Abstract
Pseudomonas aeruginosa uses quorum sensing (QS) to regulate the production of public goods such as the secreted protease elastase. P. aeruginosa requires the LasI–LasR QS circuit to induce elastase and enable growth on casein as the sole carbon and energy source. The LasI–LasR system also induces a second QS circuit, the RhlI–RhlR system. During growth on casein, LasR-mutant social cheaters emerge, and this can lead to a population collapse. In a minimal medium containing ammonium sulfate as a nitrogen source, populations do not collapse, and cheaters and cooperators reach a stable equilibrium; however, without ammonium sulfate, cheaters overtake the cooperators and populations collapse. We show that ammonium sulfate enhances the activity of the RhlI–RhlR system in casein medium and this leads to increased production of cyanide, which serves to control levels of cheaters. This enhancement of cyanide production occurs because of a trade-off in the metabolism of glycine: exogenous ammonium ion inhibits the transformation of glycine to 5,10-methylenetetrahydrofolate through a reduction in the expression of the glycine cleavage genes gcvP1 and gcvP2, thereby increasing the availability of glycine as a substrate for RhlR-regulated hydrogen cyanide synthesis. Thus, environmental ammonia enhances cyanide production and stabilizes QS in populations of P. aeruginosa.
Keywords: quorum sensing, LasR, RhlR, sociomicrobiology, hydrogen cyanide
Introduction
Bacterial quorum sensing (QS) regulates group behaviors. Pseudomonas aeruginosa uses QS to regulate the production of a variety of extracellular products, which can be shared “public goods” in population of cells (Diggle et al., 2007; Wilder et al., 2011; Stevens et al., 2012). There are two acyl-homoserine lactone (AHL) QS circuits in P. aeruginosa, the LasI–LasR circuit and the RhlI–RhlR circuit. LasI is an enzyme that catalyzes synthesis of the diffusible QS signal N-3-oxo-dodecanoyl-homoserine lactone (3OC12-HSL). At sufficient concentrations, 3OC12-HSL can bind to the transcription factor LasR, which in turn activates a constellation of genes including rhlR and rhlI. The RhlI–RhlR circuit consists of butanoyl-homoserine lactone (C4-HSL), produced by RhlI, and the C4-HSL-dependent RhlR (Schuster et al., 2003; Gilbert et al., 2009), which activates a set of genes that partially overlap with LasR-dependent genes.
Some quorum-regulated secreted products have been established as public goods that are shared among all cells in a population (Diggle et al., 2007; Pollitt et al., 2014). Growth of P. aeruginosa with casein as the sole source of carbon and energy requires production of the QS-regulated protease elastase (Diggle et al., 2007), which digests casein into peptides and amino acids that can be taken up and used by P. aeruginosa cells regardless of whether they produce elastase themselves. When P. aeruginosa is grown continuously on casein, LasR-mutant cheaters, which produce neither elastase nor other quorum-regulated factors, emerge (Sandoz et al., 2007; Dandekar et al., 2012).
Uncontrolled cheating can result in a tragedy of the commons wherein there are insufficient numbers of cooperators to maintain growth on casein (Diggle et al., 2007; Dandekar et al., 2012; Wang et al., 2015). Mechanisms for cheater control have been described and include kin-selection (Diggle et al., 2007; Allen et al., 2013), prudent regulation of costly metabolism (Xavier et al., 2011), metabolic constraints (Dandekar et al., 2012), and metabolic policing (Wang et al., 2015).
When P. aeruginosa is grown on casein as the sole carbon and energy source and ammonium sulfate as additional nitrogen source, cheaters emerge and eventually equilibrate with cooperators at 25–40% of the total population (Sandoz et al., 2007; Dandekar et al., 2012). However, when casein is the sole nitrogen as well as carbon source, cheater frequency rises to 80% or more, ultimately leading to a tragedy of the commons (Dandekar et al., 2012). Establishment of the cooperator–cheater equilibrium in media containing ammonium sulfate depends on RhlR-dependent hydrogen cyanide (HCN) production by the cooperators (Wang et al., 2015), as deletion of hcnC, a component of the HCN synthase, or rhlR results in cheaters outcompeting cooperators. The cooperators presumably regulate both cyanide production and a detoxification mechanism through rhl QS (Wang et al., 2015). The production of cyanide by cooperators in these conditions serves as a policing strategy to enforce cooperation (West et al., 2007). There is a cost (in both growth rate and final cell density) to the wildtype (w.t.) in generation of HCN (Wang et al., 2015). However, the production of HCN imparts a disproportionate fitness impact on cheaters (i.e., they are more susceptible to cyanide intoxication).
We are interested in the costs and benefits of cooperation in bacteria and asked if there is an interaction between exogenous ammonia and production of cyanide in casein medium. One possible link is through glycine degradation: P. aeruginosa converts glycine to cyanide (Castric, 1977; Devi et al., 2013); alternatively, glycine can be degraded to ammonia and CO2 by glycine dehydrogenase, which is encoded by gcvP1 and gcvP2 (Toone et al., 2002; Kai et al., 2009), in a process involving tetrahydrofolate and NADH (Goro et al., 2008). gcvP2, but not gcvP1, is also LasR-regulated (Schuster et al., 2003). We reasoned that ammonia in the environment might alter the outcome of glycine metabolism to favor cyanide production, and therefore improve the ability of P. aeruginosa to restrain cheaters in media containing ammonium sulfate. We show that ammonium sulfate enhances cyanide production by P. aeruginosa. These studies help us understand how P. aeruginosa senses its environment to stabilize QS and give insight into the development and stabilization of cooperative behavior in bacteria.
Materials and Methods
Bacterial Strains and Media Used
We used the following P. aeruginosa strains in our experiments: PAO1-UW- (Stover et al., 2000) and PAO1-UW-derived deletion mutants of rhlR, lasR, and hcnC (Wang et al., 2015). Bacteria were grown in either LB media buffered with 50 mM 3-(N-Morpholino) propane-sulfonic acid (MOPS), pH 7.0 (LB-MOPS) (Sambrook et al., 1989) or in photosynthetic medium (PM) (Kim and Harwood, 1991) supplemented with 1% sodium caseinate (Dandekar et al., 2012). The base PM-casein medium contains 1% ammonium sulfate, which we reduced or increased as described in individual experiments. Unless otherwise specified, cultures were grown in 4 ml of PM-casein medium in 16-mm test tubes with shaking at 37°C.
