Falco SC, Cigan AM. Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 2015;169:931–945. doi: 10.1104/pp.15.00793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitashev S, Schwartz C, Lenderts B, Young JK, Cigan AM. Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat Commun. 2016;7:13274. doi: 10.1038/ncomms13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot V. Saturation mutagenesis using maize transposons. Curr Opin Plant Biol. 2000;3:103–107. doi: 10.1016/S1369-5266(99)00051-5. [DOI] [PubMed] [Google Scholar]
- Waltz E. CRISPR-edited crops free to enter market, skip regulation. Nat Biotechnol. 2016;34:582–583. doi: 10.1038/nbt0616-582. [DOI] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR–Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhang S, Wang W, Xiong X, Meng F, Cui X. Efficient targeted mutagenesis in potato by the CRISPR/Cas9 system. Plant Cell Rep. 2015;34:1473–1476. doi: 10.1007/s00299-015-1816-7. [DOI] [PubMed] [Google Scholar]
- Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, Cheng AW, Trevino AE, Konermann S, Chen S, Jaenisch R. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Zhang J, Yang Y. Genome-wide prediction of highly specific guide RNA spacers for CRISPR–Cas9-mediated genome editing in model plants and major crops. Mol Plant. 2014;7:923–926. doi: 10.1093/mp/ssu009. [DOI] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014;14:327. doi: 10.1186/s12870-014-0327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadava P, Abhishek A, Singh R, Singh I, Kaul T, Pattanayak A, Agrawal PK. Advances in maize transformation technologies and development of transgenic maize. Front Plant Sci. 2017;7:1949. doi: 10.3389/fpls.2016.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liang Z, Zong Y, Wang Y, Liu J, Chen K, Qiu JL, Gao C. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun. 2016;7:12617. doi: 10.1038/ncomms12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhang C, Liu W, Gao W, Liu C, Song G, Li WX, Mao L, Chen B, Xu Y, Li X, Xie C. An alternative strategy for targeted gene replacement in plants using a dual-sgRNA/Cas9 design. Sci Rep. 2016;6:23890. doi: 10.1038/srep23890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Song N, Sun S, Yang W, Zhao H, Song W, Lai J. Efficiency and inheritance of targeted mutagenesis in maize using CRISPR–Cas9. J Genet Genomics. 2016;43:25–36. doi: 10.1016/j.jgg.2015.10.006. [DOI] [PubMed] [Google Scholar]
- Zong Y, Wang Y, Li C, Zhang R, Chen K, Ran Y, Qiu JL, Wang D, Gao C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol. 2017;35:438–440. doi: 10.1038/nbt.3811. [DOI] [PubMed] [Google Scholar]