Abstract
The signalling lymphocyte activation molecule (SLAM) family receptors play important roles in modulating immune responses. Previous studies in murine models and patients have suggested an association of the SLAM family (SLAMF) members with the development of autoimmunity, particularly systemic lupus erythematosus (SLE). Since previous investigations on CD244 expression have focussed on NK and T cells, the aim of this study was to evaluate the surface expression of major SLAMF members across monocytes and polymorphonuclear cells in an Asian SLE cohort and explore their potential associations with SLE-related disease activity and autoantibodies. Thirty-nine SLE patients and twenty-nine healthy controls (HC) were evaluated for the expression of CD150, CD84, CD229, CD48, CD244, CD352 and CD319. We determined a significantly lower expression of CD244 on monocytes in SLE patients compared to HC. Furthermore, monocyte CD244 expression was negatively associated with several serum autoantibody titres. Our findings suggest that this molecule plays an important role in immune tolerance mechanisms and should be investigated further.
Electronic supplementary material
The online version of this article (doi:10.1007/s10067-017-3698-2) contains supplementary material, which is available to authorized users.
Keywords: CD244, Monocytes, Platelets, SLAMF, SLE
Introduction
Systemic lupus erythematosus (SLE) is a chronic autoimmune disorder affecting predominantly females of childbearing age and characterised by the development of autoantibodies to nuclear antigens [1, 2]. Antinuclear antibodies (ANAs) form immune complexes with self-reactive material which deposit in tissues and promote infiltration leading to tissue destruction [3]. Both genetic and environmental factors have been implicated in SLE onset, with a syntenic region on chromosome 1 in humans and mouse models of lupus associated with disease susceptibility. In lupus-prone mice, the Sle1b locus on chromosome 1 mediates loss of tolerance and the development of highly penetrant ANAs [4]. This region encodes the highly polymorphic signalling lymphocytic activation molecule (SLAM) family genes and, in humans, the cluster of SLAM family genes are also located within the 1q23 region linked to SLE susceptibility [5, 6].
The SLAM family of receptors are expressed on cells of haematopoietic origin and play important roles in immunomodulation through predominantly homotypic interactions. The SLAM family of receptors in humans include CD150 (SLAMF1), CD48 (SLAMF2), CD229 (SLAMF3), CD244 (SLAMF4), CD84 (SLAMF5), CD352 (SLAMF6) and CD319 (SLAMF7). Receptor ligation results in signalling through immunoreceptor tyrosine-based switch motifs (ITSMs) which bind small adaptor proteins such as SLAM-associated protein (SAP) and Ewing’s sarcoma (EWS)-activated transcript-2 (EAT-2). In humans, SAP deficiency results in X-linked lymphoproliferative disorder (XLP), associated with impaired humoral immunity and defects in natural killer (NK) cells and T lymphocytes (as reviewed in [7]). An autoimmune-promoting haplotype of SLAM family members containing several polymorphisms within the SLAMF genes has been described in mice [8]. Furthermore SNPs in CD244 and CD229 have been associated with rheumatoid arthritis and SLE disease susceptibility, respectively [9, 10].
Previous studies have documented the altered expression of several SLAM family members in the context of SLE [11–14]. Increased CD229 and CD352 on T lymphocytes from SLE patients was found to promote Th17 differentiation, while loss of T cell CD244 expression is associated with decreased cytotoxic activity [11, 12]. CD244 expression on monocytes and NK cells from SLE patients was also found to be decreased [14]. Finally, increased levels of CD319 have been reported in B cells and, together with an increase in CD229, NK cells and plasmacytoid dendritic cells in patients with SLE [13, 14].
While the above studies have addressed the potential contribution of SLAM family member expression in predominantly lymphocytes and NK cells in SLE, a comprehensive analysis of their expression on myeloid lineages has yet to be addressed. We therefore investigated the cell surface expression of seven SLAM family members on circulating myeloid cells in a cohort of SLE patients in South East Asia. SLAM family receptor expression in patients was compared to levels in healthy controls (HC) and correlated with measures of SLE disease including SLE-related disease activity and serum autoantibody levels.