Evolution Experiments
Inoculation for experiments in PM-casein medium was from overnight LB-MOPS cultures (Sambrook et al., 1989). The initial optical density was 0.025 at 600 nm. Subsequently, we transferred 200 μl at 24 h intervals to fresh casein broth for days 1–3; subsequent transfers were 50 μl daily (Wang et al., 2015). Cheater abundance was determined by plating on skim milk agar, as previously described (Dandekar et al., 2012); we enumerated at least 100 colonies for each condition at each timepoint. For competition experiments, overnight cultures of PAO1 and PAO1ΔlasR were mixed at ratios of 99:1 or 9:1, and used to inoculate PM-casein medium as described above. Cheater frequency was determined daily at the time of passage.
Dialysis Membrane Competition Experiments
Strain PAO1ΔlasR was sealed in a dialysis bag in 5 ml of PM-casein medium at an initial density of 0.025. Strain PAO1 was used to inoculate 45 ml of PM-casein medium (starting OD600 of 0.025) in a 250 ml flask. The dialysis bag containing PAO1ΔlasR was placed into the PAO1 culture flask which was then incubated at 37°C with shaking (250 rpm). We verified that PAO1ΔlasR did not leak from the dialysis bag by phenotype screening (Dandekar et al., 2012).
Reporter Assays
We used lasI-gfp (pBS351) and rhlA-gfp promoter fusion constructs (pJF01) (Feltner et al., 2016) in pPROBE-GT (Gmr) (Miller et al., 2000) to measure LasR and RhlR activity, respectively. Cells from overnight cultures were electrotransformed with either plasmid after preparation with 300 mM sucrose (Choi et al., 2006). Transformants were selected on LB agar containing 100 mg ml-1 gentamicin. GFP activity was determined after 24 h of incubation by adding 200 μl culture to black 96-well plates and measuring fluorescence in a SpectraMax® i3 Plate Reader (Molecular Devices, Sunnyvale, CA, United States) with 488 nm excitation and 525 nm detection wavelengths.
RT-qPCR
Cells from either a mixed culture or the outside of the dialysis bag were pelleted by centrifugation at 16,430 × g for 3 min at 4°C to collect cells. RNA isolation and cDNA synthesis were performed as described elsewhere (Fey et al., 2004). Primers for lasR, rhlR, gcvP1, gcvP2, hcnC, and proC are shown in Table 1. qPCR reactions were carried out in 96-well plates with a CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA, United States) using SYBR premix Ex Taq II Kit (TaKaRa, Japan). After 40 cycles, a melting curve analysis was performed (60–95°C) to verify the specificity of amplicons. Each amplification was repeated three times. The specificity of the amplicons was confirmed by the presence of a single peak. The relative level of expression (Q) for a given gene was calculated based on the formula Q = 2-ΔCt. Each Ct value represents the average of three replicates, where ΔCt represents the target gene Ct minus the Ct of the housekeeping gene proC (Kim et al., 2017).
Table 1.
Primers used in this study.
| Target gene | Primer name | Sequence (5′–3′) | Amplicon (bp) | Tm (°C) | Function |
|---|---|---|---|---|---|
| lasR | qlasR-F | 5′-cagaagatggcgagcgaccttg-3′ | 102 | 59.9 | QS regulator |
| qlasR-R | 5′-cgggtagttgccgacgatgaag-3′ | 59.7 | |||
| rhlR | qrhlR-F | 5′-gcgaccagcagaacatctccag-3′ | 122 | 59.6 | QS regulator |
| qrhlR-R | 5′-cgggttggacatcagcatcgg-3′ | 59.6 | |||
| hcnC | qhcnC-F | 5′-gccagtacgccgagcacatc-3′ | 145 | 59.6 | Hydrogen cyanide synthase |
| qhcnC-R | 5′-acctggtggtcgcagaggaatt-3′ | 59.9 | |||
| gcvP1 | qgcvP1-F | 5′-cgctgctgaatttccagcaa-3′ | 142 | 55.5 | Glycine cleavage system protein P1 |
| qgcvP1-R | 5′-aagaaccggttgctcctggc-3′ | 58.6 | |||
| gcvP2 | qgcvP2-F | 5′-cgctgctgaacttccagacc-3′ | 142 | 57 | Glycine cleavage system protein P2 |
| qgcvp2-R | 5′-aatgcctggctggtgcggtt-3′ | 61 | |||
| proC | qproC-F | 5′-cgtcgtggtcctgtcggtca-3′ | 100 | 59.9 | House-keeping gene |
| qproC-R | 5′-ggcggcgatggagacgatca-3′ | 60.4 |
Reactive Oxygen Species Measurement
Strains PAO1, PAO1ΔrhlR, or PAO1ΔhcnC were mixed with strain PAO1ΔlasR at a 9:1 ratio in casein broth with or without ammonium sulfate. After 24 h, the supernatant fluid was collected after centrifugation at 16,430 × g for 3 min and 4°C. The supernatant was added to an equal volume of a PAO1ΔlasR culture in logarithmic phase (OD600 = 1.0). Cell density was measured every 2 h.
To measure ROS, 2′,7′-dichlorofluorescin diacetate (DCFH-DA) was added to the PAO1ΔlasR culture with different supernatants at a 10 mM final concentration. After incubating at 37°C for 90 min in dark, cells were washed in PBS twice and resuspended in 1 ml PBS. Reaction of DCFH-DA with ROS generates a fluorescent, oxidized derivative, DCF (F). DCF (F) levels were measured in a SpectraMax® i3 Plate Reader with 488 nm excitation wavelength and 525 nm detection wavelength. We calculated the relative ROS dividing the DCF (F)/OD600 of experimental samples by that of fresh PM-casein without supernatant.
Ammonia Accumulation
PAO1 was mixed with PAO1ΔlasR at a 9:1 ratio in casein broth with or without ammonium sulfate. We centrifuged 2 ml of culture fluid at 16,430 × g for 3 min. The abundance of ammonia in the supernatant was measured according to Standard Methods American Public Health Association (Federation and American Public Health Association [APHA], 2005). Briefly, we added 1 ml of the sample to a 50 ml cuvette and diluted to 25 ml with distilled water. Then we added 5 ml of phenol solution (10 g l-1 phenol and 0.1 g l-1 sodium nitroprusside) and 5 ml of sodium hypochlorite alkaline solution (10 g l-1 NaOH, 7.06 g l-1 KH2PO4, 31.8 g l-1 Na3PO4, 10 ml l-1 NaClO), for 1 h. Next, we added 1 ml of masking agent (200 g l-1 potassium sodium tartrate, 50 g l-1 EDTA disodium salt, and 2 g l-1 NaOH) and an additional 24 ml H2O. The absorbance was then measured at 625 nm using a spectrophotometer.
