Abstract
After intra-arterial delivery in the dystrophic dog, allogeneic muscle-derived stem cells, termed MuStem cells, contribute to long-term stabilization of the clinical status and preservation of the muscle regenerative process. However, it remains unknown whether the human counterpart could be identified, considering recent demonstrations of divergent features between species for several somatic stem cells. Here, we report that MuStem cells reside in human skeletal muscle and display a long-term ability to proliferate, allowing generation of a clinically relevant amount of cells. Cultured human MuStem (hMuStem) cells do not express hematopoietic, endothelial, or myo-endothelial cell markers and reproducibly correspond to a population of early myogenic-committed progenitors with a perivascular/mesenchymal phenotypic signature, revealing a blood vessel wall origin. Importantly, they exhibit both myogenesis in vitro and skeletal muscle regeneration after intramuscular delivery into immunodeficient host mice. Together, our findings provide new insights supporting the notion that hMuStem cells could represent an interesting therapeutic candidate for dystrophic patients.
Keywords: human adult stem cells, MuStem, DMD, skeletal muscle, cell therapy, regenerative medicine
Rouger et al. show that stem cells termed MuStem reside in healthy human muscle. Following delivery into injured mouse muscle, these early myogenic-committed cells, characterized by a perivascular/mesenchymal signature, give rise to efficient myofiber regeneration, the intensity of which correlates with CD56+ expression. These findings suggest new therapeutic options for muscular dystrophies.
Introduction
Duchenne muscular dystrophy (DMD) is an X-linked recessive muscle disorder that represents the most common form of muscular dystrophy, affecting about one in 3,500–5,500 male births.1, 2 It is caused by mutations in the dystrophin gene, which give rise to the protein lack, resulting in myofiber degeneration, followed by severe fibrosis.3 This leads to progressive muscle weakness and premature death near 30 years of age.4, 5 Currently, there is no effective treatment, despite the development of pharmacological strategies, molecular-based ones (such as viral-based transfer of short form of dystrophin or oligonucleotide-induced exon-skipping), and cell therapy.6
Concerning the last one, intramuscular (IM) injections of murine7, 8 or human9, 10 myoblasts generated encouraging results, with demonstration of cell fusion and dystrophin restoration in the murine DMD model, i.e., the mdx mice. The first clinical studies, however, produced very limited successes, failing to deliver significant levels of dystrophin and to demonstrate clinical benefit.11, 12 Later, specific conditions of cell delivery and immunosuppression, corresponding to a “high-density injection” protocol and the use of tacrolimus, were defined in suitable animal models13 to adequately take into account the acute immune rejection,14 poor survival,15, 16 and low migration17 of injected cells advanced to explain the disappointing initial results. Phase IA clinical trials designed with these appropriate conditions in DMD patients unequivocally demonstrated a significant increase of the engraftment efficiency, with up to 34.5% myofibers expressing donor-derived dystrophin at the injection sites for a long period.18, 19, 20, 21 Although myoblast transplantation could be an elective treatment for small and accessible muscles, it seemed quite inappropriate to treat numerous large ones, considering the migration of myoblasts and the potential invasiveness of the injection protocol, which prompted the search for alternative cell types.
Over the past 15 years, several cell types distinct from satellite cells (SCs) have been described as exhibiting myogenic fate after engraftment into damaged or diseased muscle (Table S1).22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99 After IM or intra-arterial (IA) injection in scid/mdx mice, blood- and muscle-derived CD133+ cells were able to participate in muscle regeneration and colonize the SC niche.30, 32 Such cells were even more effective than myoblasts when injected intramuscularly in Rag−/−/γC−/−/C5−/−mice.35 Successively, intravenous (i.v.) delivery in lethally irradiated mdx mice of total bone marrow cells or a Hoeschst 33342-stained subpopulation of bone marrow cells called side population cells resulted in cell integration into skeletal muscle and formation of up to 4% dystrophin+ myofibers.39, 40 Human mesoangioblasts (Mabs)/pericyte-derived cells crossed the vessel barrier following IA injection in scid/mdx mice and colonized host muscle, where they generated numerous dystrophin+ myofibers and replenished the SC pool.57 In addition, IA delivery of wild-type canine Mabs resulted in muscle homing, dystrophin expression recovery, and improvement of muscle function as well motility in Golden Retriever muscular dystrophy (GRMD) dogs that represent the clinically relevant DMD model.56 Following IM or IA injections in mdx mice, murine muscle-derived stem cells (MDSCs) (preplated cells that adhered between 96 and 168 hr) exhibited an improved ability to restore dystrophin expression compared to myoblasts.67 Similarly, human adipose-derived stem cells (ADSCs) reached skeletal muscle, engrafted, and expressed dystrophin after local or systemic delivery in mdx mice or GRMD dogs.81, 86 IM injection of myogenic endothelial cells in scid/mdx mice was shown to give rise to efficient myofiber regeneration and dystrophin restoration.91 Finally, PW1+ interstitial cells (PICs) were shown to generate new myofibers, SCs, and PICs following engraftment into damaged muscle.100 Together, these compelling results have opened up novel therapeutic opportunities for muscular dystrophies to face the limited efficacy of myoblast transplantation.
However, several major obstacles have hindered the development of analogous approaches in clinically relevant models or clinical trials. Analysis of muscle biopsies from a DMD patient who received bone marrow transplantation 13 years before for X-linked severe combined immune deficiency revealed a very limited ability of donor cells to integrate myofibers and produce dystrophin.42 Also, wild-type bone marrow cell transplantation did not restore dystrophin expression or improve muscle function in GRMD dogs.44 Following umbilical cord blood cell transplantation done in a DMD patient to treat chronic granulomatous disease, neither donor cell engraftment nor dystrophin expression was observed.47 A modest regenerative index was observed after IM injection of human MDSCs (preplated cells that adhered between 48 and 120 hr) in scid/mdx mice.70 In addition, a lack of demonstration of effective integration into myofibers was determined after IM injection of muscle-derived CD133+ cells in DMD patients, which nevertheless can largely be due to the fact that the graft was autologous, making the location of cells and poor number of injected cells difficult.33 Importantly, a phase I/IIa clinical trial consisting of multiple IA infusions of human leukocyte antigen (HLA)-matched donor Mabs/pericytes into immunosuppressed DMD patients pointed on a very low level of donor cell engraftment, with no or only a few dystrophin+ myofibers as well as a lack of any functional improvement.63
In 2011, we isolated MDSCs as preplated cells that adhered between 120 and 192 hr (we named them cMuStem cells) from healthy dogs and established that their systemic delivery into immunosuppressed GRMD dogs was associated with a striking and persistent clinical stabilization.96 Concomitantly, a positive histological impact was determined with long-term and diffuse dystrophin expression, increased regeneration activity characterized by a persistent presence of developmental myosin+ myofibers, and reduced endomysial space. In parallel to modifications in both lipid homeostasis and energy metabolism processes, an enhancement of the ubiquitin-dependent protein degradation pathway, the structural interactions between myofibers and the extracellular matrix, and the oxidative stress response was also shown though transcriptomic and proteomic studies.97, 98, 99 These preclinical data positioned MuStem cells as a possible attractive therapeutic avenue for DMD patients. Here, we report for the first time the isolation of human MuStem cells (hMuStem cells) based on an adaptation of the protocol initially developed for canine muscle. In addition, we provide an extensive analysis of their phenotype as well as in vitro/in vivo behavior. Overall, we demonstrate in vitro myogenesis and a muscle regenerative potential of hMuStem cells, reinforcing the fact that they could be presented as a therapeutic option in view of clinical application.
Results
hMuStem Cells Reside in Adult Skeletal Muscle and Exhibit a High Proliferation Rate
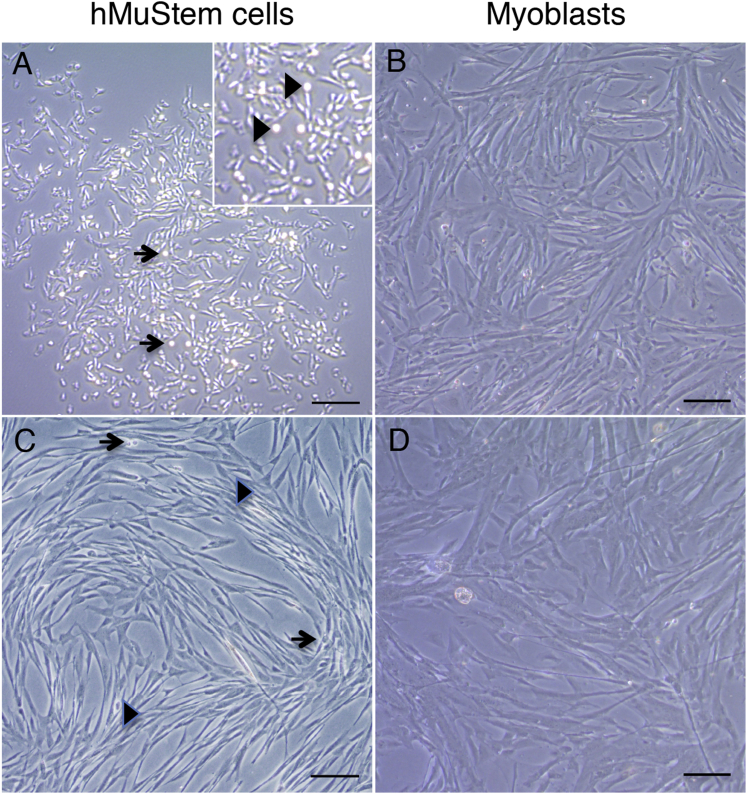
MuStem cells were initially isolated from a pool of posterior limb and postural muscles of 2.5-month-old healthy dogs.96 To analyze whether a similar muscle-resident cell subset could be identified in humans, we performed experiments using a modified version of our original protocol, as shown in Figure S1. We isolated hMuStem cells from small biopsies of postural (pP; Longissimus dorsi) muscle or locomotor (pL; Gastrocnemius, Fascia lata tensor, and Vastus lateralis) muscles (n = 3 each) collected from 15- to 51-year-old male (n = 4) or female (n = 2) subjects free from a known muscle disease. 5 days after the first plating, floating cells, corresponding to 0.9 × 105 to 1.8 × 105 cells per gram of muscle, were seeded on new gelatin-coated plastic to finally obtain a marginal fraction of poorly adherent cells, corresponding to hMuStem cells 3 days later. These cells remained as small round cells for the following 7 days independently of the source of the tissue samples from which they were collected. After that, they started to proliferate as pseudo-clonal cultures composed of poorly adherent cells (Figure 1A), some of which clearly remaining in the supernatant as refractile round cells (Figure 1A, arrows). Although most of the thin fusiform cells divided to generate spindle-shaped cells, pairs of round cells were regularly observed during the first step of proliferation (Figure 1A, inset with arrowhead), revealing their ability to divide as floating cells. In contrast, myoblasts that adhered between 24 and 96 hr after the initial plating rapidly formed lengthened and large spindle-shaped cells, for which cell outlines were sometimes difficult to define (Figure 1B). After two passages, hMuStem-cell-derived primary cultures were composed of some refractile round cells (Figure 1C, arrows) and a majority of thin spindle-shaped ones (Figure 1C, arrowhead). No spontaneous fusion event was observed despite high confluence of cultures and the presence of cell multilayers as well as alignments even after 1 month of culture (Figure 1C). In contrast, myoblasts generated cell monolayers composed of much larger and elongated cells as soon as they reached confluence (Figure 1D).
Figure 1.
Morphological Features of hMuStem Cells and Myoblasts
(A) 7 days after the end of the isolation protocol, phase contrast microscopy revealed that hMuStem cells formed a colony unit composed of round and thin cells (arrow) as well as short spindle-shaped cells. A part of the cells remained in the supernatant as floating cells, corresponding to small and highly refractile cells positioned above adherent cells (arrowhead in insert). (B) Typical lengthened and large spindle-shaped cells were observed in myoblast-derived primary culture. (C) hMuStem-cell-derived primary culture was characterized by a large majority of thin elongated cells aligned in networks (arrowhead) and a permanent presence of some refractile round cells (arrow). (D) Monolayer of spindle-shaped cells and thick multinucleated cells characterized the myoblast-derived primary cultures. Scale bars, 100 μm.
The in vitro expansion potential is an essential cell feature, especially when a therapeutic application based on whole body treatment is considered. hMuStem cells were able to generate a mean cumulative number of population doubling of 25.5 ± 3.5 over a period of 38 days, corresponding to a doubling time of 1.5 ± 0.2 days (n = 4; Figure S2). Interestingly, the rate of expansion was quite homogeneous, despite a distinct origin of the muscle, age, or gender of the donors. Also, it is in agreement with those previously described for cMuStem cells and other muscle-derived progenitor cells (see Discussion).