Materials and methods
SLE patients and serology
Thirty-nine adult patients fulfilling the American College of Rheumatology classification criteria for SLE were recruited from the Lupus Clinic at the National University Hospital (NUH), Singapore [1, 2]. Twenty-nine HC matched to the SLE patients for ethnicity and gender were recruited for comparison (control and SLE donors were 19–64 years old) and peripheral blood harvested within an hour of the SLE patient peripheral blood draw. Details of peripheral blood collection and serology analyses for this cohort have been previously described [15]. The study was approved by the local ethics committees. Written informed consent was obtained from all participants prior to recruitment.
Flow cytometry
Whole blood was stained with directly-conjugated antibodies and incubated for 30 mins at 4 °C. Following red blood cell lysis (BD FACS Lysing solution), myeloid lineages were identified using anti-CD45, anti-CD14, anti-CD11b (BD Pharmingen), anti-CD16 (eBioscience), and anti-CD62L (Beckman Coulter)(online resource 1A, B). For analysis of SLAM family receptor expression, anti-CD150, anti-CD84, anti-CD229 (eBioscience), anti-CD48, anti-CD244, anti-CD352 and anti-CD319 (Biolegend) antibodies were used. Acquisition and analysis were performed using a BD LSRFortessa™ and FlowJo v10.1 for Windows, respectively (Treestar). Median fluorescence intensity (MFI) was normalised to a mean equivalent of fluorochrome (MEF) SPHERO™ Rainbow Calibration Particles and the isotype control, as per the manufacturer’s instructions (online resource 1C).
Statistics
For flow cytometry, mean equivalent of fluorochrome (MEF) values are reported. Results are expressed as the arithmetic mean ± standard error of the mean (SEM) for a given number of values (n). Comparisons between SLE patients and HC were performed using the Mann-Whitney U test. For data analysis involving more than two groups, the Kruskal-Wallis non-parametric ANOVA followed by Dunn’s multiple comparison test was used. The Spearman rank correlation was performed for correlation analyses. Statistics and graphs were generated using GraphPad Prism v6.00 for Windows (GraphPad Software).
Results
Patient demographics are summarised in online resource 2, reproduced with permission from Rheumatology [15]. All patients included in the study were positive for ANAs and we observed significant negative correlations between serum complement C3 and C4 levels and anti-dsDNA and anti-snRNP autoantibodies, respectively (online resource 1), in the sera of SLE patients.
CD244 and CD84 were both determined to be broadly expressed on myeloid lineages in SLE patients and HC, while CD48, the ligand for CD244, is expressed only on monocytes (Table 1). In contrast, in both patients and HC, expression of CD352 and CD319 was found to be restricted to eosinophils and monocytes, respectively (Table 1). While prior studies have demonstrated that Ly108, the murine homologue of CD352, is expressed on polymorphonuclear cells (PMNs), we did not detect expression above levels of background isotype staining (data not shown) [16]. Cell surface expression of CD229 and CD150 on circulating myeloid cells was undetected in both patients and HC, potentially reflecting restricted expression on NK cells, lymphocytes and antigen presenting cells (APCs) (data not shown) [13, 17, 18]. CD150 expression on platelets has been previously described however, and this discrepancy is likely attributable to the different antibody clone used in this study [19].
Table 1.