Hydrogen Cyanide Measurements
We modified the method of Gallagher and Manoil (2001) to measure HCN. Briefly, cultures were grown for 24 h at 37°C with shaking in 16 mm tubes with a rubber stopper. Two needles were inserted into the rubber stopper. Nitrogen gas entered through one needle, the other needle served as an outflow for gas collection. The collected gas was passed through 5.0 ml of 4 M NaOH in a cyanide collection tube. Cyanide levels were assessed by comparison with KCN standards. Cyanide production is expressed as the amount (μM) of HCN per cooperator. All experiments were performed in triplicate.
Results
Addition of Ammonium Sulfate Favors Cooperators in Competition with Cheaters
When w.t. P. aeruginosa PAO1 is grown in PM with casein as the sole carbon source (see section “Materials and Methods”), LasR-mutant social cheaters emerge and come to an equilibrium with the w.t. (Sandoz et al., 2007; Dandekar et al., 2012; Wang et al., 2015). However, when casein also serves as the sole nitrogen source, LasR mutants overrun the w.t. and cause a population collapse (Dandekar et al., 2012). This phenomenon has been attributed to the heightened cost of using QS to obtain carbon, energy, and nitrogen (Dandekar et al., 2012), and we reasoned that added ammonium sulfate might interact with QS-regulated gene products in a way that determined the fates of the populations.
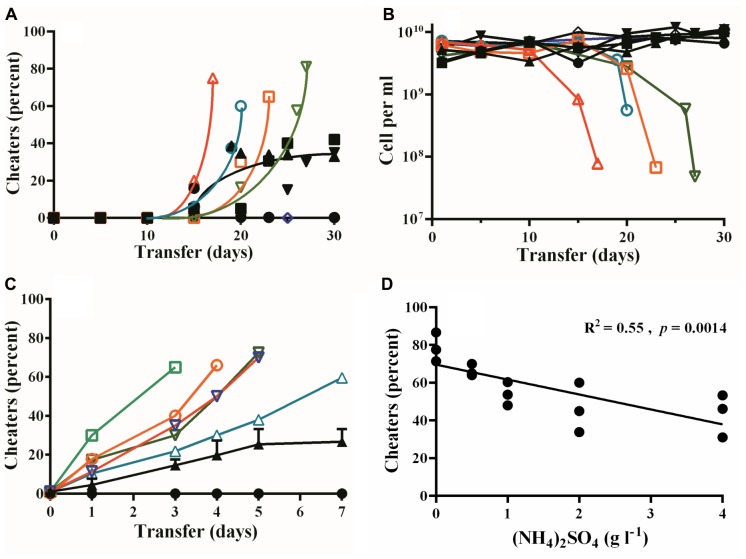
Consistent with prior publications, when we grew PAO1 in PM-casein media with or without ammonium sulfate, LasR-mutant cheaters emerged by 15 days in four out of five experiments (Figures 1A,B). In PM-casein with ammonium sulfate, cheaters and cooperators reached a stable equilibrium as previously reported (Sandoz et al., 2007; Dandekar et al., 2012; Wang et al., 2015). In PM-casein without ammonium sulfate, however, the LasR-mutant frequency exceeded 60%, after which populations did not grow upon transfer.
FIGURE 1.
Emergence of LasR-mutant cheaters and their fitness in PM-casein with or without ammonium sulfate added. (A) Percent of protease-negative cheaters in PM-casein media over time. In four of five individual experiments without added ammonium sulfate, cheaters emerged and increased (hollow symbols). In PM-casein with ammonium sulfate (black symbols), cheaters reached a frequency of 27 ± 7%. In one experiment with and one experiment without ammonium sulfate, cheaters did not emerge. (B) Cell densities in casein experiments. In the cases where the culture could not be propagated, the final cell densities fell by 90–99% (hollow symbols). Experiments in PM-casein with ammonium sulfate (black symbols) are shown for comparison. (C) LasR-mutant frequency after 7 days of passage. LasR-mutant cheater frequency in PM-casein without ammonium sulfate was rapid in all five replicates from an initial frequency of 1% (hollow symbols). An equilibrium was always established (black triangles) in media with ammonium sulfate added. When LasR mutants were not present at the beginning of the experiment, they did not emerge in 7 days PM-casein with ammonium sulfate (black circles). (D) Outcomes of w.t. – LasR-mutant competitions with increasing ammonium sulfate concentrations. w.t. PAO1 and LasR-mutant cheaters were mixed at a 1:1 ratio and the frequency of each was enumerated after 24 h. Experiments were performed in triplicate. For the data in panel D, a linear regression analysis was performed.
To further examine the influence of ammonium sulfate on competition between w.t. and LasR mutants, we performed a 7-day competition between w.t. and LasR mutants. The inocula consisted of 99% w.t. and 1% LasR mutants. Consistent with prior reports (Sandoz et al., 2007; Dandekar et al., 2012), w.t. and LasR mutants reached a stable equilibrium of about 20% cheaters in PM-casein with ammonium sulfate. Without added ammonium sulfate, the LasR-mutant frequency in all five experiments increased rapidly and four of five populations collapsed (Figure 1C). This experiment is consistent with the results in Figure 1A and our previous report (Dandekar et al., 2012) that the presence of ammonium sulfate significantly reduced the fitness of LasR mutants in competition with the w.t.
The Relative Fitness of LasR Mutants Is Dependent on the Concentration of Ammonium Sulfate
We next assessed the impact of ammonium sulfate on the cheater–cooperator equilibrium by performing 24 h competitions between the w.t. and LasR mutants. We started these competitions at initial ratios of 1:1 with variable amounts of added ammonium sulfate. After 24 h in ammonium sulfate-free PM-casein the frequency of cheaters rose to 79 ± 8%. At the maximum concentration of ammonium sulfate (4 g l-1), the frequency of cheaters decreased (albeit slightly) to 44 ± 11%. As the concentration of ammonium sulfate rose, the frequency of cheaters decreased (R2 = 0.55, p = 0.0014, Figure 1D).
Ammonium Sulfate Enhances w.t. RhlR Activity in the Presence of LasR Mutant
Our observation that ammonium sulfate favored the w.t. (cooperators) over LasR mutants (cheaters) suggested that ammonium sulfate might influence the LasIR system, the RhlIR system, or both. As discussed above, both systems have a role in cheater–cooperator dynamics: the LasIR system regulates the public good elastase (Passador et al., 1993; Park et al., 2014), and RhlR regulates production of cyanide and perhaps other factors as a policing mechanism (Smalley et al., 2015; Wang et al., 2015).