A clonogenic potential was also demonstrated in vitro for the four cell batches we tested, although it was limited because we obtained 1–14 clones (25–731 cells per clone) after 30 days from 300 initially seeded cells. Importantly, the presence of both spindle-shaped cells and round ones was detected in colonies, as described for the original primary cultures, which demonstrated atypical division modalities for the hMuStem cells.
hMuStem Cells Mainly Correspond to Early Myogenic-Committed Cells of Perivascular Origin
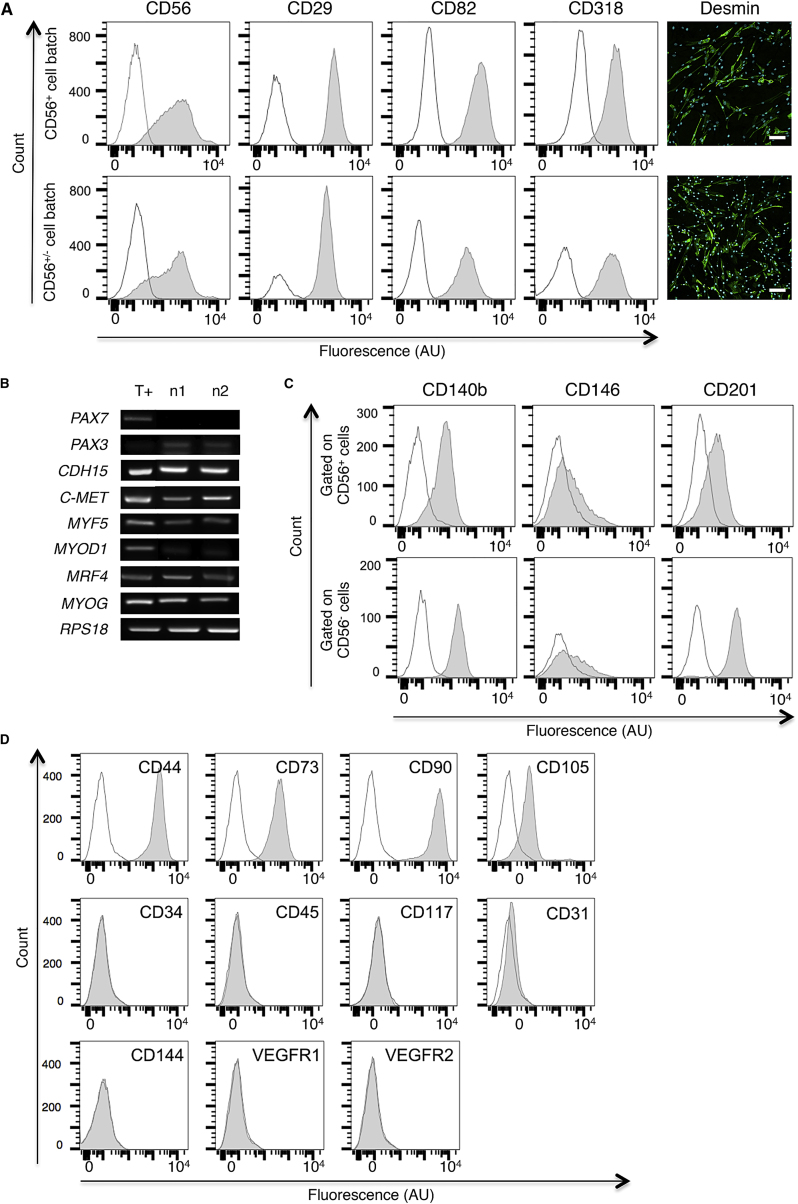
To characterize the hMuStem cell population, a large panel of lineage-specific markers was investigated using RT-PCR, flow cytometry, and immunocytochemistry analysis on cells at passage 5 (P5). First, 2 out of 4 cell batches contained more than 99% of cells positive for the SC and myoblast marker CD56 (NCAM) (CD56+ cell batch; representative profiles are shown in Figure 2A, top), whereas 69% and 73% of CD56+ cells were determined in the other cell batches (CD56+/− cell batch; Figure 2A, bottom). In contrast, CD29 (β1-integrin), recently shown to be expressed by human muscle stem cells/SCs,101, 102 was homogeneously detected in all hMuStem cell batches. The same homogeneous expression profiles were obtained for CD82, another newly identified marker of SCs103 and myogenic cells displaying high proliferation and differentiation rates104, and for CD318, also reported to be present in SCs.103 45%–93% of the cells were positive for the muscle-specific marker desmin. RT-PCR analysis showed a lack of the paired box transcription factor PAX7 mRNA in all hMuStem cell batches, whereas PAX3, M-cadherin (CDH15), and C-MET transcripts, corresponding to classical markers of SCs and myogenic cells, were systematically detected (Figure 2B). In addition, hMuStem cells expressed the myogenic regulatory factors (MRFs) MYF5, MYOD1, MRF4, and myogenin (MYOG) at the RNA level, which was implemented at the protein level by the detection of MYF5+ cells (range: 14%–40%), MYOD+ cells (less than 14%), and few myogenin+ ones (less than 5%).
Figure 2.
Cell Lineage-Specific Phenotype of hMuStem Cells
(A) Immunolabelings against CD56, CD29, CD82, CD318, and desmin were performed on hMuStem cells (P5) cultivated in growth medium. The upper line shows the profile of a representative cell batch (n1) made exclusively of CD56+ cells, whereas the lower line depicts a representative cell batch (n2) containing CD56+/− cells. Isotype control and specific signal are in white and gray, respectively. For labelings done on culture chamber slides, nuclei were counterstained with 10 μg/mL DAPI (blue). Scale bars, 100 μm. (B) Detection of transcripts specific to SC and myogenic cell markers on the two representative CD56+ (n1) and CD56+/− (n2) cell batches (P5). Myoblasts and RPS18 were used as positive control (T+) and housekeeping gene, respectively. (C) Representative flow cytometry images revealing the expression of perivascular (CD140b and CD146) and mesenchymal (CD201) markers in gated CD56+ and CD56− cells. Isotype control and specific signal are in white and gray, respectively. (D) Representative flow cytometry profiles revealing the homogeneous expression of MSC markers CD44, CD73, CD90, and CD105 by hMuStem cells and the lack of expression for the hematopoietic (CD34, CD45, and CD117) and endothelial (CD31, CD144, VEGFR1, and VEGFR2) markers. Control- and specific antibody-stained cells are shown in white and gray profiles, respectively.
Multi-labelings with CD56 and well-described perivascular cell markers evidenced that CD56+ cells contained at least 86% of CD140b+, whereas CD56− cells were all CD140b+ (Figure 2C). Both cell types exhibited similar CD146 expression patterns, with around 25% of CD146+ cells, and interestingly expressed the mesenchymal progenitor marker CD201, but with a higher intensity in the CD56+ cells. In return, the other typical mesenchymal stem cell (MSC) markers CD44, CD73, CD90, and CD105 were homogeneously observed in all hMuStem cells (Figure 2D). Lastly, hMuStem cells did not express the classical hematopoietic markers CD34, CD45, and CD117 and were consistently defined by a lin− phenotype based on the expression of CD4, CD8, CD19, CD33, and CD38 antigens (Figure S3A). In addition, they were uniformly negative for the expression of the endothelial markers CD31, CD144, vascular endothelial growth factor receptor 1 (VEGFR1), and VEGFR2 (Figure 2D). Interestingly, the same profiles were observed for CD15, CD133, and CD338, which are commonly used to identify myo-adipogenic cells, AC133+ cells, and side population ones (Figure S3B). Overall, we showed that the hMuStem cell population mainly corresponded to early myogenic-committed progenitors, with expression of several typical SC markers and a signature of perivascular mesenchymal cells evoking a pericyte origin.
hMuStem Cells Display an Oligopotent Status
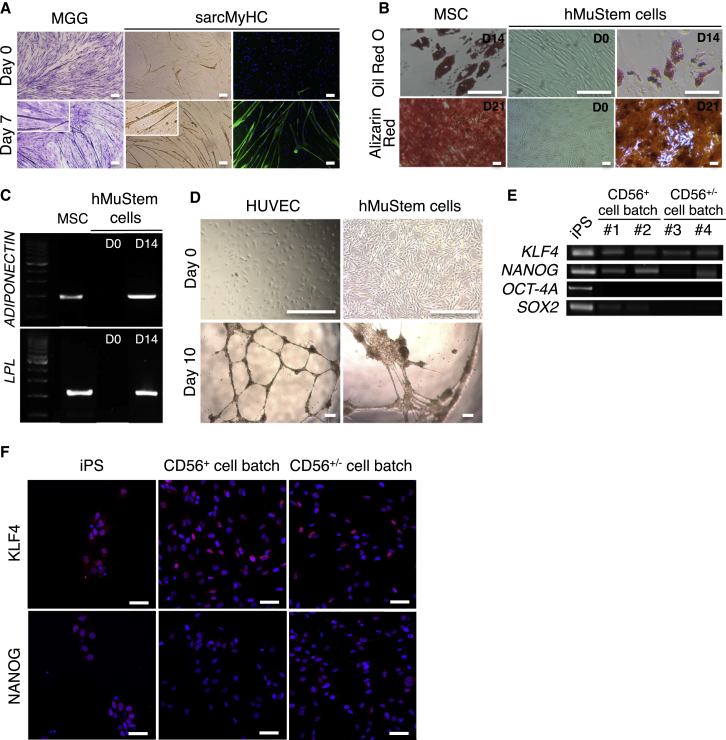
We next investigated the degree of plasticity of the hMuStem cells. First, the in vitro differentiation potential into mesodermal lineages of cells at P5 was examined. After 7 days in low serum “fusion promoting” medium, all hMuStem cell batches formed multinucleated myotubes expressing sarcomeric myosin heavy chain isoform (sarcMyHC) and displayed a fusion index (FI) of 28% ± 3.0%, revealing their ability to differentiate into the myogenic lineage (Figure 3A). Their differentiation into adipocytes was revealed with the cytoplasmic accumulation of small lipid vesicles in all hMuStem-cell-derived cultures 2 weeks after their switch in adipogenic induction media using oil red O staining (Figure 3B). Also, adiponectin and lipoprotein lipase (LPL) mRNAs were detected by RT-PCR (Figure 3C). Alizarin red staining indicated the formation of calcium deposits in all cultures when cultivated for 3 weeks in osteogenic induction media, revealing the ability of hMuStem cells to differentiate into osteogenic lineage (Figure 3B). In addition, expression of the osteogenic-specific gene, osteoprotegerin (OPG), was detected by RT-qPCR (mean cycle threshold [CT] of 26.4 ± 1.8). Finally, we showed that hMuStem cell populations cultivated in endothelial-specific medium and plated on Matrigel aligned and formed capillary-like structures (Figure 3D). In return, they did not express the endothelial markers CD31, CD144, and von Willebrand factor (VWF), demonstrating that they failed to commit into endothelial lineage (Figure S4).
Figure 3.
Multilineage Potential and Pluripotent Phenotype of Long-Term Cultured hMuStem Cells
(A) Cultured hMuStem cells were grown in low-serum medium for 7 days, then fixed and submitted to May-Grümwald Giemsa (MGG) staining and sarcMyHC immunolabeling to reveal multinuclei myotubes. hMuStem cells placed on growth medium were used as negative control. Fusion index was calculated on sarcMyHC+ myotubes. Nuclei were counterstained with 10 μg/mL DAPI (blue). Scale bars, 100 μm. (B) Top: after 14 days in adipogenic induction medium, the differentiation of hMuStem cells was assessed by detection of lipid vesicles through oil red O staining. Bottom: at 80% of confluency, hMuStem cells were placed in osteogenic differentiation medium for 21 days. Alizarin red staining revealed the formation of calcium deposits. Scale bars, 100 μm. (C) RT-PCR revealed the presence of adiponectin and LPL RNA on hMuStem cells only after the adipogenic induction. (D) hMuStem cells were cultivated in endothelial medium for 7 days before being plated with a Matrigel coating for a further 3 days. Phase contrast microscopy revealed the formation of capillary-like structures. Scale bars, 300 μm. (E) Expression of classical pluripotent marker mRNA. (F) Immunolabelings against KLF4 and NANOG were performed on hMuStem cells (P5) cultivated in growth medium. Nuclei were counterstained with 10 μg/mL DAPI (blue). Scale bars, 50 μm. MSCs, human umbilical vein endothelial cells (HUVECs) and iPSCs were used as positive controls for (B)–(D) and (E) and (F), respectively.
Finally, we addressed the expression profile of the pluripotent stem cell markers. All hMuStem cell batches expressed KLF4 and NANOG mRNAs (respective mean CT values of 26.9 ± 1.3 and 30.9 ± 1.0), whereas OCT-4A transcripts were not detected (Figure 3E). Also, SOX2 transcript was only detected in a single cell batch upon 4. At the protein level, KLF4 and NANOG were respectively expressed by 95% ± 3% and 83% ± 10% of hMuStem cells (Figure 3F). Taken together, our results indicated that hMuStem cells correspond to oligopotent cells and not to pluripotent ones.
CD56+ Subset in hMuStem Cell Population Exhibits a Higher Myogenic Commitment In Vitro
To deepen the characterization of the hMuStem cell population, CD56− cells that could be detected in some cell batches as a minority fraction were investigated. For that, CD56+ and CD56− cells (n = 4) were separated by magnetic microbeads and analyzed on the basis of their expression for MRFs and desmin as well their ability to fuse in vitro (Table 1). The MYF5+ cells represented 4.6%–27.0% of the cells among CD56+ cells and only 1.9%–3.5% in CD56− cells (Mann-Whitney; p < 0.03). In addition, less than 1% MYOD+ cells and a lack of myogenin+ cells were detected in both cell fractions. Desmin was expressed by 49.0%–98.0% of CD56+ cells compared to 8.0%–35% desmin+ cells among the CD56− ones (Mann-Whitney; p < 0.03). When switched into myogenic differentiation medium, sorted CD56+ cells gave rise to multinucleated myotubes after 7 days, which was illustrated by FI ranging from 5% to 23% (Table 1). In contrast, sorted CD56− cells only formed thin myotubes, with a lower number of nuclei, as revealed by a FI < 3.5% (Mann-Whitney; p < 0.03). Overall, these results showed that CD56+ and CD56− hMuStem cells differed in vitro in their myogenic capacities, the CD56− cells displaying a lower capacity to give rise to myogenic cells, and differentiate into myotubes compared to CD56+ cells, which correspond to more committed myogenic cells.
