Abstract
Background
The dual-specificity T-box/basic helix-loop-helix leucine zipper transcription factor MGA is part of the MAX-interacting network of proteins. In the mouse, MGA is necessary for the survival of the pluripotent epiblast cells of the peri-implantation embryo and a null, gene-trap allele MgaGt results in embryonic lethality shortly after implantation. We have used this allele to document expression of Mga in postimplantation embryos and also investigated a second, hypomorphic gene-trap allele, MgaInv.
Results
Compound heterozygotes, MgaGt/MgaInv, die prior to midgestation. The extraembryonic portion of the embryos appears to develop relatively normally while the embryonic portion, including the pluripotent cells of the epiblast, is severely retarded by E7.5. Mga expression is initially limited to the pluripotent inner cell mass of the blastocyst and epiblast, but during organogenesis it is widely expressed notably in the central nervous system and sensory organs, reproductive and excretory systems, heart, somites and limbs.
Conclusions
Widespread yet specific areas of expression of Mga during organogenesis raise the possibility that the transcription factor may play roles in controlling proliferation and potency in the progenitor cell populations of different organ systems. Documentation of these patterns sets the stage for the investigation of specific progenitor cell types.
Keywords: T-box, bHLHZip, transcription factor, mouse, pluripotency
Introduction
Mga is an unusual gene that codes for a dual specificity, T-box domain and basic helix-loop-helix leucine-zipper (bHLHZip) domain transcription factor. It is part of the MAX-interacting network of proteins that form heterodimers with MAX and bind DNA at E-box sequences to activate or repress target genes (Hurlin et al., 1999; Meroni et al., 2000; Baudino and Cleveland, 2001; Walker et al., 2005). Heterodimerization with MAX is required for MGA to bind E-box-containing promoters, whereas MGA is able to bind DNA alone on T-box binding elements (TBEs), although gene activation or repression is modulated by heterodimerization with MAX (Hurlin et al., 1999).
In early mouse development, Mga is required for the survival of the epiblast (EPI) of the peri-implantation mouse embryo (Washkowitz et al., 2015). Using a null, gene-trap allele, MgaGt, we showed that in homozygous embryos blastocyst formation and differentiation of the primitive endoderm is normal and the epiblast expresses markers of pluripotency Pou5f1 and Nanog, but that apoptosis is increased and embryos die shortly after implantation due to the failure to maintain pluripotent cells. While the original gene trap allele, MgaGt, appears to be functionally null, a derivative allele, MgaInv, that should have restored function appears hypomorphic. Approximately half of MgaInv homozygotes fail to survive to weaning, although those that do are viable and fertile. Furthermore, MgaInv cannot compensate for MgaGt, as no compound heterozygotes were observed at weaning from heterozygous matings (Washkowitz et al., 2015).
This study investigates time of death of MgaGt/MgaInv compound heterozygotes and shows that MgaGt/MgaInv embryos survive for longer than MgaGt homozygotes but that the epiblast is severely affected. Although it is expressed during preimplantation development and has been reported at midgestation in specific tissues (Hurlin et al., 1999; Yoshikawa et al., 2006; Hoffman et al., 2008; Sansom et al., 2009; Guo et al., 2010; Washkowitz et al., 2015), Mga expression has not been systematically studied. Making use of the β-geo reporter, we show detailed expression of Mga in postimplantation development that indicates Mga could have tissue-specific effects on the development of multiple tissues and organ systems. This work sets the stage for the investigation of gene function using conditional alleles.
Results
Two Mga alleles affect different stages of development
The MgaGt(E153E01)Wrst allele, referred to as MgaGt, is a gene trap allele generated by the German Gene Trap Consortium that creates a truncated fusion protein carrying a β-geo reporter under the control of the Mga promoter. This is a multipurpose allele that when exposed to FLPe recombinase inverts the inserted cassette to produce, theoretically, a functional, conditional-mutation allele, MgaInv, that splices around the inserted cassette (Fig. 1a). As compound heterozygous MgaGt/MgaInv embryos fail to survive to weaning age (Washkowitz et al., 2015), we investigated the time of their death by mating MgaGt heterozygotes with MgaInv heterozygotes or homozygotes and documenting genotypes of offspring by PCR (Table 1). Among 8 litters with 69 offspring followed from birth to weaning, no MgaGt/MgaInv pups were found (X2>30; p<0.0001). At embryonic day (E) 12.5, no MgaGt/MgaInv embryos were recovered among 24 embryos genotyped (X2=9; p<0.05), indicating early gestation lethality.