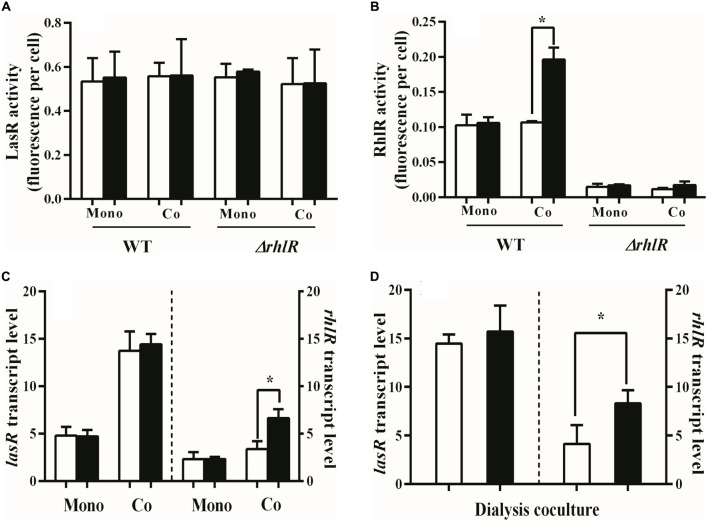
To test the hypothesis that ammonium sulfate enhances QS in the w.t., we used gfp fusion plasmids to monitor transcription from the lasI and rhlA promoters, as a proxy for LasR and RhlR activity (Feltner et al., 2016). Regulation of lasI and rhlA, unlike many QS-regulated genes, is specific to LasR and RhlR, respectively. Ammonium sulfate did not affect lasI or rhlA transcription in the w.t. when LasR mutants were not present (Figures 2A,B). Similar results were obtained with a RhlR-null mutant (Figure 2A). We reasoned that the w.t. might only increase QS in the presence of LasR mutants, as the presence of these cheaters increases the relative production of QS-regulated products by individual cooperators (Dandekar et al., 2012). Therefore, examined LasR and RhlR activity of the w.t. in the presence of 10% LasR mutants. We found that RhlR activity of the w.t. increased nearly twofold in the presence of LasR-mutant cheaters, but only in PM-casein with added ammonium sulfate (Figure 2B). The presence of LasR-mutant cheaters did not influence lasI transcription in the w.t.
FIGURE 2.
LasR and RhlR activities in w.t. P. aeruginosa. (A) GFP transcription in w.t. containing the lasI–gfp reporter plasmid pBS351. GFP is a proxy for LasR activity. Added ammonium sulfate does not alter the LasR activity of cooperators in w.t or a RhlR mutant, either in the presence or absence of cheaters. (B) GFP transcription in w.t. using the rhlA–gfp reporter plasmid pJF01. Addition of ammonium sulfate significantly increased the RhlR activity of cooperators in presence of cheaters. A RhlR mutant is shown as a reference. (C) Transcription of lasR and rhlR measured by qRT-PCR. Added ammonium sulfate significantly increased the transcript levels of rhlR but not lasR in the presence of cheaters. (D) Added ammonium sulfate significantly increased the expression of rhlR in the presence of LasR-mutant cheaters. The w.t. and LasR mutants were separated by a dialysis membrane. “Mono”: monoculture of w.t. or the RhlR mutant. “Co”: co-culture with 9:1 mixture of w.t and LasR mutants or a mixture of RhlR mutants and LasR mutants. We used proC as a reference (housekeeping) gene. White bars indicate PM-casein without ammonium sulfate and black bars indicate PM-casein with ammonium sulfate. Experiments were performed in triplicate. Data reported as the mean ± SD. ∗ indicates p < 0.05 by paired Student’s t-tests.
The reporter experiments suggested that exogenous ammonium sulfate promotes RhlR, but not LasR, activity, only in presence of LasR-mutant cheaters. To test this idea further, we employed RT-qPCR to measure lasR and rhlR transcript levels in PM-casein with or without added ammonium sulfate, with and without LasR mutants. The presence of LasR mutants resulted in increased lasR transcription in the w.t., consistent with the previous observation of increased public-goods production on a per-cooperator basis (Dandekar et al., 2012); however, lasR transcript levels were unaffected by the presence of ammonium sulfate in PM-casein (Figure 2C). The expression of rhlR was induced by the presence of LasR-mutant cheaters, but only in the presence of added ammonium sulfate (Figure 2C). Finally, to exclude the possibility, however unlikely, that the w.t. can physically detect LasR mutants and activate policing in the presence of ammonium sulfate, we isolated the w.t. and LasR mutants using a dialysis membrane. We found the same dependence on ammonium sulfate (Figure 2D). Together, these experiments suggested that the presence or absence of exogenous ammonium sulfate was a cue for activation of the policing mechanism.
Ammonium Sulfate Promotes Cyanide Production by Cooperators to Police Cheaters
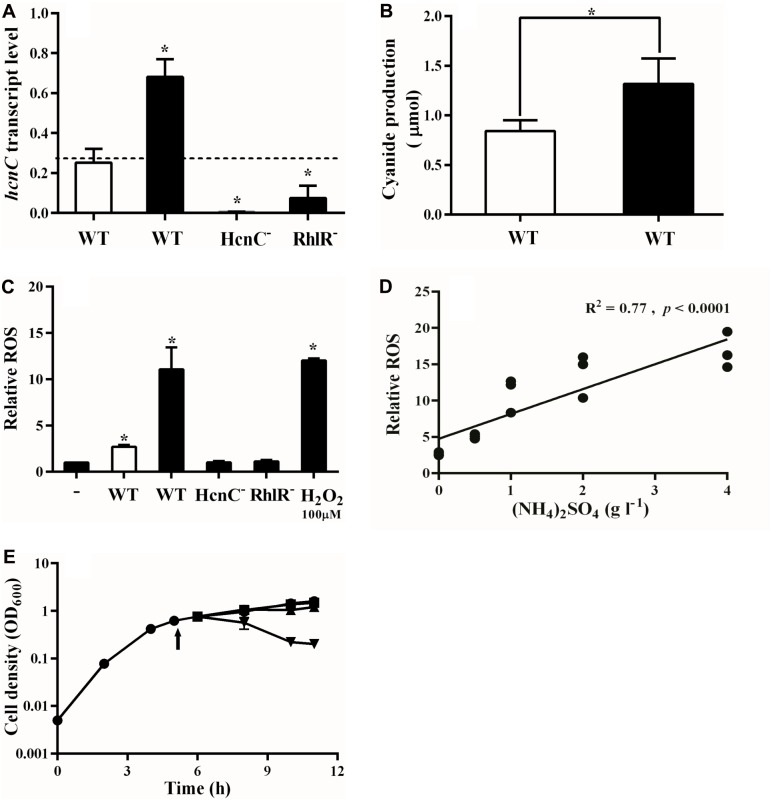
In P. aeruginosa, cooperators produce cyanide to police social cheaters, which results in a population equilibrium between cooperators and cheaters in the conditions of our experiments (Wang et al., 2015). Based on the results above, we reasoned that ammonium sulfate might alter the amount of RhlR-dependent cyanide produced. To test this possibility, we measured expression of the HCN synthase gene hcnC in mixed populations of cooperators and cheaters in PM-casein, with or without ammonium sulfate and found that added ammonium sulfate increased w.t. hcnC expression by a factor of about 2 (Figure 3A). As expected, hcnC transcription was minimal in the RhlR mutant and undetectable in an HcnC deletion mutant (Figure 3A). We also measured cyanide levels in both conditions and found increased levels of PM-casein with ammonium sulfate (Figure 3B).