Table 1.
Phenotypic Analysis of Sorted CD56 Cells Present in hMuStem Cell Population
| Sorted CD56+ Cells | Sorted CD56− Cells | |
|---|---|---|
| MYF5 | 4.6%–27% | 1.9%–3.5% |
| MYOD | 0%–1% | ND |
| Desmin | 49%–98% | 8.0%–35% |
| Fusion index | 5%–23% | 0%–3.5% |
Percentage of MYF5-, MYOD-, and desmin-positive cells was determined in sorted CD56 cells (n = 4 per fraction). Eight randomly selected fields were used to analyze at least 300 cells and determine the proportion of cells positive for each of the myogenic markers. Fusion index was determined after 7 days of myogenic differentiation by determining the percentage of nuclei within sarcMyHC+ myotubes (>2 nuclei) in two random fields per well in three replicate wells. At least 1,199 nuclei per well were considered. ND, not detected.
hMuStem Cells Participate in Regeneration of Injured Muscles
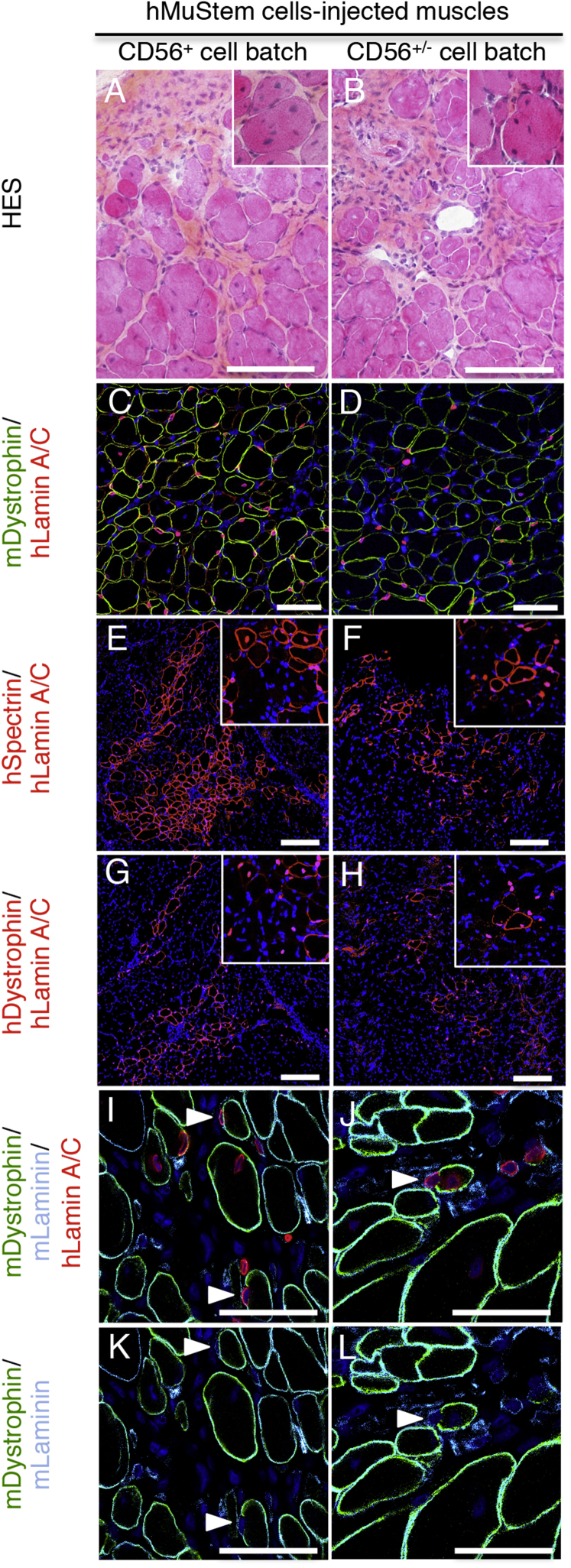
The in vivo behavior of hMuStem cells was investigated, especially in terms of contribution to muscle regenerative capacity. Two CD56+ cell batches (corresponding to n1 and n3 generated from pP and pL muscle, respectively) were considered and compared to 2 CD56+/− cell batches (n2 and n4 from pP and pL muscle, respectively) to determine if the presence of these CD56− cells, defined by a lesser in vitro myogenesis, may affect the global myogenic capacity in vivo of the hMuStem cell populations. Two hundred and fifty thousand hMuStem cells were injected into cryodamaged TA muscles of Rag2−IL2rβ− mice (n = 3 per cell batch). 3 weeks later, hematoxylin-eosin-saffron (HES)-stained sections from each injected muscle displayed numerous centronucleated myofibers (insets, Figures 4A and 4B) and large foci rich in mononucleated cells (Figures 4A and 4B). Rare fibroblasts, macrophages, and lymphocytes were also noted. The amount of endomysial tissue enriched in a lightly basophilic matrix and mixed with some collagen fibers (stained in yellow by saffron) was mildly increased. Specific human lamin A/C immunolabeling showed the presence of a similar number of donor nuclei in the different muscles injected with the four cell batches, revealing the ability of both CD56+ and CD56+/− hMuStem cells to integrate into the host injured tissue (Figures 4C and 4D). The percent of donor nuclei displaying a cytoplasmic location was 48.5% ± 9.18% and 80.1% ± 6.30% in muscles injected with cell batches made of exclusive CD56+ cells, whereas it was 20.2% ± 3.35% and 13.8% ± 6.19% for those receiving cell batches in which CD56− cells were found (Table 2; Figure S5). Interestingly, this distribution analysis of human lamin A/C+ nuclei revealed that CD56+ hMuStem cell batches contributed significantly more to myofiber formation than those exhibiting CD56− cell fraction (Kruskal-Wallis; p < 0.001). Human-specific MYF5, MYOD1, MYOG, and CDH15 mRNAs were detected in all injected muscles, revealing the ability of the hMuStem cells to differentiate in vivo into myogenic cells (Figure S6). Furthermore, the presence of cytoplasmic donor nuclei was associated with expression of the human spectrin protein and dystrophin one, although less frequently due to their late maturation (Figures 4E–4H, inset). Thus, the mean number of human spectrin+ myofibers per section was 97.7 ± 51.7 and 149 ± 36.8 in muscles injected with CD56+ cell batches compared to 58.3 ± 16.1 and 48.4 ± 19.2 for those injected with CD56+/− cell batches. For human dystrophin, the mean number of positive myofibers per section represented 29.7 ± 26.8 and 40.4 ± 26.8 for CD56+ cell batches. In comparison, it was 9.40 ± 7.04 and 5.00 ± 6.24 for CD56+/− cell batches (Table 2; Figure S5). Interestingly, the numbers of human spectrin+ and dystrophin+ myofibers per section were systematically higher in the muscles injected with exclusive CD56+ hMuStem cells, revealing a better myogenic commitment for them. In addition, confocal microscopy analysis indicated that hMuStem cell nuclei were rarely observed in the SC location, with less than 2.50% of human lamin A/C+ nuclei being found in this position in all injected muscles (Figures 4I–4L; Table 2; Figure S5). Finally, hMuStem cells with an interstitial location never expressed the endothelial marker CD31 (data not shown).
Figure 4.
Contribution of Both Long-Term Cultured CD56+ and CD56+/− hMuStem Cells to Myofiber Regeneration
hMuStem cell batches (P5) made exclusively of CD56+ cells (left) or containing a CD56− cell fraction (right) were injected into cryodamaged TA muscle of Rag2−IL2rβ− mice. (A–L) 3 weeks later, frozen sections of recipient muscle were submitted to HES staining (A and B) and co-labeled with specific Abs against human lamin A/C (red; C–J), murine dystrophin (green; C and D and I–L), murine laminin (light blue, I–L), human spectrin (red; E and F), and human dystrophin (red; G and H). (C–L) All nuclei were counterstained using DAPI (dark blue). Foci of regeneration composed of numerous centronucleated fibers (inset, A and B) and scattered mononucleated cells were observed. A large number of human nuclei were found in muscle tissue (C and D). Also, numerous human spectrin+ and dystrophin+ fibers were detected. Insets (E–H) show the presence of myofibers characterized by both detection of cytoplasmic donor nuclei and human spectrin or dystrophin expression. In return, hMuStem cells were rarely found in a satellite cell location (arrowhead, I–L). Scale bars, 100 μm (A–D), 150 μm (E–H), and 50 μm (I–L).
Table 2.
Histological Analysis of Cryodamaged TA Muscles of Rag2−IL2rβ− Mice following Intramuscular Injection of hMuStem Cells
| Cell Batch | Cell Batch Number | Mice Number | hLamin A/C+ Nuclei |
hSpectrin+ Myofibers with hLamin A/C+ Nuclei |
hDystrophin+ Myofibers with hLamin A/C+ Nuclei |
|||
|---|---|---|---|---|---|---|---|---|
| hLamin A/C+ Nuclei No. per Muscle | Tissue Distribution (%) |
No. of Myofibers per Section | ||||||
| Cytoplasmic Location | Satellite Cell Location | Interstitial Location | ||||||
| CD56+ | n1 | 1 | 9,891 | 51.3 | 3.90 | 44.9 | 89.6 | 37.0 |
| 2 | 4,181 | 38.2 | 0.70 | 61.1 | 50.5 | nd | ||
| 3 | 2,164 | 55.9 | nd | 44.2 | 153 | 52.0 | ||
| mean ± SD | 5,412 ± 4,008 | 48.5 ± 9.18 | 1.50 ± 2.08 | 50.1 ± 9.56 | 97.7 ± 51.7 | 29.7 ± 26.8 | ||
| n3 | 4 | 30,601 | 73.7 | 6.10 | 20.2 | 191 | 62.3 | |
| 5 | 17,180 | 86.3 | 0.80 | 13.0 | 124 | 48.3 | ||
| 6 | 10,152 | 80.3 | 0.60 | 19.1 | 131 | 10.5 | ||
| mean ± SD | 15,821 ± 10,986 | 80.1 ± 6.30 | 2.50 ± 3.12 | 17.4 ± 3.88 | 149 ± 36.8 | 40.4 ± 26.8 | ||
| CD56+/− | n2 | 7 | 12,169 | 23.5 | 1.50 | 75.0 | 70.4 | 6.00 |
| 8 | 50,498 | 20.2 | 0.40 | 79.4 | 64.5 | 17.5 | ||
| 9 | 21,312 | 16.8 | nd | 83.2 | 40.0 | 4.70 | ||
| mean ± SD | 27,993 ± 20,019 | 20.2 ± 3.35 | 0.60 ± 0.78 | 79.2 ± 4.10 | 58.3 ± 16.1 | 9.40 ± 7.04 | ||
| n4 | 10 | 7,142 | 20.9 | 2.70 | 76.5 | 62.8 | 3.00 | |
| 11 | 6,849 | 9.40 | 0.50 | 90.2 | 26.6 | nd | ||
| 12 | 21,686 | 11.2 | 0.30 | 88.5 | 55.8 | 12.0 | ||
| mean ± SD | 11,892 ± 8,483 | 13.8 ± 6.19 | 1.20 ± 1.33 | 85.1 ± 7.47 | 48.4 ± 19.2 | 5.00 ± 6.24 | ||
| p value (CD56+ versus CD56+/− cells) | >0.05 | <0.001* | >0.05 | <0.001* | >0.05 | >0.05 | ||
Four hMuStem cell batches were considered: two with exclusive CD56+ cells (n1 and n3) and two that contain CD56− ones (n2 and n4). Human lamin A/C+ nuclei were counted per section, and total number of human cells in the muscles was determined based on a linear density by considering the size of the hMuStem cell nuclei and the thickness of the section, as previously described.35 The cytoplasmic, satellite, and interstitial distribution was expressed as a percentage, whereas human spectrin+ and dystrophin+ myofibers were expressed as the number of counted myofibers per section. The p value was determined with a Kruskal-Wallis test. * indicates a significant result.
Taken together, these results gave evidence that the incorporation of hMuStem cell nuclei into host myofibers is effective and associated with both expression of human muscle-specific genes and production of structural proteins expressed in differentiated myofibers. hMuStem cells exhibit an in vivo muscle regenerative capacity whose intensity is directly associated with their content in CD56+ cells.