Expression of SLAMF receptors on myeloid lineages in SLE patients and healthy controls
| SLE patients mean ± SEM (n) | Controls mean ± SEM (n) | |
|---|---|---|
| CD48 (SLAMF2) | ||
| Monocytes (MEF) | 1180 ± 88 (37) | 1105 ± 76 (27) |
| CD244 (SLAMF4) | ||
| Monocytes (MEF) | 13,547 ± 1464 (37) | 17,522 ± 1775 (27) |
| Eosinophils (MEF) | 14,751 ± 1510 (29) | 12,253 ± 1176 (27) |
| Platelets (MEF) | 1471 ± 135 (37) | 2034 ± 183 (27) |
| CD84 (SLAMF5) | ||
| PMNS (%) | 33.9 ± 3.6 (39) | 31.6 ± 4.0 (29) |
| Monocytes (MEF) | 1509 ± 137 (35) | 1555 ± 203 (25) |
| Eosinophils (MEF) | 745 ± 67 (27) | 740 ± 105 (25) |
| Platelets (MEF) | 841 ± 78 (35) | 1087 ± 102 (25) |
| CD352 (SLAMF6, NTB-A) | ||
| Eosinophils (MEF) | 2262 ± 326 (18) | 1979 ± 235 (16) |
| CD319 (SLAMF7, CRACC, CS1) | ||
| Monocytes (MEF) | 460 ± 152 (37) | 309 ± 47 (27) |
MEF mean equivalent of fluorochrome, SEM standard error of the mean, n number of individuals
In agreement with a study by Kim et al., levels of CD244 on SLE patient monocytes were significantly lower as compared to HC (Fig. 1a) [14]. Furthermore, SLE patients with active disease were also found to express the lowest levels of CD244 on monocytes. Decreased CD244 expression on platelets from SLE patients was also observed and correlated strongly with decreased monocyte CD244 expression (Fig. 1b–c). While CD244 expression on platelets was noted to be approximately tenfold lower than for monocytes, expression of this SLAM family receptor in this cell type has been reported previously at the RNA level [19].
Fig. 1.
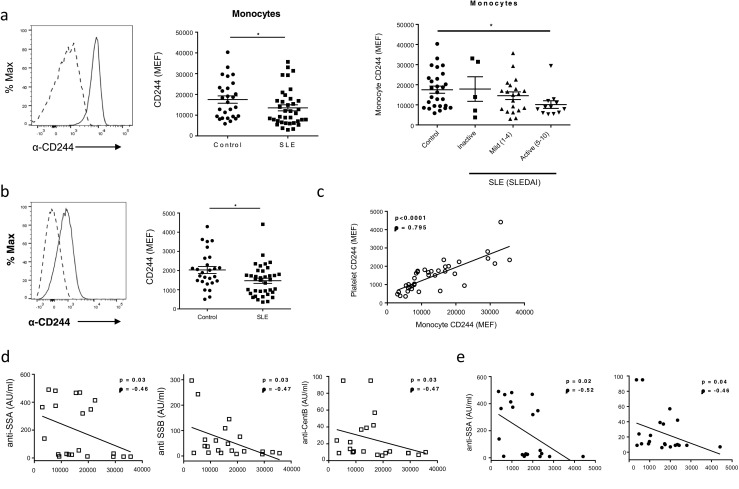
CD244 expression is decreased on monocytes and platelets in SLE patients and correlates with serum autoantibodies. a CD244 monocyte expression levels are decreased in SLE patients compared to healthy controls. Patients with active disease show lower levels of CD244. Histogram illustrates anti-CD244 staining on monocytes (dashed line represents isotype control). b Decreased platelet CD244 expression in SLE patients. Histogram illustrates anti-CD244 staining on platelets (dashed line represents isotype control). c Decreased levels of CD244 on SLE patient platelets correlate with decreased monocyte CD244 expression. d Significant correlations between SLE patient monocyte CD244 expression levels and serum autoantibodies. e Significant correlations between SLE patient platelet CD244 expression levels and serum autoantibodies. Bars represent mean ± SEM. *P ˂ 0.05
Our analysis of monocyte and platelet CD244 revealed significant correlations between decreased SLE patient CD244 expression and increased serum autoantibody levels (anti-SSA 52/60, anti-SSB and anti-CentB) (Fig. 1d, e and Table 2).