FIGURE 3.
Ammonia promotes cyanide and ROS production by cooperators. (A) hcnC transcription in w.t. increases in presence of LasR-mutant cheaters in PM-casein with ammonium. hcnC expression from either a RhlR or HcnC deletion mutant in co-culture with the LasR mutant is shown as controls. proC was used as reference gene. In panels A–C, white bars indicate media without ammonium sulfate and black bars indicate that ammonium sulfate was added. (B) Cyanide production by cooperators in PM-casein with or without ammonium sulfate. (C) ROS production from LasR mutants in LB-MOPS, combined with supernatants of strains grown in PM-casein media. “-” indicates no cells. (D) ROS production by the w.t. increases as the concentration of ammonium sulfate is increased. (E) Growth inhibition of the LasR mutant by supernatant from w.t.:LasR-mutant co-culture from PM-casein with ammonia media (inverted triangle). The supernatant from a w.t.:LasR-mutant co-culture in PM-casein media (triangle) did not inhibit the growth of LasR mutants; neither did supernatant from a HcnC-mutant: LasR-mutant co-culture (square). The arrow indicates the time of supernatant addition. Experiments were performed in triplicate. ∗ indicates p < 0.05 by paired Student’s t-tests (panels A–C and E). A linear regression analysis was performed for the data presented in panel D.
Cyanide inhibits cytochrome c oxidase in susceptible cells (Bernier et al., 2016), resulting in the production of reactive oxygen species (ROS) (Gallagher and Manoil, 2001; Hazan et al., 2016). We next asked if the cyanide-generated ROS inhibited the growth of cheaters (Garcia-Contreras et al., 2015). To do so, we analyzed effects of the supernatant from co-cultures of the w.t. and LasR mutant on the growth of LasR mutants. The ROS generated under these conditions paralleled the hcnC expression data (Figure 3C). The ROS generated by treatment of LasR mutants with supernatant from co-cultures in PM-casein with ammonium sulfate was nearly equal to that induced by addition of 100 μM of H2O2 in LB. Similar to hcnC expression, supernatant from co-cultures in PM-casein without ammonium sulfate only induced one-third of the ROS as those from where ammonium sulfate was added (Figure 3C). Is production of ROS by supernatant related to the concentration of ammonium sulfate in the medium? As the concentration of ammonium sulfate rose, so did ROS production, with a positive correlation (R2 = 0.77, p < 0.0001, Figure 3D).
As a final test of this hypothesis, we asked if co-culture supernatants could inhibit the growth of LasR-mutant cheaters. LasR-null mutants could not grow well in the supernatant from a co-culture grown in PM-casein with ammonium sulfate (Figure 3E), but the supernatant from a co-culture without ammonium sulfate did not inhibit growth of the LasR mutants. LasR-mutant cheaters also grew well in the supernatant from co-culture in which the cooperator was an HcnC mutant, in PM-casein with ammonium sulfate. From these data, we concluded that exogenous ammonium sulfate promotes cyanide production and enhances policing in a concentration-dependent manner. As such, populations of P. aeruginosa in media containing exogenous ammonium sulfate are more stable than those in PM-casein without ammonium sulfate.
Ammonium Sulfate Increases the Transformation of Glycine to Hydrogen Cyanide
In P. aeruginosa, the main pathway to production of HCN is by using HCN synthase to catabolize glycine (Castric, 1977; Lenney and Gilchrist, 2011; Nandi et al., 2017). Glycine alternately can be converted to ammonia and 5,10-methylenetetrahydrofolate (5,10-methylene-THF) by the glycine cleavage system (Campbell, 1955; Toone et al., 2002). In P. aeruginosa, the glycine dehydrogenase is encoded by gcvP1 and gcvP2 (Kai et al., 2009; Zaara et al., 2016). The favored path of glycine catabolism might be determined through the abundance of substrates (van Niel et al., 2003; Neerincx et al., 2015). We hypothesized that the presence of ammonium sulfate in PM-casein medium inhibited the glycine cleavage pathway, thereby increasing HCN synthesis.
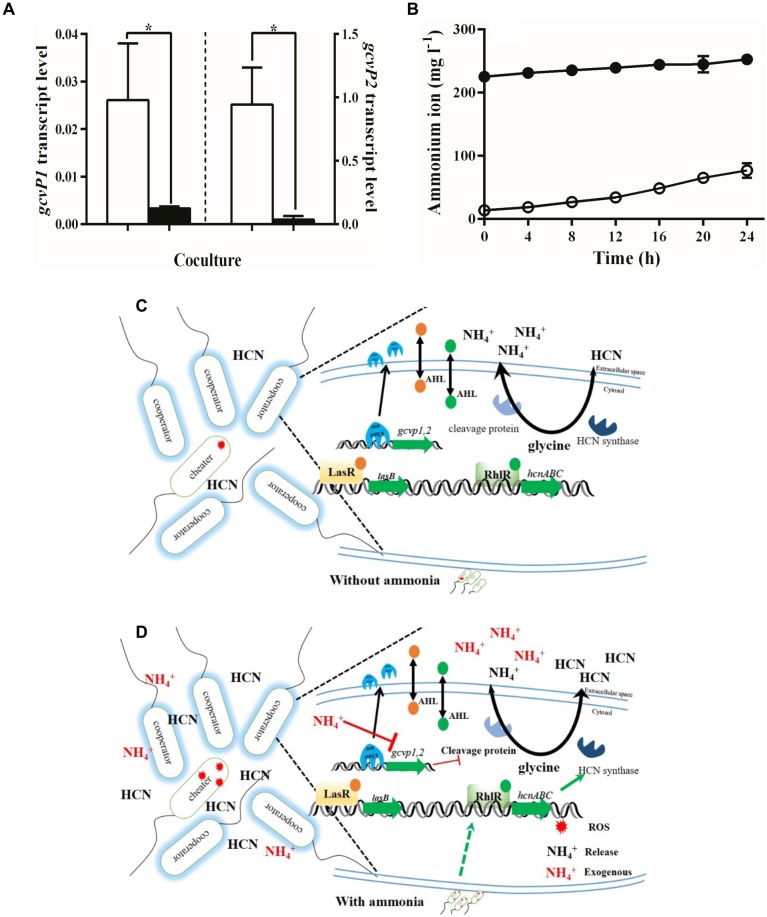
To test this possibility, we measured gcvP1 and gcvP2 transcription in the presence or absence of ammonium sulfate. We found that added ammonium sulfate decreased gcvP1 transcription by w.t. PAO1 in the presence of 10% cheaters. gcvP1 transcription in PM-casein with ammonium sulfate was only ∼12% of that in PM-casein without ammonium sulfate (Figure 4A). A similar reduction was seen in gcvP2 expression (Figure 4A). Under both conditions, gcvP2 expression was higher than gcvP1, consistent with evidence that gcvP2 is LasR-regulated (Schuster et al., 2003). However, the reduction in expression of gcvP2 in the absence of exogenous ammonium sulfate mitigates the QS-enhanced transcription of this gene. Combined with the hcnC expression and the cyanide production and relative ROS data above (Figures 3A–C), these results suggested that exogenous ammonium sulfate inhibits the glycine cleavage pathway, thereby favoring HCN synthesis.