Figure 1.
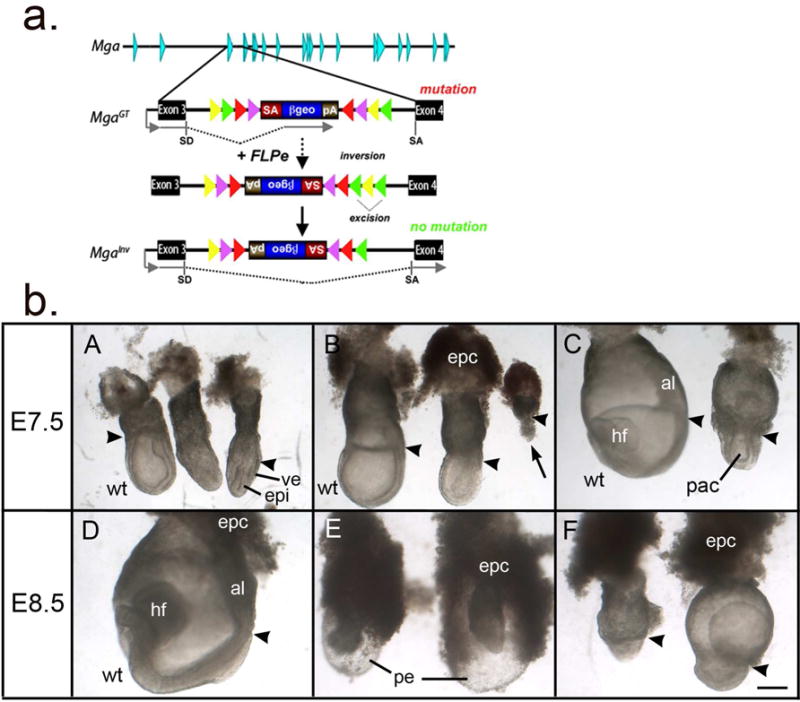
a. Diagram of the Mga locus and the gene trap alleles. The MgaGt allele orients a splice acceptor-β-galactosidase-neomycin-resistance cassette to accept the upstream exon 3 splice site of the Mga locus and create a mutant truncated reporter protein. Treatment with FLP recombinase results in inversion and excision producing the MgaInv allele, with splicing around the inserted cassette to produce a wild-type transcript (adapted from (Schnutgen et al., 2005) and (Washkowitz et al., 2015). SA, splice acceptor; SD, splice donor.
b. Embryos dissected from MgaGt/+ X MgaInv/Mga+ matings at E7.5 and E8.5 showing wild type (wt) embryos (on the left in A–C and in panel D) and grossly retarded putative MgaInv/MgaGt embryos. A–C. Embryos from litters of progressively more advanced developmental stages. A proamniotic cavity has formed with the exception of a few very retarded embryos (arrow in B), and the visceral endoderm and epiblast have differentiated in the mutants, but remain underdeveloped in the embryonic region, whereas the extraembryonic region is disproportionately large. D. A normal 3–5 somite littermate of the embryos in E and F. E. Two abnormal embryos dissected from the decidua intact, showing the distended sac formed from parietal endoderm and Reichert’s membrane and small, retarded embryos within. F. Two additional abnormal embryos further dissected to reveal a very small embryonic region and disproportionately large extraembryonic region. Arrowheads indicates the division between embryonic and extraembryonic regions. al, allantois; epc, ectoplacental cone; epi, epiblast; hf, headfold; pac, proamniotic cavity; pe, parietal endoderm; ve, visceral endoderm; wt, wild type. Scale bar= 200 microns.
Table 1.