Table 2.
Association of CD244 with autoantibody titres
| Autoantibody | Monocyte CD244 | Platelet CD244 | ||
|---|---|---|---|---|
| p | ρ | p | ρ | |
| Anti-SSB | 0.033 | −0.468 | 0.172 | −0.310 |
| Anti-SSA60 | 0.034 | −0.464 | 0.016 | −0.518 |
| Anti-SSA52 | 0.082 | −0.389 | 0.140 | −0.333 |
| Anti-Sm | 0.179 | −0.305 | 0.750 | −0.074 |
| Anti-Scl70 | 0.085 | −0.385 | 0.159 | −0.319 |
| Anti-RNP | 0.278 | −0.248 | 0.704 | −0.088 |
| Anti-RibP | 0.097 | −0.371 | 0.313 | −0.231 |
| Anti-Jo1 | 0.322 | −0.227 | 0.598 | −0.122 |
| Anti-CentB | 0.031 | −0.471 | 0.043 | −0.445 |
p probability, ρ Spearman’s rank correlation
Significance; p < 0.05 indicated in italics
Elevated levels of circulating PMNs have been previously reported in SLE; in agreement with this, we found the proportion of PMNs in peripheral blood to be significantly increased in our patient cohort as compared to HC (online resource 2) [18]. Circulating PMNs were found to express only a single SLAM family receptor, CD84, with no detectable difference in expression levels between SLE patients and HC (Table 1).
Discussion
In this study, we have demonstrated a significant reduction in CD244 expression on SLE patient monocytes and platelets compared to gender- and ethnicity-matched healthy subjects in an Asian cohort. Furthermore, we showed that monocyte CD244 levels correlated in a negative manner with serum autoantibody levels, including anti-SSA52/60, anti-SSB and anti-CentB antibodies.
Altered expression of SLAM family receptors has been reported in several autoimmune disorders, including Crohn’s disease, rheumatoid arthritis and SLE (as reviewed in [20]). While previous studies have focused predominantly on lymphocytes and NK cells, we performed a comprehensive flow cytometric analysis of SLAM family receptor expression on circulating myeloid lineages. CD150 and CD229 were undetectable on circulating myeloid cells, while monocytes were found to express all remaining SLAM family members, except for CD352. Eosinophils were demonstrated to express CD244, CD84 and CD352, and PMNs were found to express only CD84. CD84 expression was also detected on platelets, together with low levels of CD244.
Our findings on reduced monocyte CD244 levels in SLE are consistent with previous data from a mixed African-American, Hispanic and Caucasian population from the USA [14]. Furthermore, these phenotypic associations in patients are supported by murine studies which have shown an association of the CD244/Ly9/Cs1 region with ANA titres in the B6.Sle1 benign SLE model [21]. In addition, genetic ablation of CD244 on a non-autoimmune B6 background resulted in a breach of self-tolerance in 30% of female mice aged 12 months old. This study, by Sharpe and colleagues, also demonstrated that CD244-deficient mice have higher levels of anti-DNA autoantibodies following a graft-versus-host disease challenge [22].
CD244 is expressed on NK cells, γδT cells, basophils, monocytes, eosinophils and a subset of CD8 cells (reviewed in [23]). Unlike other SLAMF members, which are homotypic, CD244 binds with CD48 to trigger SAP signalling and activation of the CD244 cytoplasmic ITSM domains. Research on immunological functions to date has focused on the complex role of CD244 in enhancing and decreasing NK cell function [24]. However, it appears that the loss of tolerance in the experimental model described above is NK cell-independent [22].