FIGURE 4.
Exogenous ammonia inhibits the transformation from glycine to 5,10-methylene-THF. (A) Expression of gcvP1 and gcvP2 by the w.t. in a co-culture with 10% LasR mutants. Hollow bars indicate PM-casein without ammonium sulfate and black bars indicate PM-casein with ammonium sulfate. proC was used as a housekeeping gene. (B) PAO1 generates ammonia faster in the absence of ammonium sulfate (hollow circles) than in its presence (black circles). (C) Model of the flux between exogenous ammonia and cyanide production in P. aeruginosa. In panel C, red stars indicate ROS. The red NH4+ indicates exogenous ammonia in media, and black text indicates released ammonia by metabolism. Experiments were performed in triplicate. ∗ indicates p < 0.05 by paired Student’s t-test (panel A) or one-way ANOVA (panel B).
An implication of this hypothesis is that more ammonia would be generated (from glycine) in PM-casein without ammonium sulfate. Therefore, we measured ammonium ion generation with and without added ammonium sulfate. In casein media with ammonium sulfate, little additional ammonia accumulated. However, in the absence of ammonium sulfate, the concentration of ammonium ion increased significantly (p = 0.0085, Figure 4B).
Discussion
Quorum sensing, a cell–cell communication system, allows groups of bacteria to regulate a variety of biological processes important for adaptation and survival (Waters and Bassler, 2005; Xavier et al., 2011; Viducic et al., 2017) in response to cell density. Some of these processes are cooperative and therefore potentially vulnerable to social cheating (Joelsson et al., 2007; Wilder et al., 2011). Several mechanisms have been described that maintain cooperation in bacterial populations by which either invasion or growth of cheaters was restricted (Xavier et al., 2011; Dandekar et al., 2012; Darch et al., 2012; Pollitt et al., 2014; Wang et al., 2015; Allen et al., 2016).
When P. aeruginosa PAO1 is grown on casein as the sole carbon and energy source, LasR-mutant social cheaters emerge in the population. In our previous work, we showed that the presence of ammonium sulfate in casein broth leads to a stable frequency of cooperators and cheaters, while cheaters are uncontrolled in the absence of ammonium sulfate, leading to a tragedy of the commons (Dandekar et al., 2012). We used this system to study how environmental conditions might affect the stability of QS. We showed that the presence of ammonium sulfate results in upregulation of HCN synthesis, with the effect of policing social cheaters and stabilizing QS.
In our experiments, cheaters and cooperators co-existed stably in PM-casein with ammonium sulfate added but not in ammonium sulfate-free media (Figures 1A–C). The frequency of cheaters at the equilibrium was inversely correlated with the concentration of ammonium sulfate (Figure 1D). These finding extend our prior observation (Dandekar et al., 2012) that the absence of ammonium sulfate in the medium increases the per-cell cost of QS in P. aeruginosa to show that there is a biological effect of the ammonium sulfate in the growth medium.
RhlR activity among cooperators was significantly higher in PM-casein with ammonium sulfate than without (Figures 2B–D). Similarly, RhlR-dependent HCN production was significantly increased when ammonium sulfate was present in the media (Figures 3A,B). ROS generated as a consequence of HCN production also significantly increased (Figures 3C,D). Together, these finding suggested enhanced policing (Wang et al., 2015) by P. aeruginosa cooperators in the presence of ammonium sulfate.
The enhancement of cyanide production by ammonium sulfate appeared to involve a biochemical tradeoff. In P. aeruginosa, glycine is the main substrate for formation of cyanide (Castric, 1977); however, glycine cleavage results in ammonium ion production (Goro et al., 2008). We reasoned that the presence of exogenous ammonium sulfate reduces the need for glycine transfer to ammonia. In keeping with this hypothesis, the expression of gcvP1 and gcvP2, genes whose products are involved in glycine cleavage, was inhibited in the presence of ammonium sulfate (Figures 4A,B). Conversely, HCN synthase gene expression was enhanced (Figures 3A,B). Thus, ammonium sulfate, through its modulation of the glycine cleavage pathway (Figure 4C), enhances policing in cooperating populations of P. aeruginosa.
There are other environmental conditions that have been reported to enhance HCN synthesis. For example, microaerophilic conditions and iron excess can have a stimulatory effect on cyanogenesis in P. aeruginosa (Blumer and Haas, 2000; Pessi and Haas, 2001). As HCN is an effector of policing in cooperating populations, these environmental conditions might also enhance the ability of populations to police themselves of cheaters.
Our experiments do not uncover the selective forces that gave rise to this mechanism of cheater control in the first place. It is notable that the mechanism does not require detection of cheaters per se but rather the abundance of ammonium ion in the growth medium. Although the presence of ammonium sulfate in our experiments affects the production of HCN by P. aeruginosa (Figures 3, 4), it is most likely that this phenomenon simply reflects metabolic needs of the bacterium. However, it is possible that regulation of the HCN synthase by the QS regulator RhlR had the effect of selecting against LasR mutants in the population, providing stability to QS. Several studies have shown that cooperative behaviors (including QS) are stabilized by pleiotropy (Foster et al., 2004; Dandekar et al., 2012; Mitri and Foster, 2016); our results provide further support for this idea.