Number of embryos or offspring recovered and genotyped by PCR at different developmental stages from matings of MgaGt/+ mice with either MgaInv/MgaInv or MgaInv/+ mice.
| Genotype | |||||||
|---|---|---|---|---|---|---|---|
| Mating | Age | n | +/+ | MgaGt/+ | MgaInv/+ | MgaGt/MgaInv | ND |
| MgaGt/+ X MgaInv/+ | P0 to P21 | 69 | 321 | 182 | 19 | 0 | 0 |
| MgaGt/+ X MgaInv/+ | E12.5 | 26 | 7 | 7 | 10 | 0 | 23 |
| MgaGt/+ X MgaInv/+ | E7.5 | 22 | 1 | 4 | 7 | 9 | 14 |
| MgaGt/+ X MgaInv/MgaInv | E7.5 | 12 | – | – | 4 | 4 | 44 |
one found dead at P3
one lost at P17
resorptions with giant cells and debris
empty deciduae
To narrow down the time of death of the compound mutant embryos, we dissected litters at earlier time points. At E6.5, all embryos examined were grossly normal (n=14) apart from a single empty decidua. At E7.5, 13 MgaGt/MgaInv embryos were identified by PCR (Table 1). Without exception, compound mutants were abnormally small and retarded compared with normal littermates, including single heterozygotes and wild type, which were at the late bud to early head fold stage (Downs and Davies, 1993). Compound mutants had a disproportionately large ectoplacental cone and an underdeveloped embryonic region, although there was usually a clear differentiation of visceral endoderm and epiblast layers and the presence of a proamniotic cavity (Fig. 1A–C). Additional embryos from dissections at E7.5 and E8.5, which were not genotyped but which fit a mendelian distribution of normal and abnormal phenotypes, showed a similar pattern with normal embryos between the early bud and 4–6 somite stages (n=39/58) and putative compound heterozygous embryos (n=19/58) with disproportionately large extraembryonic regions and underdeveloped embryonic regions. Between E7.5 and E8.5, the embryonic region did not progress developmentally, whereas the extraembryonic region was expanded in some embryos. The parietal endoderm and Reichert’s membrane frequently formed extended sacs, which the abnormal embryos did not fill (Fig. 1E). In addition, there were a few extremely retarded, tiny embryos (e.g. Fig. 1B, arrow) and a number of empty decidua.
Expression of Mga during development
In our previous study, Mga expression was first detected at E3.5 by RT-PCR and was limited to the inner cell mass (ICM) or EPI of the embryo at E4.5 to E6.5 (Washkowitz et al., 2015). We used β-galactosidase activity as a reporter for Mga expression at later stages of development in both whole mounts and sections, recognizing that perdurance of β-galactosidase activity may slightly overestimate the actual domain of Mga expression. At E7.5, expression is still limited to the EPI. With the exception of the allantois, there is no expression in the extraembryonic region (Fig. 2A–C, Fig. 3A). At E8.5–E10.5 (Fig. 2 & 3), expression is widespread in a number of developing tissues and is particularly prominent in the nervous system including optic and otic vesicles, all brain segments and the neural tube throughout the length of the embryo, and in the cranial and dorsal root ganglia (Fig. 2B–E, Fig. 3B–G). Expression is also in the allantois core (Fig. 2B, C), lateral plate mesoderm, mandibular arches, atria and ventricles of the heart (Fig. 3C, D, H, I), somites, limb buds, mesoderm surrounding the trachea and hepatic diverticulum, genital ridges and mesonephric tubules and ducts (Fig. 2D, 3J) and in the peritoneal lining, particularly in the pleural cavity (Fig. 3D). In the developing heart, expression is limited to the chamber myocardium and is excluded from the outflow tract and the endocardial cushions (Fig. 3 H, I).
Figure 2.
