Abstract
The Atoh7 transcription factor catalyzes the rate-limiting step in the specification of retinal ganglion cells (RGCs). As a tool to study vertebrate retinal development, we validate an antibody that recognizes human and mouse Atoh7 polypeptide, using informative knockout and transgenic mouse tissues and overexpression experiments. The transient features of Atoh7 protein expression during retinal neurogenesis match the expected pattern at the tissue and cellular level. Further, we compare endogenous Atoh7 to established RGC markers, reporter mouse lines and cell cycle markers, demonstrating the utility of the antibody to investigate molecular mechanisms of retinal histogenesis.
Keywords: Atoh7, Math5, bHLH factor, optic nerve, retinal ganglion cell, retina, neurogenesis
INTRODUCTION
Atonal homologue 7 (Atoh7, formerly Math5) is a proneural basic helix-loop-helix (bHLH) transcription factor that is vital for genesis of retinal ganglion cells (RGCs) (Brown et al., 1998; Brown et al., 2001; Wang et al., 2001). RGCs represent 0.5% of cells of the mammalian neuroretina, but are the sole projection neurons for the eye; their axons comprise the optic nerve. Atoh7 is expressed transiently, during the terminal cell cycle, in a subset of multipotent retinal progenitor cells (RPCs), but it is only needed to specify RGC fate (Brzezinski et al., 2012). The Atoh7 gene contains a single exon, and its dynamic spatiotemporal expression is largely controlled by two cis-regulatory elements (Ghiasvand et al., 2011; Prasov et al., 2010; Riesenberg et al., 2009; Skowronska-Krawczyk et al., 2009). RGCs are the first-born retinal cell type, and thus Atoh7 transcription coincides with the onset of neurogenesis in the eye.
Atoh7 mutant mice have few RGCs and no optic nerves, and are thus blind (Brown 2001; Wang 2001; Brzezinski 2005). Similar autosomal recessive loss-of-function phenotypes have been reported in zebrafish and humans (Kay 2001; Prasov 2012b, Ghiasvand 2011). At the cellular level, Atoh7 appears to confer ganglion cell competence to RPCs; however, it is not sufficient to instruct RGC fate (Brzezinski et al., 2012; Fu et al., 2009; Prasov and Glaser, 2012). Indeed, its primary role may be to activate expression of downstream factors Isl1 and/or Pou4f2 during the terminal cell cycle, and thereby to initiate the RGC development program (Wu and Mu 2015). To date, our knowledge of Atoh7 expression dynamics and protein function has been derived from in vivo mouse experiments that rely on mRNA in situ hybridization or transgenic and knock-in reporter strains (Brown et al., 1998; Brown et al., 2001; Fu et al., 2009; Ghiasvand et al., 2011; Hufnagel et al., 2007; Wang et al., 2001). One important resource is lacking – an antibody to detect endogenous Atoh7 protein devoid of epitope tags.
Since 1998, several commercial and academic laboratories have worked to develop an antibody to detect murine Atoh7. Our group generated and tested sera to multiple Atoh7 peptide immunogens, yet found only one polyclonal reagent capable of reliably detecting human ATOH7 in transfected HEK293T cell lysates (Prasov et al., 2010); however, this antisera did not detect mouse Atoh7 in embryonic retinas or transfected HEK293T cells. Recently, a new antibody has shown promising results in human retinal organoid cultures, with expression of presumptive ATOH7 protein in a salt-and-pepper pattern (Aparicio et al., 2017), as previously observed for Atoh7 mRNA and reporters during retinal development in vivo. Moreover, a subset of these ATOH7+ cells co-expressed POU4F2 (Aparicio et al., 2017). We therefore rigorously tested this antibody with the goal of providing the vision community with a research resource to detect murine Atoh7 in future studies. We have validated this reagent using Atoh7 mutant tissue, overexpression systems, and reporter lines. Our results show that the antibody is specific to Atoh7 in cryopreserved tissue and detects both native and denatured forms. Endogenous Atoh7 protein expression follows the dynamic pattern of Atoh7 mRNA as the initial wave of neurogenesis proceeds in the optic cup, and co-localizes appropriately with transgene and knock-in reporters, and with cell cycle and RGC markers. This Atoh7 antibody is thus a valuable resource for vertebrate retinal development and disease research.