Our observation of ammonia-dependent elaboration of a QS-regulated product echoes the idea of “metabolic prudence” (Xavier et al., 2011). Part of the idea of metabolic prudence is that production of public goods should be deferred until the resources necessary for their production are abundant in the environment, thus minimizing the benefit of exploitation by cheaters. The production of rhamnolipid in P. aeruginosa occurs in this kind of prudent manner (Xavier et al., 2011). HCN production only when ammonia (the alternate fate of glycine) is abundant is similarly “prudent” for cooperators. However, in this case, the reduction in production of QS-regulated products, while possibly beneficial for cooperators in the short term, is ultimately deleterious for the population, and the production of HCN is lowest in the conditions where it is most needed (Figure 3). Although the cooperators may gain a short-term metabolic advantage when deprived of an exogenous nitrogen source, the population as a whole is prone to destabilization, ultimately resulting in the loss of cooperation. Further investigation into this mechanism might reveal methods to induce policing within or promote destabilization of a bacterial population, with ramifications for synthetic biological and clinical applications.
Author Contributions
HY, MW, AD, and DS designed the research. HY and FS performed the research. HY, MW, AD, and NL analyzed the data. HY, MW, and AD wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank E. Peter Greenberg for reviewing the manuscript.
Footnotes
Funding. This study was supported by the National Natural Science Foundation of China (Grant Nos. 31570490 and 51478432) and the Natural Science Foundation of Zhejiang Province (Grant No. LY15C010001).
References
- Allen B., Nowak M. A., Wilson E. O. (2013). Limitations of inclusive fitness. Proc. Natl. Acad. Sci. U.S.A. 110 20135–20139. 10.1073/pnas.1317588110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen R. C., Mcnally L., Popat R., Brown S. P. (2016). Quorum sensing protects bacterial co-operation from exploitation by cheats. ISME J. 10 1706–1716. 10.1038/ismej.2015.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier S. P., Workentine M. L., Li X., Magarvey N. A., O’Toole G. A., Surette M. G. (2016). Cyanide toxicity to Burkholderia cenocepacia is modulated by polymicrobial communities and environmental factors. Front. Microbiol. 7:725. 10.3389/fmicb.2016.00725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer C., Haas D. (2000). Iron regulation of the hcnABC genes encoding hydrogen cyanide synthase depends on the anaerobic regulator ANR rather than on the global activator GacA in Pseudomonas fluorescens CHA0. Microbiology 146 2417–2424. 10.1099/00221287-146-10-2417 [DOI] [PubMed] [Google Scholar]
- Campbell L. L. (1955). The oxidative degradation of glycine by a Pseudomonas. J. Biol. Chem. 217 669–674. [PubMed] [Google Scholar]
- Castric P. A. (1977). Glycine metabolism by Pseudomonas aeruginosa: hydrogen cyanide biosynthesis. J. Bacteriol. 130 826–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K. H., Kumar A., Schweizer H. P. (2006). A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64 391–397. 10.1016/j.mimet.2005.06.001 [DOI] [PubMed] [Google Scholar]
- Dandekar A. A., Chugani S., Greenberg E. P. (2012). Bacterial quorum sensing and metabolic incentives to cooperate. Science 338 264–266. 10.1126/science.1227289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darch S. E., West S. A., Winzer K., Diggle S. P. (2012). Density-dependent fitness benefits in quorum-sensing bacterial populations. Proc. Natl. Acad. Sci. U.S.A. 109 8259–8263. 10.1073/pnas.1118131109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi K. K., Deepika S., Bhaduri A., Kothamasi D. (2013). Polymorphism in hcnAB gene in Pseudomonas species reveals ecologically distinct hydrogen cyanide-producing populations. Geomicrobiol. J. 30 131–139. 10.1080/01490451.2011.654377 [DOI] [Google Scholar]
- Diggle S. P., Griffin A. S., Campbell G. S., West S. A. (2007). Cooperation and conflict in quorum-sensing bacterial populations. Nature 450 411–414. 10.1038/nature06279 [DOI] [PubMed] [Google Scholar]
- Federation and American Public Health Association [APHA] (2005). Standard Methods for the Examination of Water and Wastewater. Washington, DC: American Public Health Association (APHA). [Google Scholar]
- Feltner J. B., Wolter D. J., Pope C. E., Groleau M. C., Smalley N. E., Greenberg E. P., et al. (2016). LasR variant cystic fibrosis isolates reveal an adaptable quorum-sensing hierarchy in Pseudomonas aeruginosa. mBio 7:e01513-16. 10.1128/mBio.01513-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey A., Eichler S., Flavier S., Christen R., Höfle M. G., Guzmán C. A. (2004). Establishment of a real-time PCR-based approach for accurate quantification of bacterial RNA targets in water, using Salmonella as a model organism. Appl. Environ. Microbiol. 70 3618–3623. 10.1128/AEM.70.6.3618-3623.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster K. R., Shaulsky G., Strassmann J. E., Queller D. C., Thompson C. R. L. (2004). Pleiotropy as a mechanism to stabilize cooperation. Nature 431 693–696. 10.1038/nature02894 [DOI] [PubMed] [Google Scholar]
- Gallagher L. A., Manoil C. (2001). Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183 6207–6214. 10.1128/JB.183.21.6207-6214.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Contreras R., Nunez-Lopez L., Jasso-Chavez R., Kwan B. W., Belmont J. A., Rangel-Vega A., et al. (2015). Quorum sensing enhancement of the stress response promotes resistance to quorum quenching and prevents social cheating. ISME J. 9 115–125. 10.1038/ismej.2014.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert K. B., Kim T. H., Gupta R., Greenberg E. P., Schuster M. (2009). Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol. Microbiol. 73 1072–1085. 10.1111/j.1365-2958.2009.06832.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goro K., Yutaro M., Tadashi Y., Koichi H. (2008). Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 84 246–263. 10.2183/pjab.84.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan R., Que Y. A., Maura D., Strobel B., Majcherczyk P. A., Hopper L. R., et al. (2016). Auto poisoning of the respiratory chain by a quorum-sensing-regulated molecule favors biofilm formation and antibiotic tolerance. Curr. Biol. 26 195–206. 10.1016/j.cub.2015.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joelsson A., Kan B., Zhu J. (2007). Quorum sensing enhances the stress response in Vibrio cholerae. Appl. Environ. Microbiol. 73 3742–3746. 10.1128/AEM.02804-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai T., Tateda K., Kimura S., Ishii Y., Ito H., Yoshida H., et al. (2009). A low concentration of azithromycin inhibits the mRNA expression of N-acyl homoserine lactone synthesis enzymes, upstream of lasI or rhlI, in Pseudomonas aeruginosa. Pulm. Pharmacol. Ther. 22 483–486. 10.1016/j.pupt.2009.04.004 [DOI] [PubMed] [Google Scholar]