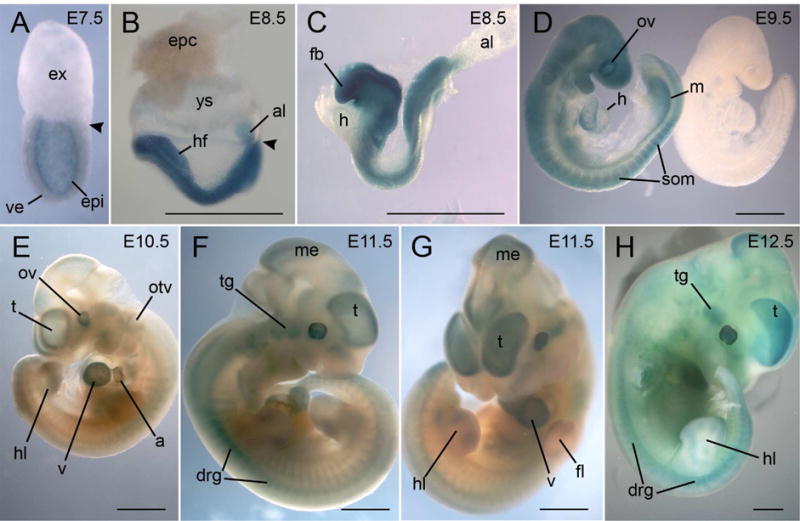
Whole mount X-gal staining of MgaGt/+ embryos from E7.5 to E12.5. A. At E7.5, expression is limited to the epiblast. Arrowhead indicates the division between embryonic and extraembryonic regions. B–D. At E8.5, the only extraembryonic tissue showing expression is the core of the allantois. At E8.5 and E9.5, expression is throughout the neural tube, in the somites, heart and mesonephric ducts. A control embryo without the transgene is shown in D. E–H. Between E10.5 and E12.5, expression is prominent in different regions of the brain, in the cranial and dorsal root ganglia, in the optic and otic vesicles, in the limb buds, and throughout the heart. a, atria; al, allantois; drg, dorsal root ganglia; epc, ectoplacental cone; epi, epiblast; ex, extraembryonic region; fb, forebrain; fl, forelimb; h, heart; hf, headfolds; hl, hindlimb; m, mesonephric duct; me, mesencephalon; otv, otic vesicle; ov, optic vesicle; som, somite; t, telencephalon; tg, trigeminal ganglion; v, ventricle; ve, visceral endoderm; ys, yolk sac. Scale bar=1mm.
Figure 3.
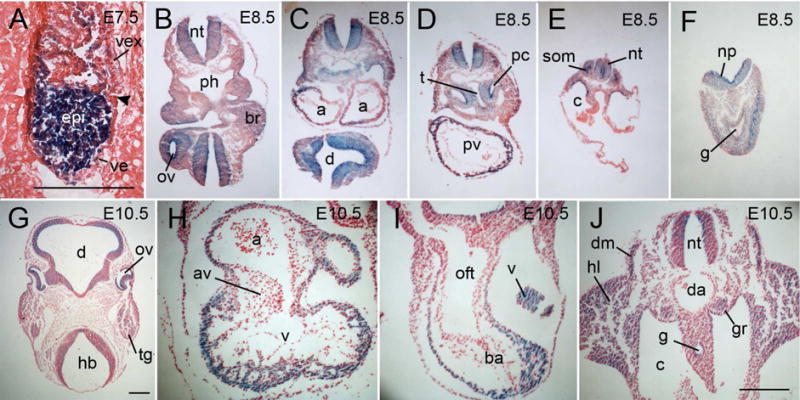
Sections of MgaGt/+ embryos between E7.5 and E10.5 stained for β-galactosidase activity. A. An E7.5 embryo sectioned in the uterus and then stained. Expression is limited to the epiblast and excluded from the visceral endoderm. B–F. An E8.5 embryo stained and then sectioned shows expression throughout the brain and neural tube, in the branchial arches, the heart, mesenchyme surrounding the trachea, somites and in the mesoderm of the tail (F). G–J. Details of expression from an E10.5 embryo showing expression in the brain and neural tube, optic vesicles, trigeminal ganglia, atria and ventricles of the heart but excluding the atrioventricular valve (H) and the outflow tract (I), the genital ridge, the dermamyotome of the somites, and the limbs (J). a, atria, av, atrioventriclualar valve; ba, bulbus arteriosus, br, branchial arch; c, coelom; d, diencephalon; da, dorsal aorta; dm, dermamyotome; epi, epiblast; g, gut; gr, genital ridge; hb, hindbrain; hl, hindlimb; np, neural plate; nt, neural tube; oft, outflow tract; ov, optic vesicle; pc, pleural cavity; ph, pharynx; pv, primitive ventricle; som, somite; tg, trigeminal ganglia; tr, trachea; v, ventricle; ve, visceral endoderm; vex, visceral extraembryonic endoderm, Scale bars= 200 microns; scale bar in J is for panels B–F and H–J.