RESULTS
Detection of murine Atoh7
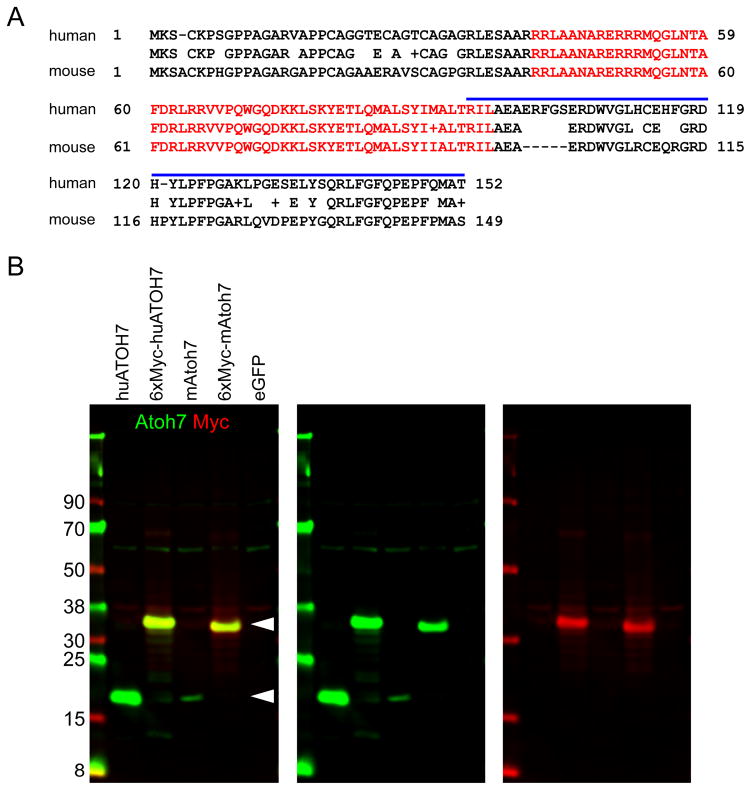
The most conserved region of Atoh7 is the 56-amino acid bHLH DNA-binding domain (Fig 1A). Outside the bHLH domain, there are multiple conserved segments with predicted antigenicity, which may be suitable as inter-species antibody targets (Jameson and Wolf, 1988; Prasov et al., 2010). Based on these parameters, we considered the antibody from Novus Biologicals, which was uses to detect ATOH7 in human retinal organoids (Aparicio et al., 2017), as potentially reactive with native Atoh7 in mouse tissue. This polyclonal rabbit antibody was generated using a recombinant peptide immunogen corresponding to the 60 C-terminal amino acids of human ATOH7 (Fig 1A). Human ATOH7 is 82% identical to mouse Atoh7, and the immunogen is 70% identical overall, with several short sequences of complete identity. As a polyclonal antibody, the sera may react avidly with multiple epitopes, each 4–6 residues long, within the C-terminal segment. As a first step to test whether the antibody cross-reacts with mouse and human proteins, we performed Western analysis on HEK293T cells transfected with wild-type Atoh7 vectors containing or lacking a poly-myc epitope tag (Prasov et al. 2010). In these experiments, the antibody recognized a 17 kDa protein in lysates expressing untagged mouse or human Atoh7 and a larger protein, migrating at 32 kDa, in lysates expressing myc-tagged Atoh7, consistent with the predicted size of 27 kDa (Fig 1B). This larger protein was detected by both anti-Atoh7 and anti-Myc reagents, as expected. The Novus Atoh7 antibody reacts well with both human and mouse proteins in denaturing SDS polyacrylamide gels.
Fig. 1. Detection of human and mouse Atoh7.
[A] Alignment of human and mouse Atoh7 polypeptides. The bHLH domain (red, 56 amino acids) and human recombinant peptide immunogen (blue bar, 59 amino acids) are indicated. The human and mouse sequences are 82% identical overall, 98% identical in the bHLH domain, and 70% identical in the C-terminal segment. [B] Western analysis of HEK293T cells transfected with human (huATOH7) or mouse (mAtoh7) expression plasmids ± six myc epitope tags. Arrowheads indicate the positions of Atoh7 (17 kDa) and 6xMyc-Atoh7 (predicted size, 27 kDa; actual size, ~32 kDa) proteins. The pCS2-mAtoh7 transfection was notably less efficient than other samples. The Atoh7 antibody cross-reacts weakly with a 60 kDa human protein in HEK293T.