- Kim M. K., Harwood C. S. (1991). Regulation of benzoate-CoA ligase in Rhodopseudomonas palustris. FEMS Microbiol. Lett. 83 199–203. 10.1111/j.1574-6968.1991.tb04440.x-i1 [DOI] [Google Scholar]
- Kim T. S., Ham S. Y., Park B. B., Byun Y., Park H. D. (2017). Lauroyl arginate ethyl blocks the iron signals necessary for Pseudomonas aeruginosa biofilm development. Front. Microbiol. 8:970. 10.3389/fmicb.2017.00970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenney W., Gilchrist F. J. (2011). Pseudomonas aeruginosa and cyanide production. Eur. Respir. J. 37 482–483. 10.1183/09031936.00122810 [DOI] [PubMed] [Google Scholar]
- Miller W. G., Leveau J. H., Lindow S. E. (2000). Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant Microbe Interact. 13 1243–1250. 10.1094/MPMI.2000.13.11.1243 [DOI] [PubMed] [Google Scholar]
- Mitri S., Foster K. R. (2016). Pleiotropy and the low cost of individual traits promote cooperation. Evolution 70 488–494. 10.1111/evo.12851 [DOI] [PubMed] [Google Scholar]
- Nandi M., Selin C., Brawerman G., Fernando W. G. D., Kievit T. D. (2017). Hydrogen cyanide, which contributes to Pseudomonas chlororaphis strain PA23 biocontrol, is upregulated in the presence of glycine. Biol. Control 108 47–54. 10.1016/j.biocontrol.2017.02.008 [DOI] [Google Scholar]
- Neerincx A. H., Mandon J., Van Ingen J., Arslanov D. D., Mouton J. W., Harren F. J., et al. (2015). Real-time monitoring of hydrogen cyanide (HCN) and ammonia (NH3) emitted by Pseudomonas aeruginosa. J. Breath Res. 9:027102. 10.1088/1752-7155/9/2/027102 [DOI] [PubMed] [Google Scholar]
- Park S. J., Kim S. K., So Y. I., Park H. Y., Li X. H., Yeom D. H., et al. (2014). Protease IV, a quorum sensing-dependent protease of Pseudomonas aeruginosa modulates insect innate immunity. Mol. Microbiol. 94 1298–1314. 10.1111/mmi.12830 [DOI] [PubMed] [Google Scholar]
- Passador L., Cook J. M., Gambello M. J., Rust L., Iglewski B. H. (1993). Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260 1127–1130. 10.1126/science.8493556 [DOI] [PubMed] [Google Scholar]
- Pessi G., Haas D. (2001). Dual control of hydrogen cyanide biosynthesis by the global activator GacA in Pseudomonas aeruginosa PAO1. FEMS Microbiol. Lett. 200 73–78. 10.1111/j.1574-6968.2001.tb10695.x [DOI] [PubMed] [Google Scholar]
- Pollitt E. J. G., West S. A., Crusz S. A., Burtonchellew M. N., Diggle S. P. (2014). Cooperation, quorum sensing, and evolution of virulence in Staphylococcus aureus. Infect. Immun. 82 1045–1051. 10.1128/IAI.01216-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual. New York, NY: Cold Spring Harbor Press. [Google Scholar]
- Sandoz K. M., Mitzimberg S. M., Schuster M. (2007). Social cheating in Pseudomonas aeruginosa quorum sensing. Proc. Natl. Acad. Sci. U.S.A. 104 15876–15881. 10.1073/pnas.0705653104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Lostroh C. P., Ogi T., Greenberg E. P. (2003). Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185 2066–2079. 10.1128/JB.185.7.2066-2079.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley N. E., An D., Parsek M. R., Chandler J. R., Dandekar A. A. (2015). Quorum sensing protects Pseudomonas aeruginosa against cheating by other species in a laboratory coculture model. J. Bacteriol. 197 3154–3159. 10.1128/JB.00482-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens A. M., Schuster M., Rumbaugh K. P. (2012). Working together for the common good: cell-cell communication in bacteria. J. Bacteriol. 194 2131–2141. 10.1128/JB.00143-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover C. K., Pham X. Q., Erwin A. L., Mizoguchi S. D., Warrener P., Hickey M. J., et al. (2000). Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406 959–964. 10.1038/35023079 [DOI] [PubMed] [Google Scholar]
- Toone J. R., Applegarth D. A., Kure S., Coulter-Mackie M. B., Sazegar P., Kojima K., et al. (2002). Novel mutations in the P-protein (glycine decarboxylase) gene in patients with glycine encephalopathy (non-ketotic hyperglycinemia). Mol. Genet. Metab. 76 243–249. 10.1016/S1096-7192(02)00041-0 [DOI] [PubMed] [Google Scholar]
- van Niel E. W., Claassen P. A., Stams A. J. (2003). Substrate and product inhibition of hydrogen production by the extreme thermophile, Caldicellulosiruptor saccharolyticus. Biotechnol. Bioeng. 81 255–262. 10.1002/bit.10463 [DOI] [PubMed] [Google Scholar]
- Viducic D., Murakami K., Amoh T., Ono T., Miyake Y. (2017). Role of the interplay between quorum sensing regulator VqsR and the Pseudomonas quinolone signal in mediating carbapenem tolerance in Pseudomonas aeruginosa. Res. Microbiol. 168 450–460. 10.1016/j.resmic.2017.02.007 [DOI] [PubMed] [Google Scholar]
- Wang M. Z., Schaefer A. L., Dandekar A. A., Greenberg E. P. (2015). Quorum sensing and policing of Pseudomonas aeruginosa social cheaters. Proc. Natl. Acad. Sci. U.S.A. 112 2187–2191. 10.1073/pnas.1500704112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters C. M., Bassler B. L. (2005). Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21 319–346. 10.1146/annurev.cellbio.21.012704.131001 [DOI] [PubMed] [Google Scholar]
- West S. A., Griffin A. S., Gardner A. (2007). Evolutionary explanations for cooperation. Curr. Biol. 17 R661–R672. 10.1016/j.cub.2007.06.004 [DOI] [PubMed] [Google Scholar]
- Wilder C. N., Diggle S. P., Schuster M. (2011). Cooperation and cheating in Pseudomonas aeruginosa: the roles of the las, rhl and pqs quorum-sensing systems. ISME J. 5 1332–1343. 10.1038/ismej.2011.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier J. B., Kim W., Foster K. R. (2011). A molecular mechanism that stabilizes cooperative secretions in Pseudomonas aeruginosa. Mol. Microbiol. 79 166–179. 10.1111/j.1365-2958.2010.07436.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaara S., Lundgren B. R., Grassa M. T., Wang M. X., Megan G., Moffat J. F., et al. (2016). Erratum for Sarwar et al., GcsR, a TyrR-like enhancer-binding protein, regulates expression of the glycine cleavage system in Pseudomonas aeruginosa PAO1. mSphere 1:e00200-16. 10.1128/mSphere.00020-16 [DOI] [PMC free article] [PubMed] [Google Scholar]