Discussion
Over the last few years, several adult stem cell populations were presented experimentally as having a myogenic potential in the muscular dystrophy context, suggesting that their clinical use could allow surmounting of the major limitations of the myoblast transplantation. However, when protocols were developed with these cells of human origin60, 70, 80 using a clinically relevant animal model78, 86 or in a clinical trial63, the initially described regenerative potential was unfortunately not confirmed. These works pointed out the difficulties of translating from preclinical studies to clinical trials and the need to carefully define standardized experimental protocols.
hMuStem Cells Could Be Isolated from Skeletal Muscle Tissue of a Different Source
Canine MuStem cells have been previously obtained from large samples of young healthy dog limb muscles.96 Based on an adaptation of this protocol, we report here that human MuStem cells could be successfully isolated from small biopsies of postural or locomotor muscles of 15- to 51-year-old women and men donors. Like cMuStem cells, all hMuStem cell batches generated a polyclonal population morphologically composed of predominantly spindle-shaped flat cells and some round cells. hMuStem cell-derived primary cultures were able to produce more than 20 population doublings in 38 days, revealing their extended proliferation capacity in vitro. This result is in agreement with those previously presented for cMuStem cells as well as for human myosphere-derived progenitor cells (MDPCs) and freshly isolated muscle AC133+ cells that could undergo more than 40 population doublings before the onset of senescence105 and 25 population doublings in 50 days of culture,35 respectively. Whether the age and the gender of donors influence the morphological and behavioral features of hMuStem cells still needs to be addressed by considering a higher number of cell batch because these parameters were previously shown to affect the features of other adult-derived stem cells, such as MDSCs,106 ADSCs, and MSCs.107, 108
hMuStem Cells Predominantly Correspond to Early Myogenic-Committed Progenitors with a Perivascular Origin
Based on a panel of SC and muscle-specific markers, we established that cultured hMuStem cells are mainly early myogenic-committed progenitors, as previously shown for canine cells.96 Consistently, hMDSCs defined as preplated cells adhering between 48 and 120 hr70 or after 96 hr71 were characterized as being CD56+ and expressing MYF5, MYOD1, or MYOG at the RNA level. In the last few years, Peault’s group58, 109 proposed that adult stem cells identified in multiple human organs could be related to a common ancestor, corresponding to perivascular cells expressing MSC markers. Like long-term cultured human perivascular cells, we determined that in vitro expanded hMuStem cells express the pericyte markers CD146 and CD140b, the recognized MSC markers, but not the markers of hematopoietic and endothelial cells. These results evoke a possible blood vessel wall origin for the hMuStem cells. The high expression we observed for the MSC markers is consistent with the description done for hMDPCs and hMDSCs isolated by a similar preplating technique from muscle biopsies of subjects with no known muscle disease.70, 71 Based on their broad expression of endothelial markers, it was hypothesized that hMDPC subsets may originate from blood vessels110 and correspond to myo-endothelial cells.92 Our present results clearly indicate that it is not the case for the hMuStem cells. Also, a major difference between hMuStem cells and human pericytes concerns their myogenic expression pattern. Although freshly isolated or long-term cultured muscle-derived perivascular cells neither expressed the CD56 protein nor the PAX7, MYF5, MYOG, and M-CDH15 transcripts,57, 58 cultured hMuStem cells were defined by their expression of several muscle-specific markers. Interestingly, human muscle-derived cell (MDC) preparations obtained as previously described for pericytes were defined as containing a CD56+ cell subpopulation (ranged from 1.5% to 47%) and expressing PAX3, MYF5, and MYOD1 mRNAs.60 Only few preparations expressed PAX7. In agreement with these data, we detected PAX3, MYF5, and MYOD1 mRNAs in all hMuStem cell batches, whereas we did not observe PAX7 expression. In return, the fraction of CD56+ cells is lower in the MDCs and much more heterogeneous than the one obtained for the hMuStem cell populations (69%–99%). Similarly, 76% of CD56+ cells were detected in the hMDPCs.71 Considering the concomitant high expression of several robust markers of human SCs, such as CD56 and CD29,102 we determined for the hMuStem cells, it is quite intriguing to note a lack of the canonical SC transcription factor PAX7. It is noteworthy that the expression of Pax7 like Myf5 and MyoD was shown to decrease with time in culture in MDC preparation.60 Also, in myogenic cell cultures derived from human fetal muscle, PAX7 expression was shown to be low or undetectable at the proliferative myoblast stage.111 Thus, the lack of Pax7 expression might be related to the high proliferative status of hMuStem cells. Interestingly, the existence of a small fraction of PAX7− SCs was previously described, revealing that PAX7 is not absolutely expressed in all human SCs.104, 111 A hypothesis might be that the hMuStem cells correspond to these marginal SCs that would have a distant perivascular affiliation.
Based on the lack of CD15/SSEA-1 antigen expression, we additionally found that hMuStem cells do not correspond to the adipogenic CD56−/CD15+ cells, which are progeny of the myo-adipogenic CD56+/CD15+ progenitors.112 In vitro, the proliferation of these bipotent cells gave rise to CD56+/CD15− progenitors that express MYF5, but not the SC marker PAX7, as we determined here for the hMuStem cells. Intriguingly, RT-PCR analysis showed that CD56+/CD15− cells express PW1/PEG3, which was also detected in all hMuStem cell preparations. Considering that CD56+/CD15− cells, which were found in the interstitial compartment of human muscle,94 were defined as positive for the MSC markers CD44, CD49, CD90, and CD146 and negative for the lineage markers CD45, CD106, CD117, and CD133, we cannot exclude that hMuStem cells could belong to the CD56+/CD15− cell fraction.
hMuStem Cells Contribute to Muscle Regeneration
In agreement with our previous results obtained with cMuStem cells after IM injection in GRMD dogs,96 we showed that hMuStem cells can engraft into host tissue, where they efficiently participate in myofiber regeneration. Indeed, many hundreds of hMuStem cell nuclei fused with host myofibers were detected, which were associated with production of human proteins. These data distinguished hMuStem cells from hMDSCs that were characterized by a modest regenerative index after transplantation in scid/mdx mice muscles.70 One possible element to explain the difference between these two cell types may be that the hMDSCs corresponded to cells adhering between 48 and 120 hr, whereas the hMuStem cells were defined as cells adhering after 120 hr. Human muscle-derived AC133+ cells also contributed to extensive muscle regeneration after grafting into irradiated and cryodamaged TA muscles of Rag2−/γ chain×/C5 mice.35, 37 These cells efficiently participated in SC formation, with 25% of human nuclei corresponding to SCs, which contrasts with the modest contribution we determined for the hMuStem cells. Meng et al.37 demonstrated that AC133+ cells that had undergone greater in vitro expansion contributed less efficiently to muscle regeneration in vivo. How this parameter might influence hMuStem cell features will have to be addressed in conditions compliant with cell therapy. We also found that hMuStem cells adopt an interstitial location, with a higher rate than cMuStem cells. We can postulate that the lower ability of hMuStem cells to fuse with host myofibers compared to cMuStem cells is related not to their intrinsic properties but to the type of damaged muscle they were injected in. hMuStem cells were injected in a mouse muscle that was exposed to a single experimental injury and still contained a normal SC pool. In contrast, cMuStem cells were placed in a GRMD dog muscle characterized by continuous cycles of degeneration/regeneration and architectural alterations with inflammation, fibrosis, and deposition of non-myogenic material,113, 114 which could contribute to numerical and/or functional loss of SCs, as has been described in DMD patients,115, 116, 117 mdx mice models,118, 119, 120 and GRMD dogs.121 To investigate whether the grafted hMuStem cells with interstitial location are able to generate new human myofibers and so display a myogenic potential, it could be informative to perform on transplanted mice a second injury protocol a few weeks following initial cell transplantation, as previously assessed.102 Intriguingly, a similar ability to give rise to interstitial cells had been described for the PICs that exhibited a broad range of gene expression common to MSCs,122 did not express PAX7, displayed myogenic potential in vitro, and formed myofibers after engraftment in damaged mice muscle,100 as we describe here for the hMuStem cells. Considering that all hMuStem cell batches tested express PW1 mRNA, further experiments may be developed to determine in what manner they share other biological properties with these populations.
Concerning the hMuStem cell behavior after IM injection, it is noteworthy that the higher percentages of fused donor nuclei and human spectrin+ myofibers were found in muscles injected with the hMuStem cell batches containing homogeneous CD56 expression, revealing that the muscle regenerative potential mainly relies on CD56+ cells. These data, generated on four healthy donor MDC batches, are in agreement with those obtained for CD56+ and CD56− MDCs isolated as previously described for pericytes from a DMD patient.60 One difference found between these CD56+ and CD56− MDCs was their expression of desmin and MHC, which suggests a more differentiated status of CD56+ MDCs. Concerning hMuStem cells, a similar difference was observed in the percentage of desmin+ cells between CD56+ and CD56− cell fractions, corroborating the notion of a more pronounced myogenic commitment for the CD56+ cells.
We are currently isolating clinical grade hMuStem cells. Depending on the pattern of CD56 expression, we will generate an extended analysis of fractions if required and analyze their ability of muscle homing after the vascular route. Then, we will measure their contribution to muscle repair, which is of major interest because disappointing results were reported after systemic delivery of promising human AC133+ cells and MDCs isolated as previously described for pericytes in immunodeficient injured mice models.37, 60
In conclusion, our results demonstrate that hMuStem cells resemble cMuStem cells and provide the proof of concept that they display myogenic features after transplantation in vivo. Thus, hMuStem cells may represent a promising agent for clinical application in the DMD context.
Materials and Methods
Human Skeletal Muscle Tissue
Samples were obtained from pP muscle corresponding to Longissimus dorsi of two females (20 and 22 year old) and one 15-year-old male patient. pL muscles corresponding to Gastrocnemius, Fascia lata tensor, and Vastus lateralis of 38-, 45-, and 51-year-old male patients were also sampled. These muscles were chosen to determine if MuStem cells could be isolated from different tissue origins. Patients were free of known muscular disease and operated at the Department of Pediatric Surgery of the Centre Hospitalier Universitaire (CHU) de Nantes (France). All patients gave written informed consent. All protocols were approved by the Clinical Research Department of the CHU (Nantes, France) according to the rules of the French Regulatory Health Authorities (permit number: MESR/DC-2010-1199). The biological sample bank was constituted in compliance with the national guidelines regarding the use of human tissue for research (permit number: CPP/29/10).
Animals
Immunodeficient Rag2−IL2rβ− mice123, 124 were housed in the specific pathogen-free animal facilities at the Faculty of Medicine of the University Pierre and Marie Curie (UPMC, Paris, France). Mice were fed ad libitum and allowed continuous access to tap water. Twelve 5- to 6-month-old mice were used as recipients for human cell implantation. All experiments were carried out in accordance with the guidelines from the French National Research Council for the Care and Use of Laboratory Animals (Referenced number: 2016071316136141).
Isolation of Human MuStem Cells
Freshly surgically taken muscle tissues were placed in cold PBS (PAA, Les Rumeaux, France) supplemented with 2% UI/mL penicillin, 0.1 mg streptomycin, and 0.25 mg/mL amphotericin B (PSF) (Sigma-Aldrich, Saint Quentin-Fallavier, France) and transferred to the laboratory. They were weighed, washed several times in PBS/2% PSF, carefully minced into 1-mm3 pieces using forceps and scalpel and enzymatically digested (15 min, 37°C) by a mix of collagenase type VIII (2,000 U/g of tissue, Sigma-Aldrich) and 0.2% hyaluronidase type 1S (Sigma-Aldrich). The pre-digested tissue was centrifuged (100 × g, 5 min) and the supernatant was collected and neutralized with 20% (v/v) fetal calf serum (FCS) (Sigma-Aldrich) while the pellet was digested (30 min, 37°C) with 0.125% Pronase E (Sigma-Aldrich). After centrifugation (100 × g, 5 min), the supernatant was collected, pooled with those obtain after the first enzymatic digestion, and submitted to successive centrifugation (300 × g, 15 min) and sequential filtering through 100, 70, and 40 μm pore-diameter nylon mesh (BD Biosciences; Franklin Lakes, NJ, USA). Muscle-derived cells (MDCs) were resuspended in PBS (PAA) supplemented with 2% FCS (Sigma-Aldrich) and 1% PSF (Sigma-Aldrich), and viability was assessed using 0.1% trypan blue staining (VWR, Strasbourg, France). hMuStem cells were isolated using a modified version of the preplating technique previously described.96 A flow chart is shown in Figure S1. Briefly, skeletal MDCs were seeded at 1 × 105 cells/cm2 on uncoated tissue culture plastic flasks, whereas gelatin-coated ones (Sigma-Aldrich) have been used in the previous protocol. After 1 hr, non-adherent cells were placed at 5 × 104 cells/cm2 on gelatin-coated flasks under standard conditions (37°C in 95% humidified air and 5% CO2) for 1 day. Then, non-adherent cells were collected and transferred on other coated flasks for 3 days, whereas the procedure has been repeated daily for 3 days in the previous protocol. After that, floating cells were placed at 2 × 104 cells/cm2 in freshly coated flasks for 1 day again. Finally, non-adherent cells were seeded at the same density and maintained for 3 days to isolate hMuStem cells as poorly adhering cells between 5 and 8 days post-plating. They were amplified in growth medium (Macopharma, Mouvaux, France) containing 10% FCS, 1% PSF, 10 ng/mL human recombinant basic fibroblast growth factor, and 25 ng/mL human recombinant epidermal growth factor (PromoCell, Heidelberg, Germany). Growth medium was replaced every 4 days. At confluence, cell layers were dissociated with trypsin-EDTA at 0.25% v/v (Invitrogen, Cergy-Pontoise, France). hMuStem cells were then resuspended in growth medium and seeded at 1 × 104 cells/cm2 (passage 1 [P1]). Experiments were performed with the six cell samples obtained from pP and pL muscles. Myoblasts, corresponding to a pool of cells that adhered between 24 and 96 hr after the initial plating, were plated at 2 × 104 viable cells/cm2 to gelatin-coated plastic flasks and grown in HAM F12 (Invitrogen) supplemented with 15% FCS and 1% PSF.