Innate immune mechanisms play a critical role in the development of SLE and disease severity (reviewed in [25]). Under resting conditions, normal monocytes have little capability for antigen presentation; however, in the permitting inflammatory environment, they can differentiate into both macrophages and dendritic cells (DCs). SLE patient monocytes have been shown to have enhanced DC-like function [26]. In contrast, the phagocytic ability of macrophages to engulf apoptotic material is reduced in SLE [27, 28]. The contribution of CD244 to monocyte, DC, or macrophage cell function is unknown and is an area of future investigation.
PMNs are the most abundant cell type in peripheral blood, and we observed a significant increase in the proportion of circulating PMNs in SLE patients when compared to HC. Mice deficient in Ly108 (CD352) exhibit defects in PMN function leading to increased susceptibility to bacterial infection [16]. We only detected SLAMF member CD84 on PMNs in our cohort however and did not find any associations with disease.
In summary, we describe significant decreases in platelet and monocyte CD244 expression in SLE patients which correlates with ANA titre. Investigating the role of monocyte CD244 may prove to be important in mechanisms of tolerance and autoimmunity.
Electronic supplementary material
(DOCX 186 kb)
Acknowledgments
We thank the clinical liaison team at the National University Hospital, Singapore, and the flow cytometry core at SIgN, A*STAR.
Compliance with ethical standards
Funding
This study was supported by funding from an A*STAR-JJSI Joint Grant 1218226002 and core funding from the Singapore Immunology Network (SIgN) to AMF and the Institute for Molecular and Cell Biology to JEC at the Agency for Science, Technology and Research (A*STAR), Singapore.
Ethics approval
The study was approved by the NHG Domain Specific Review Board (DSRB-E/11/061) and the National University of Singapore Institutional Review Board (NUS-IRB-09-256).
Disclosures
None.
Footnotes
Anselm Mak and Susannah I Thornhill contributed equally to this work
Electronic supplementary material
The online version of this article (doi:10.1007/s10067-017-3698-2) contains supplementary material, which is available to authorized users.
References
- 1.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40(9):1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 2.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, Schaller JG, Talal N, Winchester RJ. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25(11):1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 3.Fairhurst AM, Wandstrat AE, Wakeland EK. Systemic lupus erythematosus: multiple immunological phenotypes in a complex genetic disease. Adv Immunol. 2006;92:1–69. doi: 10.1016/S0065-2776(06)92001-X. [DOI] [PubMed] [Google Scholar]
- 4.Wandstrat AE, Nguyen C, Limaye N, Chan AY, Subramanian S, Tian XH, Yim YS, Pertsemlidis A, Garner HR, Jr, Morel L, Wakeland EK. Association of extensive polymorphisms in the SLAM/CD2 gene cluster with murine lupus. Immunity. 2004;21(6):769–780. doi: 10.1016/j.immuni.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Moser KL, Neas BR, Salmon JE, Yu H, Gray-McGuire C, Asundi N, Bruner GR, Fox J, Kelly J, Henshall S, Bacino D, Dietz M, Hogue R, Koelsch G, Nightingale L, Shaver T, Abdou NI, Albert DA, Carson C, Petri M, Treadwell EL, James JA, Harley JB. Genome scan of human systemic lupus erythematosus: evidence for linkage on chromosome 1q in African-American pedigrees. Proc Natl Acad Sci U S A. 1998;95(25):14869–14874. doi: 10.1073/pnas.95.25.14869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsao BP, Cantor RM, Grossman JM, Kim SK, Strong N, Lau CS, Chen CJ, Shen N, Ginzler EM, Goldstein R, Kalunian KC, Arnett FC, Wallace DJ, Hahn BH. Linkage and interaction of loci on 1q23 and 16q12 may contribute to susceptibility to systemic lupus erythematosus. Arthritis Rheum. 2002;46(11):2928–2936. doi: 10.1002/art.10590. [DOI] [PubMed] [Google Scholar]