At E11.5 and E12.5, whole mounts show the prominent expression in brain, cranial and dorsal root ganglia, heart, and the margins of the limbs (Fig. 2F–H). This pattern of expression is confirmed in sections of E12.5 embryos (Fig. 4), which also reveal regionalization of expression in the brain to the inner layers and the choroid plexus (Fig. 4A, B), whereas expression in the developing spinal cord is in the basal region (Fig. 4C–E). Expression is also seen in the lens of the eye (Fig. 4G), the inner ear including cochlea (Fig. 4H), the nasal epithelium and the vomeronasal organ (Fig. 4I), lung mesenchyme (Fig. 4D), endoderm of the stomach and gut, in the developing pancreas (Fig. 4K), mesonephric duct, metanephros (Fig. 4L), developing gonads, branchial pouches and clefts. Expression in the heart is throughout the atria and ventricles excluding the endocardial cushions and developing valves (Fig. 4J).
Figure 4.
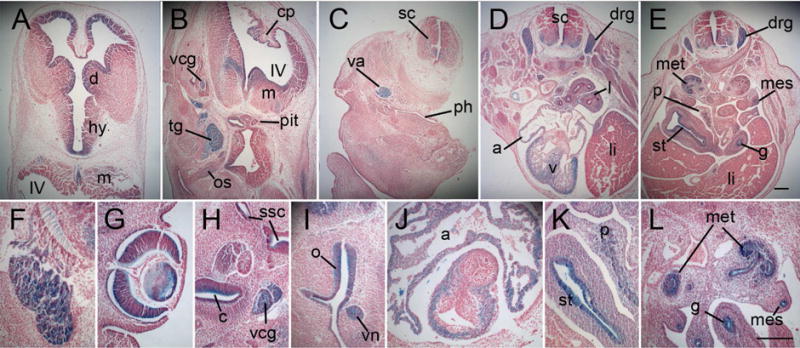
Sections of MgaGt/+ embryos at E12.5 stained for β-galactosidase activity. A. Section through the diencephalon, showing expression primarily in the ependymal layer, and myelencephalon (at bottom) showing expression in the basal region. B. Section through the hindbrain and developing pituitary gland showing expression in the choroid plexus, myelencephalon and cranial ganglia. C. Section through the spinal cord and neck region showing expression in the vagus nerve. D. Section through the thorax showing expression in the dorsal root ganglia and the basal part of the spinal cord, the lung buds, the atria and ventricles of the heart. E. Section through the abdomen showing expression in the basal spinal cord, the dorsal root ganglia, the metanephros, the mesonephric duct, the stomach and gut and the developing pancreas. F–L. Higher magnification of specific structures with expression including the trigeminal ganglion (F), the optic cup and lens (G), the inner ear, cochlea and vestibulocochlear ganglion (H), the nasal epithelium and vomeronasal organ (I), the atria and myocardium of the ventricles, excluding the developing valves (J), the stomach and developing pancreas (K), and the metanephros, mesonephric ducts and gut epithelium (L). a, atria; c, cochlea; cp, choroid plexus; d, diencephalon; drg, dorsal root ganglion; g, gut; hy, hypothalamus; l, lung; li, liver; m, myelencephalon; met, metanephros; mes, mesonephric ducts; o, olfactory epithelium; os, optic stalk; p, pancreas; ph, pharyngeal portion of the foregut; pit, developing pituitary gland; sc, spinal cord; ssc, semicircular canals; st, stomach; tg, trigeminal ganglion; v, ventricle; va, vagus nerve; vcg, vestibulocholear ganglion; vn, vomeronasal organ. IV, fourth ventricle; Scale bar= 200 microns; scale bar in E is for panels A–E; scale bar in L is for panels F–L.