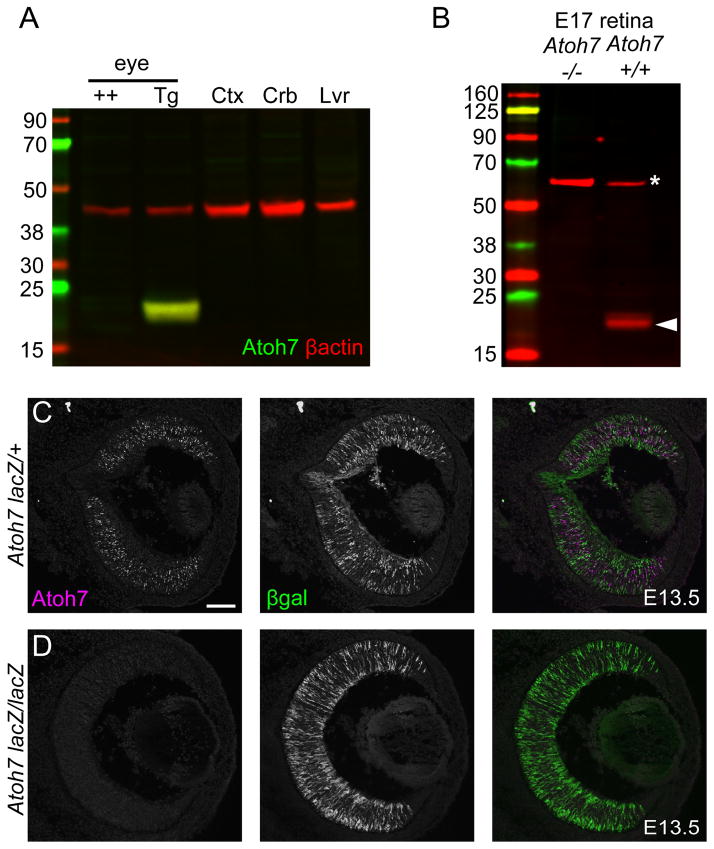
To validate detection of endogenous mouse Atoh7, we used three methods and two different mouse lines. First, we tested protein extracts from adult eyes in which mouse Atoh7 expression is driven ectopically within the Crx (cone-rod homeobox) lineage (Crx>Atoh7-IRES-Cre BAC line 60) (Prasov and Glaser, 2012). In these mice, Atoh7 is continuously expressed in photoreceptors and bipolar cells, comprising 80% of the neuroretina, long after expression of the native Atoh7 gene in RPCs has ceased (Brown et al. 1998). These samples were compared to Atoh7-negative tissues, including non-transgenic adult eyes, liver and cerebral cortex, and cerebellum, which expresses low levels of Atoh7 in granule cells (Saul et al. 2008). Only one sample, the Crx>Atoh7 transgenic eye lysate, had an immunoreactive 17 kDa protein (Fig 2A).
Fig. 2. Detection of endogenous mouse Atoh7.
[A] Western analysis of adult mouse tissues – wild-type (++) or Crx>Atoh7 BAC transgenic (Tg) eyes, and wild-type cerebral cortex (Ctx), cerebellum (Crb) and liver (Lvr). Atoh7 (green, 17 kDa) is present in transgenic eyes, which express Atoh7 robustly in the Crx lineage, with β-actin (red, 42 kDa) as a loading control. [B] Western analysis of E17 Atoh7 −/− and +/+ retinal extracts. Atoh7 is present in wild-type (arrowhead, 17 kDa) but not in mutant embryos. The 60 kDa protein (asterisk) in both lanes reacts nonspecifically with the Atoh7 antibody, and serves as a loading control. [C] Immunofluorescence of E13.5 retinas showing Atoh7+ cells in Atoh7 heterozygotes (lacZ/+) but no signal in Atoh7 mutants (lacZ/lacZ). βgal immunoreactivity is present in both, but stronger in the homozygote, with two lacZ genes.