In Vitro Proliferation Analysis
hMuStem cell-derived primary cultures (n = 4) were plated in a gelatin-coated flask at a density of 1 × 104 cells/cm2 in growth medium and placed under standard conditions. Cell population growth was monitored over a period of 38 days. The population doubling level and the doubling time were calculated at each passage performed when cultures reach 80% confluence, as previously described.125
Clonogenicity Assay
From a seeding density of 1 × 102 cells/gelatin-coated Petri dish, the colony-forming unit of the hMuStem cells (n = 4; P5) was evaluated over a period of 30 days. The colonies were submitted to May-Grümwald Giemsa staining and counted, and the colony-forming efficiency was expressed as the number of colonies for plated cells.
Flow Cytometry
Cultured hMuStem cell samples (n = 4; P5 corresponding to 10.4–12.7 population doublings) were resuspended in cold PBS/2% human serum and 1 × 105 cells were incubated (30 min, 4°C) in the dark with fluorochrome-conjugated antibodies (Abs) at saturating concentration (Table S2). Isotype-matched Ab and fluorescence minus control samples were used as negative controls for gating and analyses. When applicable, 7-amino-actinomycin D (7-AAD; BD Biosciences) was added to evaluate cell viability. Samples were acquired using a FACS Aria flow cytometer (BD Biosciences) and data were analyzed using FlowJo software (FlowJo, Ashland, OR, USA). For simple labeling, at least 15 × 103 viable cells were considered, whereas 3 × 104 cells were acquired for multicolor labeling.
Cell Sorting
Cultured hMuStem cell samples with a CD56− cell fraction (n = 4; P5) were sorted using CD56 MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer’s instructions. Briefly, cells were resuspended in wash buffer and incubated (15 min, 4°C, in the dark) with microbead-conjugated CD56 Abs at a saturating concentration. After washing and centrifugation, the supernatant was discarded, the cells were resuspended in buffer, and magnetic separation was done using the MS MACS Column device (Miltenyi Biotec).
Gene Expression
Total RNA was extracted from either dry pellets of 2 × 106 cells or muscle tissue samples using RNeasy mini kit and RNeasy Fibrous kit (QIAGEN, Santa Clara, CA, USA), respectively, following the manufacturer’s instructions. Total RNA was treated with DNase (Ambion, Austin, TX, USA), quantified using a NanoDrop spectrophotometer (Labtech, Wilmingtom, DE, USA), and processed for reverse transcriptase using the standard protocol. Briefly, RT reactions were carried out on 0.5 μg total RNA using the GoScript reverse transcriptase (Promega, Madison, WI, USA). Gene-specific oligonucleotide primers were designed using Oligo Primer Analysis Software v.7 (Molecular Biology Insights, Colorado Springs, CO, USA) and synthesized by MWG Operon (Eurofins, Ebersberg, Germany). PCR amplifications were performed on 0.5 μL cDNA with the following program: initial denaturation (12 min, 95°C), followed by 35 cycles (30 s, 94°C; 30 s, 60°C; and 30 s, 72°C), and a final extension (10 min, 72°C). The PCR products were migrated and visualized on a 2% agarose gel with GelRed staining. For RT-qPCR, all cDNA amplifications were performed in triplicate using 1/20th of the reverse transcription products with the MESA BLUE qPCR kit (Eurogentec, Seraing, Belgium). qPCR was run on the Thermocycler CFX96 (Biorad, Hercules, CA, USA) with the following parameters: initial denaturation step (5 min, 95°C) and a total of 40 cycles (15 s, 95°C; 1 min, 60°C per cycle). RPS18 was selected as an endogenous control, and the relative expression levels were calculated by the 2-ΔΔCt method. The primer’s sequences are supplied in Table S3.
Immunofluorescence Analysis
hMuStem cells (whole population or CD56-sorted fractions) and myoblasts (P5) were plated on gelatin-coated Lab-Tek culture chamber slides (Nalge-Nunc, Rochester, NY, USA) at 2 × 104 cells/cm2 in growth medium during 3 days. Induced pluripotent stem cells (iPSCs) were used as a positive control for pluripotent stem cell marker labeling. Cells were fixed in 2% paraformaldehyde (PFA) in PBS (10 min, 4°C or at room temperature [RT]), permeabilized with 0.3%–0.5% Triton X-100 (4°C, 20–30 min), and incubated (60 min, RT) in blocking buffer (5% goat serum in PBS). Cells were then incubated with the Abs listed in Table S4. Cells were finally counterstained (15 min, 37°C) with DAPI fluorescent-cell-permeable DNA probe (Life Technologies, Paisley, UK). More than 300 cells were counted per sample by using Fiji image analysis software v.2.126 Data were presented as mean ± SD.
In Vitro Differentiation Potential Assay
For myogenic differentiation, hMuStem cells (P5; whole population or CD56 cell-sorted fractions) were plated at 3 × 104 cells/cm2 and maintained in growth medium for 1 day, and then 10% FCS was replaced by 2% horse serum. After 7 days, differentiation was assessed based on cell morphology and expression of the sarcomeric myosin heavy chain isoform (sarcMyHC). Cultures were fixed in 4% PFA, treated with 0.5% Triton X-100/20% (w/v) goat serum in PBS, and incubated (1 hr, 37°C) with anti-human sarcMyHC Abs (1:500, Developmental Studies Hybridoma Bank [DSHB], Iowa City, IA, USA). Specific Ab binding was revealed using either AlexaFluor 488-coupled (1:500, Invitrogen) or biotinylated secondary goat-anti-mouse Abs (1:300, Dako), followed by peroxidase-coupled streptavidin (1:300, Dako). The nuclei were finally counterstained with DRAQ5 (Biostatus). FI was determined by determining the percentage of nuclei within sarcMyHC+ myotubes (>2 nuclei) in two random fields per well in three replicate wells. At least 1,199 nuclei per well were considered. For adipogenic and osteogenic differentiations, hMuStem cells (P5) were plated at 1 × 104 cells/cm2, maintained in growth medium until 80% confluence, and then incubated in specific cell-type differentiation media, as previously described.127 For endothelial differentiation, hMuStem cells were amplified in EGM-2 medium (Lonza, Basel, Swiss) for 7 days. Then, the cell layer was dissociated with trypsin and cells were seeded on Matrigel coating (3 g/mL, BD Biosciences) in EGM-2 medium. Differentiation was assessed 3 days later based on capillary-like formation. RT-PCR analysis of lineage-specific genes was performed using primers listed in Table S3.
In Vivo Experiment and Histological Analysis
Rag2−IL2rβ− mice were anesthetized with an intra-peritoneal injection (0.1 mL per 20 g body weight) of a solution containing 100 mg/mL ketamine (Merial, Lyon, France) and 20 mg/mL Rompun 2% (Bayer, Puteaux, France) in PBS. As post-operative analgesia, mice received a subcutaneous injection of 0.3 mg/mL buprenorphine (50 μg/kg body weight; Axience, Pantin, France). Prior to cell implantation, cryolesion of Tibialis anterior (TA) muscle was induced to stimulate the implanted cells to fuse and form new myofibers. After skin incision and muscle exposition, TA muscle was subjected to three freeze cycles of 15 s each by applying a liquid nitrogen cooled metallic rod, as previously described.128 Cell layers of hMuStem-cell-derived primary cultures (P5, n = 4) were then dissociated, cells were pelleted and resuspended (2.5 × 105 cells in 15 μL PBS), and each was injected in a single site in the left cryodamaged TA muscle of the host (n = 3 mice per cell batch) using a 50-μL syringe with a 22G needle. The skin was then closed with fine sutures. 3 weeks later, TA muscles were removed, frozen in isopentane cooled in liquid nitrogen, and entirely cut into 10-μm sections. For every 350 μm along the complete length of the muscle, 8 transverse sections corresponding to an 80-μm length were used for quantitative analysis. Histological analysis was done on HES-stained sections of cryodamaged muscle receiving exclusive CD56+ hMuStem cells or CD56+/− ones, with non-injected muscle being used as the control. The number of human nuclei or human spectrin+/dystrophin+ myofibers in each section examined was counted, and the total values were determined for each TA muscle investigated, as previously described.35
To quantify cytoplasmic and interstitial hMuStem cell nuclei, a triple labeling was performed. Sections were treated with Triton X-100 0.3% detergent (10 min, RT) and blocking buffer (10% goat serum in PBS; 30 min, RT) before being incubated (60 min, 37°C) with a solution of mouse anti-human lamin A/C primary Abs (1:50, Leica Biosystems, Nussloch, Germany), followed by Alexa 555 rabbit anti-mouse secondary Abs (1:300, Invitrogen) in a humid chamber (60 min, RT). The sections were then treated with goat blocking serum (30 min, RT) before incubation with a solution containing both rabbit anti-mouse dystrophin Abs (1:50, Chemicon, Rolling Meadows, IL, USA) and rat anti-mouse laminin Abs (1:100, Sigma-Aldrich; 60 min, 37°C). Secondary Abs corresponding to a mixture of Alexa 488 goat anti-rabbit Abs (1:300, Invitrogen) and Alexa 555 goat anti-rat Abs (1:300, Invitrogen) were finally added (60 min, RT). Nuclei were then counterstained by DRAQ5 (1:1,000, Invitrogen; 15 min, RT). Finally, sections were mounted in Mowiol Medium (Calbiochem EMD Biosciences). To visualize human dystrophin+ and spectrin+ myofibers, double labeling against human proteins and human lamin A/C were realized by the same protocol using monoclonal mouse anti-human Ab NCL-Dys3 and NCL-Spect1 (1:20; Leica Biosystems; 60 min, 37°C), respectively. The number of hMuStem cells and human protein+ myofibers in the injected muscles were determined on the images acquired with the laser confocal scanning microscope LSM780 Zeiss (Carl Zeiss Microscopy, Iena, Germany) at x20 magnificence and by using Fiji image analysis software v.2. Non-injected mice muscle and human muscle were used as negative and positive controls, respectively (Figure S7).
Statistical Analysis
Data are reported as mean ± SD. Statistical analysis was performed in GraphPad Prism software v6.0f using the Mann-Whitney test for the MRF expression, FI, and myogenic differentiation of the sorted CD56+ and CD56− cell fractions. Kruskal-Wallis test was used for the in vivo analysis.
Author Contributions
J.L., C. Saury, and B.L. performed the experiments, collection and/or assembly of data, data analysis, interpretation, manuscript writing, and final approval of the manuscript; C. Schleder, F.R., and E.N. performed the experiments, data analysis, and interpretation; I.L., L.C., S.V., M.L., C.B., and L.D. performed the experiments; A.H. and A.M. performed provision of study material and the experiments; L.G., B.D., Y.P., G.B.-B., and V.M. performed conception and design; K.R. performed conception and design, provision of study material, collection and/or assembly of data, data analysis and interpretation, manuscript writing, and final approval of the manuscript.
Acknowledgments
This research was supported by funds from the “Association Française contre les Myopathies” (AFM No. 15990) and the FEDER (Fonds Européens de Développement Régional) (37085 and 38436). It was carried out in the context of the IHU-Cesti project, which received French government financial support managed by the National Research Agency via investment for the future programme ANR-10-IBHU-005. The IHU-Cesti project is also supported by Nantes Metropole and the Pays de la Loire Region. The recipient of all these grants was K.R. We thank Christian Pinset (I-Stem, Evry, France) and Laurent David (iPSC platform, IRS, Nantes, France) for the supply of iPSCs. We also thank Thibaut Larcher (INRA UMR 703 PAnTher, Oniris, Nantes, France) for the HE analysis.
Footnotes
Supplemental Information includes seven figures and four tables and can be found with this article online at https://doi.org/10.1016/j.ymthe.2017.10.013.