- 7.Tangye SG. XLP: clinical features and molecular etiology due to mutations in SH2D1A encoding SAP. J Clin Immunol. 2014;34(7):772–779. doi: 10.1007/s10875-014-0083-7. [DOI] [PubMed] [Google Scholar]
- 8.Soni C, Domeier PP, Wong EB, Shwetank KTN, Elias MJ, Schell SL, Lukacher AE, Cooper TK, Rahman ZS. Distinct and synergistic roles of FcgammaRIIB deficiency and 129 strain-derived SLAM family proteins in the development of spontaneous germinal centers and autoimmunity. J Autoimmun. 2015;63:31–46. doi: 10.1016/j.jaut.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki A, Yamada R, Kochi Y, Sawada T, Okada Y, Matsuda K, Kamatani Y, Mori M, Shimane K, Hirabayashi Y, Takahashi A, Tsunoda T, Miyatake A, Kubo M, Kamatani N, Nakamura Y, Yamamoto K. Functional SNPs in CD244 increase the risk of rheumatoid arthritis in a Japanese population. Nat Genet. 2008;40(10):1224–1229. doi: 10.1038/ng.205. [DOI] [PubMed] [Google Scholar]
- 10.Cunninghame Graham DS, Vyse TJ, Fortin PR, Montpetit A, Cai YC, Lim S, McKenzie T, Farwell L, Rhodes B, Chad L, Hudson TJ, Sharpe A, Terhorst C, Greenwood CM, Wither J, Rioux JD. Association of LY9 in UK and Canadian SLE families. Genes Immun. 2008;9(2):93–102. doi: 10.1038/sj.gene.6364453. [DOI] [PubMed] [Google Scholar]
- 11.Chatterjee M, Rauen T, Kis-Toth K, Kyttaris VC, Hedrich CM, Terhorst C, Tsokos GC. Increased expression of SLAM receptors SLAMF3 and SLAMF6 in systemic lupus erythematosus T lymphocytes promotes Th17 differentiation. J Immunol. 2012;188(3):1206–1212. doi: 10.4049/jimmunol.1102773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kis-Toth K, Comte D, Karampetsou MP, Kyttaris VC, Kannan L, Terhorst C, Tsokos GC. Selective loss of signaling lymphocytic activation molecule family member 4-positive CD8+ T cells contributes to the decreased cytotoxic cell activity in systemic lupus erythematosus. Arthritis Rheumatol. 2016;68(1):164–173. doi: 10.1002/art.39410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagberg N, Theorell J, Schlums H, Eloranta ML, Bryceson YT, Ronnblom L. Systemic lupus erythematosus immune complexes increase the expression of SLAM family members CD319 (CRACC) and CD229 (LY-9) on plasmacytoid dendritic cells and CD319 on CD56(dim) NK cells. J Immunol. 2013;191(6):2989–2998. doi: 10.4049/jimmunol.1301022. [DOI] [PubMed] [Google Scholar]
- 14.Kim JR, Mathew SO, Patel RK, Pertusi RM, Mathew PA. Altered expression of signalling lymphocyte activation molecule (SLAM) family receptors CS1 (CD319) and 2B4 (CD244) in patients with systemic lupus erythematosus. Clin Exp Immunol. 2010;160(3):348–358. doi: 10.1111/j.1365-2249.2010.04116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thornhill SI, Mak A, Lee B, Lee HY, Poidinger M, Connolly JE, Fairhurst AM (2017) Monocyte Siglec-14 expression is upregulated in patients with systemic lupus erythematosus and correlates with lupus disease activity. Rheumatology (Oxford). doi:10.1093/rheumatology/kew498 [DOI] [PMC free article] [PubMed]