Discussion
The MAX-interacting network of genes is thought to play crucial roles in development, in particular in the maintenance of the pluripotent ICM of the blastocyst and EPI of the early postimplantation embryo (Grandori et al., 2000; Hurlin and Huang, 2006). Embryos lacking either of the dimerization partners MAX or MGA die soon after implantation (Shen-Li et al., 2000; Washkowitz et al., 2015). In embryonic stem cells (ESCs), a heterodimer of MAX and MGA has been shown to stably recruit components of the Polycomb repressive complex 1 (PRC1) to target loci to repress expression of germ-cell related genes and maintain the self-renewal of ESCs (Endoh et al., 2017; Zhao et al., 2017). Regulation of the cellular polyamine pool through transcriptional regulation of a key enzyme gene, ornithine decarboxylase-1 (Odc1), which has E-box sites in its promoter (Bello-Fernandez et al., 1993), is thought to be a major role for MGA in peri-implantation development. ODC catalyzes the decarboxylation of ornithine to putrescine and is a rate-limiting step in the polyamine synthesis pathway. Embryos lacking Mga show reduced levels of ODC and failure of the ICM to develop either in vivo or in vitro, whereas supplementation with putrescine partially rescues this phenotype (Washkowitz et al., 2015). The c-MYC/MAX/MAD network has been shown to be able to regulate Odc1 by interacting with the E-box elements present in the promoter, suggesting a possible role for MGA in this context (Pena et al., 1993; Auvinen et al., 2003). Although direct regulation of Odc1 by MGA via the bHLHZip domain is likely, it is not known whether the T-box domain of MGA plays any role at this or later stages of development.
In this study, we performed an initial characterization of a second Mga allele, MgaInv. Homozygotes for MgaInv have reduced survival to weaning, although those that survive are fertile and apparently normal (Washkowitz et al., 2015). Here we show that compound heterozygotes with the null allele, MgaGt/MgaInv, die during early gestation with greatly reduced development of the pluripotent cells of the embryonic region. These embryos survive approximately two days longer than MgaGt/MgaGt homozygotes, which die before E5.5 (Washkowitz et al., 2015). Similar to the homozygous null embryos, it appears that the EPI of compound heterozygotes is the primary tissue affected as this appears smaller and less developed compared with a relatively normal-sized ectoplacental cone, extraembryonic ectoderm, and distal endoderm. These results are compatible with the hypothesis that MgaInv represents a hypomorphic allele that may reduce the level of ODC to a lesser extent than the null allele but below a level compatible with maintenance of the pluripotent cells of the embryo. The eventual death of the embryo including the extraembryonic tissues would follow the loss of the pluripotent EPI. Validation of this hypothesis will require additional experimentation and characterization of the compound mutants.
Mga expression is detected in the pluripotent ICM cells of the preimplantation blastocyst (Yoshikawa et al., 2006; Washkowitz et al., 2015) although single cell gene expression analysis by RT-PCR indicates expression throughout preimplantation development (Guo et al., 2010). Expression of both mRNA and protein is present in embryonic stem cells (ESCs) (Hu et al., 2009; van den Berg et al., 2010). Although a systematic analysis of Mga expression at later stages has not been previously reported, several studies address expression at specific times and in specific tissues. In situ hybridization (ISH) of whole embryos at E9.5–10.5 indicated widespread expression, particularly prominent in the branchial arches and limb buds (Hurlin et al., 1999). However, no negative controls were provided and it is possible that there were high levels of background staining. Using a similar probe, we were unable to obtain specific staining (personal observations). High-throughput ISH of transcription factor genes found no expression of Mga at E13.5 or P0 with the caveat that nonexpression could be due to limits of sensitivity of the screen (Gray et al., 2004). Analysis of images from GenePaint.org showed expression in the ventricular zone of E14.5 mouse cerebral cortex (Sansom et al., 2009). Serial analysis of gene expression (SAGE) libraries of pancreas development from E10.5 to E18.5 identified Mga expression, and examination of GenePaint images localized weak expression in the pancreatic epithelium at E14.5 (Hoffman et al., 2008). GUDMAP (McMahon et al., 2008) indicates Mga expression in the kidney, ovaries and testes at E15.5, which disappears by E17.5.