Supplemental Information
References
- 1.Emery A.E.H. Population frequencies of inherited neuromuscular diseases--a world survey. Neuromuscul. Disord. 1991;1:19–29. doi: 10.1016/0960-8966(91)90039-u. [DOI] [PubMed] [Google Scholar]
- 2.Moat S.J., Bradley D.M., Salmon R., Clarke A., Hartley L. Newborn bloodspot screening for Duchenne muscular dystrophy: 21 years experience in Wales (UK) Eur. J. Hum. Genet. 2013;21:1049–1053. doi: 10.1038/ejhg.2012.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffman E.P., Brown R.H., Jr., Kunkel L.M. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 4.Eagle M., Bourke J., Bullock R., Gibson M., Mehta J., Giddings D., Straub V., Bushby K. Managing Duchenne muscular dystrophy--the additive effect of spinal surgery and home nocturnal ventilation in improving survival. Neuromuscul. Disord. 2007;17:470–475. doi: 10.1016/j.nmd.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Kohler M., Clarenbach C.F., Bahler C., Brack T., Russi E.W., Bloch K.E. Disability and survival in Duchenne muscular dystrophy. J. Neurol. Neurosurg. Psychiatry. 2009;80:320–325. doi: 10.1136/jnnp.2007.141721. [DOI] [PubMed] [Google Scholar]
- 6.Berardi E., Annibali D., Cassano M., Crippa S., Sampaolesi M. Molecular and cell-based therapies for muscle degenerations: a road under construction. Front. Physiol. 2014;5:119. doi: 10.3389/fphys.2014.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Partridge T.A., Morgan J.E., Coulton G.R., Hoffman E.P., Kunkel L.M. Conversion of mdx myofibres from dystrophin-negative to -positive by injection of normal myoblasts. Nature. 1989;337:176–179. doi: 10.1038/337176a0. [DOI] [PubMed] [Google Scholar]
- 8.Kinoshita I., Vilquin J.T., Guérette B., Asselin I., Roy R., Tremblay J.P. Very efficient myoblast allotransplantation in mice under FK506 immunosuppression. Muscle Nerve. 1994;17:1407–1415. doi: 10.1002/mus.880171210. [DOI] [PubMed] [Google Scholar]
- 9.Huard J., Tremblay G., Verreault S., Labrecque C., Tremblay J.P. Utilization of an antibody specific for human dystrophin to follow myoblast transplantation in nude mice. Cell Transplant. 1993;2:113–118. doi: 10.1177/096368979300200204. [DOI] [PubMed] [Google Scholar]
- 10.Huard J., Verreault S., Roy R., Tremblay M., Tremblay J.P. High efficiency of muscle regeneration after human myoblast clone transplantation in SCID mice. J. Clin. Invest. 1994;93:586–599. doi: 10.1172/JCI117011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gussoni E., Pavlath G.K., Lanctot A.M., Sharma K.R., Miller R.G., Steinman L., Blau H.M. Normal dystrophin transcripts detected in Duchenne muscular dystrophy patients after myoblast transplantation. Nature. 1992;356:435–438. doi: 10.1038/356435a0. [DOI] [PubMed] [Google Scholar]
- 12.Mendell J.R., Kissel J.T., Amato A.A., King W., Signore L., Prior T.W., Sahenk Z., Benson S., McAndrew P.E., Rice R. Myoblast transfer in the treatment of Duchenne’s muscular dystrophy. N. Engl. J. Med. 1995;333:832–838. doi: 10.1056/NEJM199509283331303. [DOI] [PubMed] [Google Scholar]
- 13.Skuk D., Goulet M., Roy B., Tremblay J.P. Efficacy of myoblast transplantation in nonhuman primates following simple intramuscular cell injections: toward defining strategies applicable to humans. Exp. Neurol. 2002;175:112–126. doi: 10.1006/exnr.2002.7899. [DOI] [PubMed] [Google Scholar]
- 14.Guérette B., Asselin I., Vilquin J.T., Roy R., Tremblay J.P. Lymphocyte infiltration following allo- and xenomyoblast transplantation in mice. Transplant. Proc. 1994;26:3461–3462. [PubMed] [Google Scholar]
- 15.Guérette B., Asselin I., Skuk D., Entman M., Tremblay J.P. Control of inflammatory damage by anti-LFA-1: increase success of myoblast transplantation. Cell Transplant. 1997;6:101–107. doi: 10.1177/096368979700600203. [DOI] [PubMed] [Google Scholar]
- 16.Fan Y., Maley M., Beilharz M., Grounds M. Rapid death of injected myoblasts in myoblast transfer therapy. Muscle Nerve. 1996;19:853–860. doi: 10.1002/(SICI)1097-4598(199607)19:7<853::AID-MUS7>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 17.Skuk D., Roy B., Goulet M., Tremblay J.P. Successful myoblast transplantation in primates depends on appropriate cell delivery and induction of regeneration in the host muscle. Exp. Neurol. 1999;155:22–30. doi: 10.1006/exnr.1998.6973. [DOI] [PubMed] [Google Scholar]
- 18.Skuk D., Roy B., Goulet M., Chapdelaine P., Bouchard J.P., Roy R., Dugré F.J., Lachance J.G., Deschênes L., Hélène S. Dystrophin expression in myofibers of Duchenne muscular dystrophy patients following intramuscular injections of normal myogenic cells. Mol. Ther. 2004;9:475–482. doi: 10.1016/j.ymthe.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 19.Skuk D., Goulet M., Roy B., Chapdelaine P., Bouchard J.P., Roy R., Dugré F.J., Sylvain M., Lachance J.G., Deschênes L. Dystrophin expression in muscles of duchenne muscular dystrophy patients after high-density injections of normal myogenic cells. J. Neuropathol. Exp. Neurol. 2006;65:371–386. doi: 10.1097/01.jnen.0000218443.45782.81. [DOI] [PubMed] [Google Scholar]
- 20.Skuk D., Goulet M., Roy B., Piette V., Côté C.H., Chapdelaine P., Hogrel J.Y., Paradis M., Bouchard J.P., Sylvain M. First test of a “high-density injection” protocol for myogenic cell transplantation throughout large volumes of muscles in a Duchenne muscular dystrophy patient: eighteen months follow-up. Neuromuscul. Disord. 2007;17:38–46. doi: 10.1016/j.nmd.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Skuk D., Tremblay J.P. Confirmation of donor-derived dystrophin in a duchenne muscular dystrophy patient allotransplanted with normal myoblasts. Muscle Nerve. 2016;54:979–981. doi: 10.1002/mus.25129. [DOI] [PubMed] [Google Scholar]
- 22.Beauchamp J.R., Morgan J.E., Pagel C.N., Partridge T.A. Dynamics of myoblast transplantation reveal a discrete minority of precursors with stem cell-like properties as the myogenic source. J. Cell Biol. 1999;144:1113–1122. doi: 10.1083/jcb.144.6.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hawke T.J., Garry D.J. Myogenic satellite cells: physiology to molecular biology. J. Appl. Physiol. 2001;91:534–551. doi: 10.1152/jappl.2001.91.2.534. [DOI] [PubMed] [Google Scholar]
- 24.Zammit P.S., Golding J.P., Nagata Y., Hudon V., Partridge T.A., Beauchamp J.R. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J. Cell Biol. 2004;166:347–357. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zammit P.S., Relaix F., Nagata Y., Ruiz A.P., Collins C.A., Partridge T.A., Beauchamp J.R. Pax7 and myogenic progression in skeletal muscle satellite cells. J. Cell Sci. 2006;119:1824–1832. doi: 10.1242/jcs.02908. [DOI] [PubMed] [Google Scholar]
- 26.Skuk D., Goulet M., Tremblay J.P. Transplanted myoblasts can migrate several millimeters to fuse with damaged myofibers in nonhuman primate skeletal muscle. J. Neuropathol. Exp. Neurol. 2011;70:770–778. doi: 10.1097/NEN.0b013e31822a6baa. [DOI] [PubMed] [Google Scholar]
- 27.Skuk D., Tremblay J.P. First study of intra-arterial delivery of myogenic mononuclear cells to skeletal muscles in primates. Cell Transplant. 2014;23(Suppl 1):S141–S150. doi: 10.3727/096368914X685032. [DOI] [PubMed] [Google Scholar]
- 28.Negroni E., Gidaro T., Bigot A., Butler-Browne G.S., Mouly V., Trollet C. Invited review: stem cells and muscle diseases: advances in cell therapy strategies. Neuropathol. Appl. Neurobiol. 2015;41:270–287. doi: 10.1111/nan.12198. [DOI] [PubMed] [Google Scholar]
- 29.Skuk D., Tremblay J.P. Cell therapy in myology: dynamics of muscle precursor cell death after intramuscular administration in non-human primates. Mol. Ther. Methods Clin. Dev. 2017;5:232–240. doi: 10.1016/j.omtm.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torrente Y., Belicchi M., Sampaolesi M., Pisati F., Meregalli M., D’Antona G., Tonlorenzi R., Porretti L., Gavina M., Mamchaoui K. Human circulating AC133(+) stem cells restore dystrophin expression and ameliorate function in dystrophic skeletal muscle. J. Clin. Invest. 2004;114:182–195. doi: 10.1172/JCI20325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gavina M., Belicchi M., Rossi B., Ottoboni L., Colombo F., Meregalli M., Battistelli M., Forzenigo L., Biondetti P., Pisati F. VCAM-1 expression on dystrophic muscle vessels has a critical role in the recruitment of human blood-derived CD133+ stem cells after intra-arterial transplantation. Blood. 2006;108:2857–2866. doi: 10.1182/blood-2006-04-018564. [DOI] [PubMed] [Google Scholar]
- 32.Benchaouir R., Meregalli M., Farini A., D’Antona G., Belicchi M., Goyenvalle A., Battistelli M., Bresolin N., Bottinelli R., Garcia L. Restoration of human dystrophin following transplantation of exon-skipping-engineered DMD patient stem cells into dystrophic mice. Cell Stem Cell. 2007;1:646–657. doi: 10.1016/j.stem.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 33.Torrente Y., Belicchi M., Marchesi C., D’Antona G., Cogiamanian F., Pisati F., Gavina M., Giordano R., Tonlorenzi R., Fagiolari G. Autologous transplantation of muscle-derived CD133+ stem cells in Duchenne muscle patients. Cell Transplant. 2007;16:563–577. doi: 10.3727/000000007783465064. [DOI] [PubMed] [Google Scholar]
- 34.Marchesi C., Belicchi M., Meregalli M., Farini A., Cattaneo A., Parolini D., Gavina M., Porretti L., D’Angelo M.G., Bresolin N. Correlation of circulating CD133+ progenitor subclasses with a mild phenotype in Duchenne muscular dystrophy patients. PLoS One. 2008;3:e2218. doi: 10.1371/journal.pone.0002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Negroni E., Riederer I., Chaouch S., Belicchi M., Razini P., Di Santo J., Torrente Y., Butler-Browne G.S., Mouly V. In vivo myogenic potential of human CD133+ muscle-derived stem cells: a quantitative study. Mol. Ther. 2009;17:1771–1778. doi: 10.1038/mt.2009.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belicchi M., Erratico S., Razini P., Meregalli M., Cattaneo A., Jacchetti E., Farini A., Villa C., Bresolin N., Porretti L. Ex vivo expansion of human circulating myogenic progenitors on cluster-assembled nanostructured TiO2. Biomaterials. 2010;31:5385–5396. doi: 10.1016/j.biomaterials.2010.03.021. [DOI] [PubMed] [Google Scholar]
- 37.Meng J., Chun S., Asfahani R., Lochmüller H., Muntoni F., Morgan J. Human skeletal muscle-derived CD133(+) cells form functional satellite cells after intramuscular transplantation in immunodeficient host mice. Mol. Ther. 2014;22:1008–1017. doi: 10.1038/mt.2014.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meng J., Bencze M., Asfahani R., Muntoni F., Morgan J.E. The effect of the muscle environment on the regenerative capacity of human skeletal muscle stem cells. Skelet. Muscle. 2015;5:11. doi: 10.1186/s13395-015-0036-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferrari G., Cusella-De Angelis G., Coletta M., Paolucci E., Stornaiuolo A., Cossu G., Mavilio F. Muscle regeneration by bone marrow-derived myogenic progenitors. Science. 1998;279:1528–1530. doi: 10.1126/science.279.5356.1528. [DOI] [PubMed] [Google Scholar]
- 40.Gussoni E., Soneoka Y., Strickland C.D., Buzney E.A., Khan M.K., Flint A.F., Kunkel L.M., Mulligan R.C. Dystrophin expression in the mdx mouse restored by stem cell transplantation. Nature. 1999;401:390–394. doi: 10.1038/43919. [DOI] [PubMed] [Google Scholar]
- 41.Ferrari G., Stornaiuolo A., Mavilio F. Failure to correct murine muscular dystrophy. Nature. 2001;411:1014–1015. doi: 10.1038/35082631. [DOI] [PubMed] [Google Scholar]
- 42.Gussoni E., Bennett R.R., Muskiewicz K.R., Meyerrose T., Nolta J.A., Gilgoff I., Stein J., Chan Y.M., Lidov H.G., Bönnemann C.G. Long-term persistence of donor nuclei in a Duchenne muscular dystrophy patient receiving bone marrow transplantation. J. Clin. Invest. 2002;110:807–814. doi: 10.1172/JCI16098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.LaBarge M.A., Blau H.M. Biological progression from adult bone marrow to mononucleate muscle stem cell to multinucleate muscle fiber in response to injury. Cell. 2002;111:589–601. doi: 10.1016/s0092-8674(02)01078-4. [DOI] [PubMed] [Google Scholar]
- 44.Dell’Agnola C., Wang Z., Storb R., Tapscott S.J., Kuhr C.S., Hauschka S.D., Lee R.S., Sale G.E., Zellmer E., Gisburne S. Hematopoietic stem cell transplantation does not restore dystrophin expression in Duchenne muscular dystrophy dogs. Blood. 2004;104:4311–4318. doi: 10.1182/blood-2004-06-2247. [DOI] [PubMed] [Google Scholar]