The most rigorous test for antibody specificity is loss of signal in protein-null genetic mutants (Saper and Sawchenko 2003). To verify specificity of this antibody, we used Atoh7 knockout mice, in which the Atoh7 locus has been disrupted by insertion of the E. coli lacZ gene, and assayed embryonic retinal tissue, where Atoh7 is normally expressed. In immunoblots of E17 retinal extracts, we detected a protein of the correct size (17 kDa) in wild type, but not in Atoh7 −/− mice (Fig 2B). We then assayed E13.5 retinal cryosections from Atoh7lacZ/+ (+/−) and Atoh7lacZ/lacZ (−/−) littermates by indirect immunofluorescence (Fig. 2C). We observed Atoh7 immunostaining in a “salt and pepper” pattern similar to that reported for Atoh7 mRNA (Brown et al., 1998). All nuclear labeling was lost in Atoh7 −/− retinal tissue. No signal was observed in the otic vesicle or cochlear expression domains for Atoh1 (Math1), a closely related bHLH paralog (not shown). We were also unable to detect Atoh7 in chick (d3/d4), zebrafish (36 hpf) or Xenopus (stage 30/33) embryonic retinal sections using standard immunofluorescence methods (not shown). The Novus reagent thus specifically reacts with native Atoh7 protein in developing mouse (and human) retina.
Mouse Atoh7 protein dynamics recapitulate mRNA expression in vivo
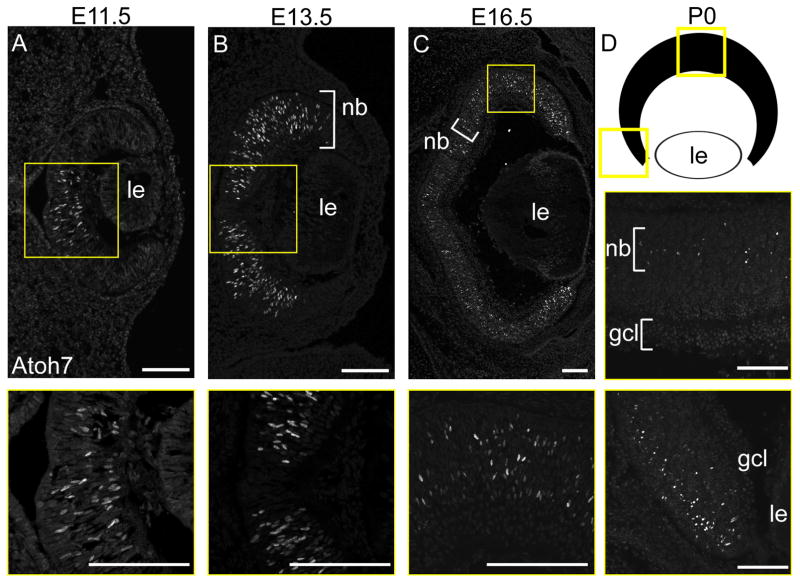
Atoh7 transcription normally initiates at E11.0, peaks at E14.5, rapidly decreases after E16.5, and is essentially absent after postnatal day 2 (P2) (Brown et al., 1998; Brzezinski et al., 2012; Skowronska-Krawczyk et al., 2009). Concurrently, the spatial distribution of Atoh7 mRNA extends progressively across the optic cup. Atoh7 mRNA first appears in the central dorsal portion of the optic cup, and spreads outward to the periphery during the initial wave of neurogenesis. At later time points (E16.5 to P0) Atoh7 mRNA is present sporadically throughout the central retina, within the neuroblast layer, whereas the peripheral retina maintains a higher proportion of Atoh7+ cells, consistent with its younger developmental status. These previous observations have been based solely on mRNA or transgene reporter data (Brown et al., 1998; Brown et al., 2001; Fu et al., 2009; Ghiasvand et al., 2011; Hufnagel et al., 2007). To evaluate Atoh7 protein dynamics, we analyzed wild-type retinas from E11.5 to E16.5 by immunofluorescence, and observed a similar spatiotemporal progression (Fig 3). Atoh7 protein was restricted to the central retina at E11.5, expanded toward the periphery at E13.5, and reached the edge of the optic cup by E16.5. At P0, we observed sparse Atoh7 labeling in the proliferative layer of the central retina, and moderate labeling at the periphery (Fig 3D).
Fig. 3. Dynamic Atoh7 protein expression during retinal neurogenesis.