- 16.Howie D, Laroux FS, Morra M, Satoskar AR, Rosas LE, Faubion WA, Julien A, Rietdijk S, Coyle AJ, Fraser C, Terhorst C. Cutting edge: the SLAM family receptor Ly108 controls T cell and neutrophil functions. J Immunol. 2005;174(10):5931–5935. doi: 10.4049/jimmunol.174.10.5931. [DOI] [PubMed] [Google Scholar]
- 17.Romero X, Benitez D, March S, Vilella R, Miralpeix M, Engel P. Differential expression of SAP and EAT-2-binding leukocyte cell-surface molecules CD84, CD150 (SLAM), CD229 (Ly9) and CD244 (2B4) Tissue Antigens. 2004;64(2):132–144. doi: 10.1111/j.1399-0039.2004.00247.x. [DOI] [PubMed] [Google Scholar]
- 18.de la Fuente MA, Tovar V, Villamor N, Zapater N, Pizcueta P, Campo E, Bosch J, Engel P. Molecular characterization and expression of a novel human leukocyte cell-surface marker homologous to mouse Ly-9. Blood. 2001;97(11):3513–3520. doi: 10.1182/blood.V97.11.3513. [DOI] [PubMed] [Google Scholar]
- 19.Nanda N, Andre P, Bao M, Clauser K, Deguzman F, Howie D, Conley PB, Terhorst C, Phillips DR. Platelet aggregation induces platelet aggregate stability via SLAM family receptor signaling. Blood. 2005;106(9):3028–3034. doi: 10.1182/blood-2005-01-0333. [DOI] [PubMed] [Google Scholar]
- 20.Cannons JL, Tangye SG, Schwartzberg PL. SLAM family receptors and SAP adaptors in immunity. Annu Rev Immunol. 2011;29:665–705. doi: 10.1146/annurev-immunol-030409-101302. [DOI] [PubMed] [Google Scholar]
- 21.Wong EB, Soni C, Chan AY, Domeier PP, Shwetank AT, Limaye N, Khan TN, Elias MJ, Chodisetti SB, Wakeland EK, Rahman ZS. B cell-intrinsic CD84 and Ly108 maintain germinal center B cell tolerance. J Immunol. 2015;194(9):4130–4143. doi: 10.4049/jimmunol.1403023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown DR, Calpe S, Keszei M, Wang N, McArdel S, Terhorst C, Sharpe AH. Cutting edge: an NK cell-independent role for Slamf4 in controlling humoral autoimmunity. J Immunol. 2011;187(1):21–25. doi: 10.4049/jimmunol.1100510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McArdel SL, Terhorst C, Sharpe AH. Roles of CD48 in regulating immunity and tolerance. Clin Immunol. 2016;164:10–20. doi: 10.1016/j.clim.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waggoner SN, Kumar V. Evolving role of 2B4/CD244 in T and NK cell responses during virus infection. Front Immunol. 2012;3:377. doi: 10.3389/fimmu.2012.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan VS, Nie YJ, Shen N, Yan S, Mok MY, Lau CS. Distinct roles of myeloid and plasmacytoid dendritic cells in systemic lupus erythematosus. Autoimmun Rev. 2012;11(12):890–897. doi: 10.1016/j.autrev.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Blanco P, Palucka AK, Gill M, Pascual V, Banchereau J. Induction of dendritic cell differentiation by IFN-alpha in systemic lupus erythematosus. Science. 2001;294(5546):1540–1543. doi: 10.1126/science.1064890. [DOI] [PubMed] [Google Scholar]
- 27.Herrmann M, Voll RE, Zoller OM, Hagenhofer M, Ponner BB, Kalden JR. Impaired phagocytosis of apoptotic cell material by monocyte-derived macrophages from patients with systemic lupus erythematosus. Arthritis Rheum. 1998;41(7):1241–1250. doi: 10.1002/1529-0131(199807)41:7<1241::AID-ART15>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 28.Ren Y, Tang J, Mok MY, Chan AW, Wu A, Lau CS. Increased apoptotic neutrophils and macrophages and impaired macrophage phagocytic clearance of apoptotic neutrophils in systemic lupus erythematosus. Arthritis Rheum. 2003;48(10):2888–2897. doi: 10.1002/art.11237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 186 kb)