- 45.Lapidos K.A., Chen Y.E., Earley J.U., Heydemann A., Huber J.M., Chien M., Ma A., McNally E.M. Transplanted hematopoietic stem cells demonstrate impaired sarcoglycan expression after engraftment into cardiac and skeletal muscle. J. Clin. Invest. 2004;114:1577–1585. doi: 10.1172/JCI23071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuhr C.S., Lupu M., Storb R. Hematopoietic cell transplantation directly into dystrophic muscle fails to reconstitute satellite cells and myofibers. Biol. Blood Marrow Transplant. 2007;13:886–888. doi: 10.1016/j.bbmt.2007.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang P.B., Lidov H.G., White A.J., Mitchell M., Balasubramanian A., Estrella E., Bennett R.R., Darras B.T., Shapiro F.D., Bambach B.J. Inefficient dystrophin expression after cord blood transplantation in Duchenne muscular dystrophy. Muscle Nerve. 2010;41:746–750. doi: 10.1002/mus.21702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Asakura A., Seale P., Girgis-Gabardo A., Rudnicki M.A. Myogenic specification of side population cells in skeletal muscle. J. Cell Biol. 2002;159:123–134. doi: 10.1083/jcb.200202092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bachrach E., Li S., Perez A.L., Schienda J., Liadaki K., Volinski J., Flint A., Chamberlain J., Kunkel L.M. Systemic delivery of human microdystrophin to regenerating mouse dystrophic muscle by muscle progenitor cells. Proc. Natl. Acad. Sci. USA. 2004;101:3581–3586. doi: 10.1073/pnas.0400373101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bachrach E., Perez A.L., Choi Y.H., Illigens B.M., Jun S.J., del Nido P., McGowan F.X., Li S., Flint A., Chamberlain J. Muscle engraftment of myogenic progenitor cells following intraarterial transplantation. Muscle Nerve. 2006;34:44–52. doi: 10.1002/mus.20560. [DOI] [PubMed] [Google Scholar]
- 51.Frank N.Y., Kho A.T., Schatton T., Murphy G.F., Molloy M.J., Zhan Q., Ramoni M.F., Frank M.H., Kohane I.S., Gussoni E. Regulation of myogenic progenitor proliferation in human fetal skeletal muscle by BMP4 and its antagonist Gremlin. J. Cell Biol. 2006;175:99–110. doi: 10.1083/jcb.200511036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uezumi A., Ojima K., Fukada S., Ikemoto M., Masuda S., Miyagoe-Suzuki Y., Takeda S. Functional heterogeneity of side population cells in skeletal muscle. Biochem. Biophys. Res. Commun. 2006;341:864–873. doi: 10.1016/j.bbrc.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 53.Motohashi N., Uezumi A., Yada E., Fukada S., Fukushima K., Imaizumi K., Miyagoe-Suzuki Y., Takeda S. Muscle CD31(-) CD45(-) side population cells promote muscle regeneration by stimulating proliferation and migration of myoblasts. Am. J. Pathol. 2008;173:781–791. doi: 10.2353/ajpath.2008.070902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cossu G., Bianco P. Mesoangioblasts--vascular progenitors for extravascular mesodermal tissues. Curr. Opin. Genet. Dev. 2003;13:537–542. doi: 10.1016/j.gde.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 55.Sampaolesi M., Torrente Y., Innocenzi A., Tonlorenzi R., D’Antona G., Pellegrino M.A., Barresi R., Bresolin N., De Angelis M.G., Campbell K.P. Cell therapy of alpha-sarcoglycan null dystrophic mice through intra-arterial delivery of mesoangioblasts. Science. 2003;301:487–492. doi: 10.1126/science.1082254. [DOI] [PubMed] [Google Scholar]
- 56.Sampaolesi M., Blot S., D’Antona G., Granger N., Tonlorenzi R., Innocenzi A., Mognol P., Thibaud J.L., Galvez B.G., Barthélémy I. Mesoangioblast stem cells ameliorate muscle function in dystrophic dogs. Nature. 2006;444:574–579. doi: 10.1038/nature05282. [DOI] [PubMed] [Google Scholar]
- 57.Dellavalle A., Sampaolesi M., Tonlorenzi R., Tagliafico E., Sacchetti B., Perani L., Innocenzi A., Galvez B.G., Messina G., Morosetti R. Pericytes of human skeletal muscle are myogenic precursors distinct from satellite cells. Nat. Cell Biol. 2007;9:255–267. doi: 10.1038/ncb1542. [DOI] [PubMed] [Google Scholar]
- 58.Crisan M., Yap S., Casteilla L., Chen C.W., Corselli M., Park T.S., Andriolo G., Sun B., Zheng B., Zhang L. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3:301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 59.Tedesco F.S., Hoshiya H., D’Antona G., Gerli M.F., Messina G., Antonini S., Tonlorenzi R., Benedetti S., Berghella L., Torrente Y. Stem cell-mediated transfer of a human artificial chromosome ameliorates muscular dystrophy. Sci. Transl. Med. 2011;3:96ra78. doi: 10.1126/scitranslmed.3002342. [DOI] [PubMed] [Google Scholar]
- 60.Meng J., Adkin C.F., Xu S.W., Muntoni F., Morgan J.E. Contribution of human muscle-derived cells to skeletal muscle regeneration in dystrophic host mice. PLoS One. 2011;6:e17454. doi: 10.1371/journal.pone.0017454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dellavalle A., Maroli G., Covarello D., Azzoni E., Innocenzi A., Perani L., Antonini S., Sambasivan R., Brunelli S., Tajbakhsh S. Pericytes resident in postnatal skeletal muscle differentiate into muscle fibres and generate satellite cells. Nat. Commun. 2011;2:499. doi: 10.1038/ncomms1508. [DOI] [PubMed] [Google Scholar]
- 62.Corselli M., Crisan M., Murray I.R., West C.C., Scholes J., Codrea F., Khan N., Péault B. Identification of perivascular mesenchymal stromal/stem cells by flow cytometry. Cytometry A. 2013;83:714–720. doi: 10.1002/cyto.a.22313. [DOI] [PubMed] [Google Scholar]
- 63.Cossu G., Previtali S.C., Napolitano S., Cicalese M.P., Tedesco F.S., Nicastro F., Noviello M., Roostalu U., Natali Sora M.G., Scarlato M. Intra-arterial transplantation of HLA-matched donor mesoangioblasts in Duchenne muscular dystrophy. EMBO Mol. Med. 2015;7:1513–1528. doi: 10.15252/emmm.201505636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qu Z., Balkir L., van Deutekom J.C., Robbins P.D., Pruchnic R., Huard J. Development of approaches to improve cell survival in myoblast transfer therapy. J. Cell Biol. 1998;142:1257–1267. doi: 10.1083/jcb.142.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jankowski R.J., Haluszczak C., Trucco M., Huard J. Flow cytometric characterization of myogenic cell populations obtained via the preplate technique: potential for rapid isolation of muscle-derived stem cells. Hum. Gene Ther. 2001;12:619–628. doi: 10.1089/104303401300057306. [DOI] [PubMed] [Google Scholar]
- 66.Torrente Y., Tremblay J.P., Pisati F., Belicchi M., Rossi B., Sironi M., Fortunato F., El Fahime M., D’Angelo M.G., Caron N.J. Intraarterial injection of muscle-derived CD34(+)Sca-1(+) stem cells restores dystrophin in mdx mice. J. Cell Biol. 2001;152:335–348. doi: 10.1083/jcb.152.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qu-Petersen Z., Deasy B., Jankowski R., Ikezawa M., Cummins J., Pruchnic R., Mytinger J., Cao B., Gates C., Wernig A. Identification of a novel population of muscle stem cells in mice: potential for muscle regeneration. J. Cell Biol. 2002;157:851–864. doi: 10.1083/jcb.200108150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deasy B.M., Gharaibeh B.M., Pollett J.B., Jones M.M., Lucas M.A., Kanda Y., Huard J. Long-term self-renewal of postnatal muscle-derived stem cells. Mol. Biol. Cell. 2005;16:3323–3333. doi: 10.1091/mbc.E05-02-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Urish K.L., Vella J.B., Okada M., Deasy B.M., Tobita K., Keller B.B., Cao B., Piganelli J.D., Huard J. Antioxidant levels represent a major determinant in the regenerative capacity of muscle stem cells. Mol. Biol. Cell. 2009;20:509–520. doi: 10.1091/mbc.E08-03-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chirieleison S.M., Feduska J.M., Schugar R.C., Askew Y., Deasy B.M. Human muscle-derived cell populations isolated by differential adhesion rates: phenotype and contribution to skeletal muscle regeneration in Mdx/SCID mice. Tissue Eng. Part A. 2012;18:232–241. doi: 10.1089/ten.tea.2010.0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H., Usas A., Poddar M., Chen C.W., Thompson S., Ahani B., Cummins J., Lavasani M., Huard J. Platelet-rich plasma promotes the proliferation of human muscle derived progenitor cells and maintains their stemness. PLoS One. 2013;8:e64923. doi: 10.1371/journal.pone.0064923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Simmons P.J., Torok-Storb B. Identification of stromal cell precursors in human bone marrow by a novel monoclonal antibody, STRO-1. Blood. 1991;78:55–62. [PubMed] [Google Scholar]
- 73.Yoon Y.S., Wecker A., Heyd L., Park J.S., Tkebuchava T., Kusano K., Hanley A., Scadova H., Qin G., Cha D.H. Clonally expanded novel multipotent stem cells from human bone marrow regenerate myocardium after myocardial infarction. J. Clin. Invest. 2005;115:326–338. doi: 10.1172/JCI22326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gonçalves M.A., de Vries A.A., Holkers M., van de Watering M.J., van der Velde I., van Nierop G.P., Valerio D., Knaän-Shanzer S. Human mesenchymal stem cells ectopically expressing full-length dystrophin can complement Duchenne muscular dystrophy myotubes by cell fusion. Hum. Mol. Genet. 2006;15:213–221. doi: 10.1093/hmg/ddi438. [DOI] [PubMed] [Google Scholar]
- 75.Rider D.A., Nalathamby T., Nurcombe V., Cool S.M. Selection using the alpha-1 integrin (CD49a) enhances the multipotentiality of the mesenchymal stem cell population from heterogeneous bone marrow stromal cells. J. Mol. Histol. 2007;38:449–458. doi: 10.1007/s10735-007-9128-z. [DOI] [PubMed] [Google Scholar]
- 76.Gang E.J., Bosnakovski D., Figueiredo C.A., Visser J.W., Perlingeiro R.C. SSEA-4 identifies mesenchymal stem cells from bone marrow. Blood. 2007;109:1743–1751. doi: 10.1182/blood-2005-11-010504. [DOI] [PubMed] [Google Scholar]
- 77.da Silva Meirelles L., Caplan A.I., Nardi N.B. In search of the in vivo identity of mesenchymal stem cells. Stem Cells. 2008;26:2287–2299. doi: 10.1634/stemcells.2007-1122. [DOI] [PubMed] [Google Scholar]
- 78.Nitahara-Kasahara Y., Hayashita-Kinoh H., Ohshima-Hosoyama S., Okada H., Wada-Maeda M., Nakamura A., Okada T., Takeda S. Long-term engraftment of multipotent mesenchymal stromal cells that differentiate to form myogenic cells in dogs with Duchenne muscular dystrophy. Mol. Ther. 2012;20:168–177. doi: 10.1038/mt.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zucconi E., Vieira N.M., Bueno C.R., Jr., Secco M., Jazedje T., Costa Valadares M., Fussae Suzuki M., Bartolini P., Vainzof M. Preclinical studies with umbilical cord mesenchymal stromal cells in different animal models for muscular dystrophy. J. Biomed. Biotechnol. 2011;2011:715251. doi: 10.1155/2011/715251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shang Y.C., Wang S.H., Xiong F., Peng F.N., Liu Z.S., Geng J., Zhang C. Activation of Wnt3a signaling promotes myogenic differentiation of mesenchymal stem cells in mdx mice. Acta Pharmacol. Sin. 2016;37:873–881. doi: 10.1038/aps.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rodriguez A.M., Pisani D., Dechesne C.A., Turc-Carel C., Kurzenne J.Y., Wdziekonski B., Villageois A., Bagnis C., Breittmayer J.P., Groux H. Transplantation of a multipotent cell population from human adipose tissue induces dystrophin expression in the immunocompetent mdx mouse. J. Exp. Med. 2005;201:1397–1405. doi: 10.1084/jem.20042224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu Y., Yan X., Sun Z., Chen B., Han Q., Li J., Zhao R.C. Flk-1+ adipose-derived mesenchymal stem cells differentiate into skeletal muscle satellite cells and ameliorate muscular dystrophy in mdx mice. Stem Cells Dev. 2007;16:695–706. doi: 10.1089/scd.2006.0118. [DOI] [PubMed] [Google Scholar]
- 83.Zannettino A.C., Paton S., Arthur A., Khor F., Itescu S., Gimble J.M., Gronthos S. Multipotential human adipose-derived stromal stem cells exhibit a perivascular phenotype in vitro and in vivo. J. Cell. Physiol. 2008;214:413–421. doi: 10.1002/jcp.21210. [DOI] [PubMed] [Google Scholar]
- 84.Vieira N.M., Bueno C.R., Jr., Brandalise V., Moraes L.V., Zucconi E., Secco M., Suzuki M.F., Camargo M.M., Bartolini P., Brum P.C. SJL dystrophic mice express a significant amount of human muscle proteins following systemic delivery of human adipose-derived stromal cells without immunosuppression. Stem Cells. 2008;26:2391–2398. doi: 10.1634/stemcells.2008-0043. [DOI] [PubMed] [Google Scholar]