We made use of the gene trap allele to track Mga expression between E7.5 and E12.5. Our results corroborate the reported expression in the brain, limb buds, kidney, gonads and the pancreas and additionally, identify widespread expression in neural tissues including the germinal layer of the brain. In each of these areas of expression, it will be interesting to determine whether MGA has a role in progenitor cells similar to its role in the pluripotent cells of the early embryo and also which of the DNA binding domains is important. The conditional allele of Mga will be useful for this purpose.
Materials and Methods
Mice and genotyping
The gene trap allele MgaGt(E153E01)Wrst, referred to as MgaGt, obtained from the German Gene Trap Consortium, and its derivative FPL-recombinase inverted gene trap allele, MgaInv (Washkowitz et al., 2015), were maintained in mice of mixed genetic background including ICR (Taconic Biosciences). Embryos were dissected from timed matings with noon of the day of the plug considered E0.5. Mice were genotyped by PCR from tail tips and embryos were genotyped by PCR of yolk sac or embryonic tissue as before (Washkowitz et al., 2015). Use of mice for embryo production was approved by the Columbia University Medical Center Institutional Animal Care and Use Committee.
β-galactosidase activity assay
Following fixation in 4% paraformaldehyde at 4° C, embryos were assayed for β-galactosidase activity either by whole mount X-gal staining followed by paraffin embedding, sectioning and counterstaining with neutral fast red or eosin, or embedded for cryosectioning followed by X-gal staining as described previously (Washkowitz et al., 2015). Extended incubation time of up to 48 hours was used for advanced embryos.
Acknowledgments
We thank Akiko DeSantis, Qingxue Lu, Thomas McCord and Tracy Wu for technical assistance. Funding was provided by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health (NIH) grant R37 HD033082 (VEP). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Auvinen M, Jarvinen K, Hotti A, Okkeri J, Laitinen J, Janne OA, Coffino P, Bergman M, Andersson LC, Alitalo K, Holtta E. Transcriptional regulation of the ornithine decarboxylase gene by c-Myc/Max/Mad network and retinoblastoma protein interacting with c-Myc. Int J Biochem Cell Biol. 2003;35:496–521. doi: 10.1016/s1357-2725(02)00305-9. [DOI] [PubMed] [Google Scholar]
- Baudino TA, Cleveland JL. The Max network gone mad. Mol Cell Biol. 2001;21:691–702. doi: 10.1128/MCB.21.3.691-702.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello-Fernandez C, Packham G, Cleveland JL. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci U S A. 1993;90:7804–7808. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs KM, Davies T. Staging of gastrulating mouse embryos by morphological landmarks in the dissecting microscope. Development. 1993;118:1255–1266. doi: 10.1242/dev.118.4.1255. [DOI] [PubMed] [Google Scholar]
- Endoh M, Endo TA, Shinga J, Hayashi K, Farcas A, Ma KW, Ito S, Sharif J, Endoh T, Onaga N, Nakayama M, Ishikura T, Masui O, Kessler BM, Suda T, Ohara O, Okuda A, Klose R, Koseki H. PCGF6-PRC1 suppresses premature differentiation of mouse embryonic stem cells by regulating germ cell-related genes. Elife. 2017;6 doi: 10.7554/eLife.21064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- Gray PA, Fu H, Luo P, Zhao Q, Yu J, Ferrari A, Tenzen T, Yuk DI, Tsung EF, Cai Z, Alberta JA, Cheng LP, Liu Y, Stenman JM, Valerius MT, Billings N, Kim HA, Greenberg ME, McMahon AP, Rowitch DH, Stiles CD, Ma Q. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Hoffman BG, Zavaglia B, Witzsche J, Ruiz de Algara T, Beach M, Hoodless PA, Jones SJ, Marra MA, Helgason CD. Identification of transcripts with enriched expression in the developing and adult pancreas. Genome Biol. 2008;9:R99. doi: 10.1186/gb-2008-9-6-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes and Development. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlin P, Huang J. The MAX-interacting transcription factor network. Seminars in Cancer Biology. 2006;16:265–274. doi: 10.1016/j.semcancer.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Hurlin PJ, Steingrimsson E, Copeland NG, Jenkins NA, Eisenman RN. Mga, a dual-specificity transcription factor that interacts with Max and contains a T-domain DNA-binding motif. European Molecular Biology Organization Journal. 1999;18:7019–7028. doi: 10.1093/emboj/18.24.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon AP, Aronow BJ, Davidson DR, Davies JA, Gaido KW, Grimmond S, Lessard JL, Little MH, Potter SS, Wilder EL, Zhang P. GUDMAP: the genitourinary developmental molecular anatomy project. J Am Soc Nephrol. 2008;19:667–671. doi: 10.1681/ASN.2007101078. [DOI] [PubMed] [Google Scholar]
- Meroni G, Cairo S, Merla G, Messali S, Brent R, Ballabio A, Reymond A. Mlx, a new Max-like bHLHZip family member: the center stage of a novel transcription factors regulatory pathway? Oncogene. 2000;19:3266–3277. doi: 10.1038/sj.onc.1203634. [DOI] [PubMed] [Google Scholar]
- Pena A, Reddy CD, Wu S, Hickok NJ, Reddy EP, Yumet G, Soprano DR, Soprano KJ. Regulation of human ornithine decarboxylase expression by the c-Myc. Max protein complex. J Biol Chem. 1993;268:27277–27285. [PubMed] [Google Scholar]
- Sansom SN, Griffiths DS, Faedo A, Kleinjan DJ, Ruan Y, Smith J, van Heyningen V, Rubenstein JL, Livesey FJ. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 2009;5:e1000511. doi: 10.1371/journal.pgen.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnutgen F, De-Zolt S, Van Sloun P, Hollatz M, Floss T, Hansen J, Atschmied J, Seisenberger C, Ghyselinck NB, Ruiz P, Chambon P, Wurst W, von Melchner H. Genomewide production of multipurpose alleles for the functional analysis of the mouse genome. PNAS. 2005;102:7221–7226. doi: 10.1073/pnas.0502273102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Li H, O’Hagan RC, Hou JH, Horner IJW, Lee H-W, DePinho RA. Essential role for Max in early embryonic growth and development. Genes and Development. 2000;14:17–22. [PMC free article] [PubMed] [Google Scholar]
- van den Berg D, Snoek T, Mullin N, Yates A, Bezstarosti K, Demmers J, Chambers I, Poot R. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell. 2010;6:369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker W, Zhou ZQ, Ota S, Wynshaw-Boris A, Hurlin PJ. Mnt-Max to Myc-Max complex switching regulates cell cycle entry. J Cell Biol. 2005;169:405–413. doi: 10.1083/jcb.200411013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washkowitz AJ, Schall C, Zhang K, Wurst W, Floss T, Mager J, Papaioannou VE. Mga is essential for the survival of pluripotent cells during peri-implantation development. Development. 2015;142:31–40. doi: 10.1242/dev.111104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T, Piao Y, Zhong J, Matoba R, Carter MG, Wang Y, Goldberg I, Ko MSH. High-throughput screen for genes predominantly expressed in the ICM of mouse blastocysts by whole mount in situ hybridization. Gene Expression Patterns. 2006;6 doi: 10.1016/j.modgep.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Tong H, Huang Y, Yan Y, Teng H, Xia Y, Jiang Q, Qin J. Essential Role for Polycomb Group Protein Pcgf6 in Embryonic Stem Cell Maintenance and a Noncanonical Polycomb Repressive Complex 1 (PRC1) Integrity. J Biol Chem. 2017;292:2773–2784. doi: 10.1074/jbc.M116.763961. [DOI] [PMC free article] [PubMed] [Google Scholar]