- 85.Goudenege S., Pisani D.F., Wdziekonski B., Di Santo J.P., Bagnis C., Dani C., Dechesne C.A. Enhancement of myogenic and muscle repair capacities of human adipose-derived stem cells with forced expression of MyoD. Mol. Ther. 2009;17:1064–1072. doi: 10.1038/mt.2009.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vieira N.M., Valadares M., Zucconi E., Secco M., Bueno C.R., Jr., Brandalise V., Assoni A., Gomes J., Landini V., Andrade T. Human adipose-derived mesenchymal stromal cells injected systemically into GRMD dogs without immunosuppression are able to reach the host muscle and express human dystrophin. Cell Transplant. 2012;21:1407–1417. doi: 10.3727/096368911X. [DOI] [PubMed] [Google Scholar]
- 87.Pelatti M.V., Gomes J.P., Vieira N.M., Cangussu E., Landini V., Andrade T., Sartori M., Petrus L., Zatz M. Transplantation of human adipose mesenchymal stem cells in non-immunosuppressed GRMD dogs is a safe procedure. Stem Cell Rev. 2016;12:448–453. doi: 10.1007/s12015-016-9659-3. [DOI] [PubMed] [Google Scholar]
- 88.Tamaki T., Akatsuka A., Ando K., Nakamura Y., Matsuzawa H., Hotta T., Roy R.R., Edgerton V.R. Identification of myogenic-endothelial progenitor cells in the interstitial spaces of skeletal muscle. J. Cell Biol. 2002;157:571–577. doi: 10.1083/jcb.200112106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tamaki T., Akatsuka A., Okada Y., Matsuzaki Y., Okano H., Kimura M. Growth and differentiation potential of main- and side-population cells derived from murine skeletal muscle. Exp. Cell Res. 2003;291:83–90. doi: 10.1016/s0014-4827(03)00376-8. [DOI] [PubMed] [Google Scholar]
- 90.Tamaki T., Okada Y., Uchiyama Y., Tono K., Masuda M., Wada M., Hoshi A., Akatsuka A. Synchronized reconstitution of muscle fibers, peripheral nerves and blood vessels by murine skeletal muscle-derived CD34(-)/45 (-) cells. Histochem. Cell Biol. 2007;128:349–360. doi: 10.1007/s00418-007-0331-5. [DOI] [PubMed] [Google Scholar]
- 91.Zheng B., Cao B., Crisan M., Sun B., Li G., Logar A., Yap S., Pollett J.B., Drowley L., Cassino T. Prospective identification of myogenic endothelial cells in human skeletal muscle. Nat. Biotechnol. 2007;25:1025–1034. doi: 10.1038/nbt1334. [DOI] [PubMed] [Google Scholar]
- 92.Chen C.W., Corselli M., Péault B., Huard J. Human blood-vessel-derived stem cells for tissue repair and regeneration. J. Biomed. Biotechnol. 2012;2012:597439. doi: 10.1155/2012/597439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen W.C., Park T.S., Murray I.R., Zimmerlin L., Lazzari L., Huard J., Péault B. Cellular kinetics of perivascular MSC precursors. Stem Cells Int. 2013;2013:983059. doi: 10.1155/2013/983059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vauchez K., Marolleau J.P., Schmid M., Khattar P., Chapel A., Catelain C., Lecourt S., Larghéro J., Fiszman M., Vilquin J.T. Aldehyde dehydrogenase activity identifies a population of human skeletal muscle cells with high myogenic capacities. Mol. Ther. 2009;17:1948–1958. doi: 10.1038/mt.2009.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rouger K., Fornasari B., Armengol V., Jouvion G., Leroux I., Dubreil L., Feron M., Guevel L., Cherel Y. Progenitor cell isolation from muscle-derived cells based on adhesion properties. J. Histochem. Cytochem. 2007;55:607–618. doi: 10.1369/jhc.6A6954.2007. [DOI] [PubMed] [Google Scholar]
- 96.Rouger K., Larcher T., Dubreil L., Deschamps J.Y., Le Guiner C., Jouvion G., Delorme B., Lieubeau B., Carlus M., Fornasari B. Systemic delivery of allogenic muscle stem cells induces long-term muscle repair and clinical efficacy in duchenne muscular dystrophy dogs. Am. J. Pathol. 2011;179:2501–2518. doi: 10.1016/j.ajpath.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Robriquet F., Lardenois A., Babarit C., Larcher T., Dubreil L., Leroux I., Zuber C., Ledevin M., Deschamps J.Y., Fromes Y. Differential gene expression profiling of dystrophic dog muscle after MuStem cell transplantation. PLoS One. 2015;10:e0123336. doi: 10.1371/journal.pone.0123336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Robriquet F., Babarit C., Larcher T., Dubreil L., Ledevin M., Goubin H., Rouger K., Guével L. Identification in GRMD dog muscle of critical miRNAs involved in pathophysiology and effects associated with MuStem cell transplantation. BMC Musculoskelet. Disord. 2016;17:209. doi: 10.1186/s12891-016-1060-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lardenois A., Jagot S., Lagarrigue M., Guével B., Ledevin M., Larcher T., Dubreil L., Pineau C., Rouger K., Guével L. Quantitative proteome profiling of dystrophic dog skeletal muscle reveals a stabilized muscular architecture and protection against oxidative stress after systemic delivery of MuStem cells. Proteomics. 2016;16:2028–2042. doi: 10.1002/pmic.201600002. [DOI] [PubMed] [Google Scholar]
- 100.Mitchell K.J., Pannérec A., Cadot B., Parlakian A., Besson V., Gomes E.R., Marazzi G., Sassoon D.A. Identification and characterization of a non-satellite cell muscle resident progenitor during postnatal development. Nat. Cell Biol. 2010;12:257–266. doi: 10.1038/ncb2025. [DOI] [PubMed] [Google Scholar]
- 101.Charville G.W., Cheung T.H., Yoo B., Santos P.J., Lee G.K., Shrager J.B., Rando T.A. Ex vivo expansion and in vivo self-renewal of human muscle stem cells. Stem Cell Reports. 2015;5:621–632. doi: 10.1016/j.stemcr.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xu X., Wilschut K.J., Kouklis G., Tian H., Hesse R., Garland C., Sbitany H., Hansen S., Seth R., Knott P.D. Human satellite cell transplantation and regeneration from diverse skeletal muscles. Stem Cell Reports. 2015;5:419–434. doi: 10.1016/j.stemcr.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Uezumi A., Nakatani M., Ikemoto-Uezumi M., Yamamoto N., Morita M., Yamaguchi A., Yamada H., Kasai T., Masuda S., Narita A. Cell-surface protein profiling identifies distinctive markers of progenitor cells in human skeletal muscle. Stem Cell Reports. 2016;7:263–278. doi: 10.1016/j.stemcr.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Alexander M.S., Rozkalne A., Colletta A., Spinazzola J.M., Johnson S., Rahimov F., Meng H., Lawlor M.W., Estrella E., Kunkel L.M. CD82 is a marker for prospective isolation of human muscle satellite cells and is linked to muscular dystrophies. Cell Stem Cell. 2016;19:800–807. doi: 10.1016/j.stem.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wei Y., Li Y., Chen C., Stoelzel K., Kaufmann A.M., Albers A.E. Human skeletal muscle-derived stem cells retain stem cell properties after expansion in myosphere culture. Exp. Cell Res. 2011;317:1016–1027. doi: 10.1016/j.yexcr.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 106.Deasy B.M., Lu A., Tebbets J.C., Feduska J.M., Schugar R.C., Pollett J.B., Sun B., Urish K.L., Gharaibeh B.M., Cao B. A role for cell sex in stem cell-mediated skeletal muscle regeneration: female cells have higher muscle regeneration efficiency. J. Cell Biol. 2007;177:73–86. doi: 10.1083/jcb.200612094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Beane O.S., Fonseca V.C., Cooper L.L., Koren G., Darling E.M. Impact of aging on the regenerative properties of bone marrow-, muscle-, and adipose-derived mesenchymal stem/stromal cells. PLoS One. 2014;9:e115963. doi: 10.1371/journal.pone.0115963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shu W., Shu Y.T., Dai C.Y., Zhen Q.Z. Comparing the biological characteristics of adipose tissue-derived stem cells of different persons. J. Cell. Biochem. 2012;113:2020–2026. doi: 10.1002/jcb.24070. [DOI] [PubMed] [Google Scholar]
- 109.Park T.S., Gavina M., Chen C.W., Sun B., Teng P.N., Huard J., Deasy B.M., Zimmerlin L., Péault B. Placental perivascular cells for human muscle regeneration. Stem Cells Dev. 2011;20:451–463. doi: 10.1089/scd.2010.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Péault B., Rudnicki M., Torrente Y., Cossu G., Tremblay J.P., Partridge T., Gussoni E., Kunkel L.M., Huard J. Stem and progenitor cells in skeletal muscle development, maintenance, and therapy. Mol. Ther. 2007;15:867–877. doi: 10.1038/mt.sj.6300145. [DOI] [PubMed] [Google Scholar]
- 111.Reimann J., Brimah K., Schröder R., Wernig A., Beauchamp J.R., Partridge T.A. Pax7 distribution in human skeletal muscle biopsies and myogenic tissue cultures. Cell Tissue Res. 2004;315:233–242. doi: 10.1007/s00441-003-0833-y. [DOI] [PubMed] [Google Scholar]
- 112.Pisani D.F., Clement N., Loubat A., Plaisant M., Sacconi S., Kurzenne J.Y., Desnuelle C., Dani C., Dechesne C.A. Hierarchization of myogenic and adipogenic progenitors within human skeletal muscle. Stem Cells. 2010;28:2182–2194. doi: 10.1002/stem.537. [DOI] [PubMed] [Google Scholar]
- 113.Valentine B.A., Cooper B.J., Cummings J.F., de Lahunta A. Canine X-linked muscular dystrophy: morphologic lesions. J. Neurol. Sci. 1990;97:1–23. doi: 10.1016/0022-510x(90)90095-5. [DOI] [PubMed] [Google Scholar]
- 114.Collins C.A., Morgan J.E. Duchenne’s muscular dystrophy: animal models used to investigate pathogenesis and develop therapeutic strategies. Int. J. Exp. Pathol. 2003;84:165–172. doi: 10.1046/j.1365-2613.2003.00354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Blau H.M., Webster C., Pavlath G.K., Chiu C.P. Evidence for defective myoblasts in Duchenne muscular dystrophy. Adv. Exp. Med. Biol. 1985;182:85–110. doi: 10.1007/978-1-4684-4907-5_7. [DOI] [PubMed] [Google Scholar]
- 116.Decary S., Hamida C.B., Mouly V., Barbet J.P., Hentati F., Butler-Browne G.S. Shorter telomeres in dystrophic muscle consistent with extensive regeneration in young children. Neuromuscul. Disord. 2000;10:113–120. doi: 10.1016/s0960-8966(99)00093-0. [DOI] [PubMed] [Google Scholar]
- 117.Mouly V., Aamiri A., Bigot A., Cooper R.N., Di Donna S., Furling D., Gidaro T., Jacquemin V., Mamchaoui K., Negroni E. The mitotic clock in skeletal muscle regeneration, disease and cell mediated gene therapy. Acta Physiol. Scand. 2005;184:3–15. doi: 10.1111/j.1365-201X.2005.01417.x. [DOI] [PubMed] [Google Scholar]
- 118.Sacco A., Mourkioti F., Tran R., Choi J., Llewellyn M., Kraft P., Shkreli M., Delp S., Pomerantz J.H., Artandi S.E. Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell. 2010;143:1059–1071. doi: 10.1016/j.cell.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Villalta S.A., Nguyen H.X., Deng B., Gotoh T., Tidball J.G. Shifts in macrophage phenotypes and macrophage competition for arginine metabolism affect the severity of muscle pathology in muscular dystrophy. Hum. Mol. Genet. 2009;18:482–496. doi: 10.1093/hmg/ddn376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Villalta S.A., Rinaldi C., Deng B., Liu G., Fedor B., Tidball J.G. Interleukin-10 reduces the pathology of mdx muscular dystrophy by deactivating M1 macrophages and modulating macrophage phenotype. Hum. Mol. Genet. 2011;20:790–805. doi: 10.1093/hmg/ddq523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.La Rovere R.M., Quattrocelli M., Pietrangelo T., Di Filippo E.S., Maccatrozzo L., Cassano M., Mascarello F., Barthélémy I., Blot S., Sampaolesi M. Myogenic potential of canine craniofacial satellite cells. Front. Aging Neurosci. 2014;6:90. doi: 10.3389/fnagi.2014.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pannérec A., Formicola L., Besson V., Marazzi G., Sassoon D.A. Defining skeletal muscle resident progenitors and their cell fate potentials. Development. 2013;140:2879–2891. doi: 10.1242/dev.089326. [DOI] [PubMed] [Google Scholar]
- 123.Kuwajima S., Sato T., Ishida K., Tada H., Tezuka H., Ohteki T. Interleukin 15-dependent crosstalk between conventional and plasmacytoid dendritic cells is essential for CpG-induced immune activation. Nat. Immunol. 2006;7:740–746. doi: 10.1038/ni1348. [DOI] [PubMed] [Google Scholar]
- 124.Vosshenrich C.A., Ranson T., Samson S.I., Corcuff E., Colucci F., Rosmaraki E.E., Di Santo J.P. Roles for common cytokine receptor gamma-chain-dependent cytokines in the generation, differentiation, and maturation of NK cell precursors and peripheral NK cells in vivo. J. Immunol. 2005;174:1213–1221. doi: 10.4049/jimmunol.174.3.1213. [DOI] [PubMed] [Google Scholar]
- 125.Blasco M.A., Lee H.W., Hande M.P., Samper E., Lansdorp P.M., DePinho R.A., Greider C.W. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 126.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Delorme B., Charbord P. Culture and characterization of human bone marrow mesenchymal stem cells. Methods Mol. Med. 2007;140:67–81. doi: 10.1007/978-1-59745-443-8_4. [DOI] [PubMed] [Google Scholar]
- 128.Riederer I., Negroni E., Bigot A., Bencze M., Di Santo J., Aamiri A., Butler-Browne G., Mouly V. Heat shock treatment increases engraftment of transplanted human myoblasts into immunodeficient mice. Transplant. Proc. 2008;40:624–630. doi: 10.1016/j.transproceed.2008.01.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.