Immunofluorescence staining of +/+ albino eyes showing Atoh7 expression [A] beginning in the central retina and [B,C] extending peripherally as neurogenesis proceeds. Insets (yellow) are magnified in lower panels. Atoh7 is localized in the proliferative neuroblast layer (nb) from E13-P0 and [D] at the margins of the retinal cup at P0. The diagram shows the orientation of views in the P0 horizontal section. Panel C is a composite of 4 images collected at 100X magnification, assembled using Leica LASX software. le, lens; gcl, ganglion cell layer. Scale bars, 100 μm
Correlated expression of Atoh7 and other markers of RGC development
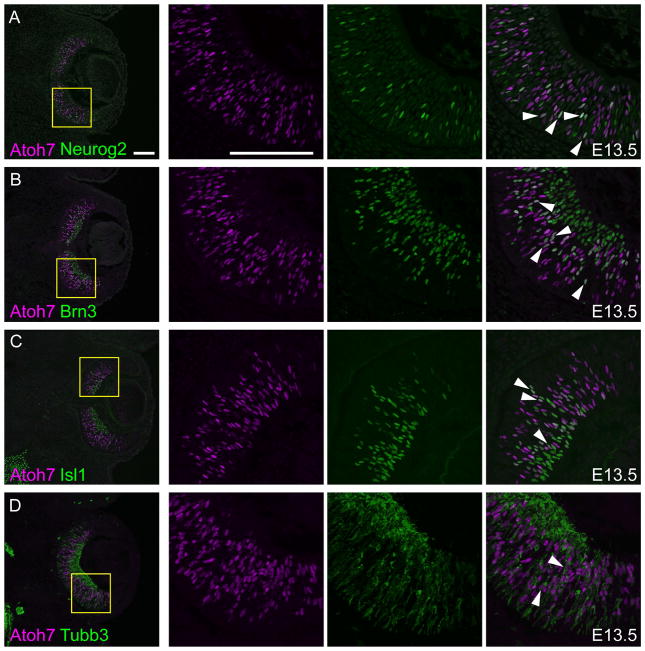
A discrete set of transcription factors controls RGC genesis. In addition to Atoh7, these include Neurogenin 2 (Neurog2), POU domain class 4F factors 1–3 (Pou4f1-3, formerly Brn3a-c), and Lim domain factor insulin gene enhancer protein 1 (Isl1). Neurog2 influences the timing of the initial neurogenic wave whereas Pou4f1-3 and Isl1 act downstream of Atoh7 to effect the RGC differentiation program (Hufnagel et al., 2010; Hutcheson and Vetter, 2001; Mu et al., 2005; Wu et al., 2015). We therefore tested the overlap between Atoh7 and these factors at E13.5, when a large number of RPCs are differentiating into RGCs (Fig 4). As expected, Neurog2 and Atoh7 double positive cells (15 ± 2.8% concordance) were detected in the neuroblast layer, but not in differentiated RGCs, which at E13.5 have migrated to the nascent ganglion cell layer (GCL) (Fig 4A). Pou4f (Brn3) and Isl1 were transiently expressed with Atoh7 (32 ± 4.9% and 30 ± 1.1% concordance, respectively) during RGC migration and maintained in the GCL (Fig 4B,C), consistent with previous observations (Fu et al., 2009; Mu et al., 2005; Wu et al., 2015). Tubb3 (class III β-tubulin) marks differentiated neurons, and mainly localized to the GCL, but was coexpressed with Atoh7 (28 ± 5.3% concordance) in some migrating cells apical to this layer (Fig 4D). These results support the proposed relationships between Atoh7 and other transcription factors in the RGC gene regulatory network (Hufnagel et al., 2010; Hutcheson and Vetter, 2001; Mu et al., 2005; Wu et al., 2015).
Fig. 4. Atoh7 overlaps established markers of retinal ganglion cell differentiation.
Comparison of Atoh7 (magenta) with [A] Neurog2, [B] pan-Brn3 (Pou4f), [C] Isl1 and [D] Tubb3 (green) in E13.5 retinal sections. Co-localized nuclear signals appear white in merged views (arrowheads). In each row, the three right panels show single-channel and merged views at higher magnification (yellow boxes). Scale bars, 100 μm.
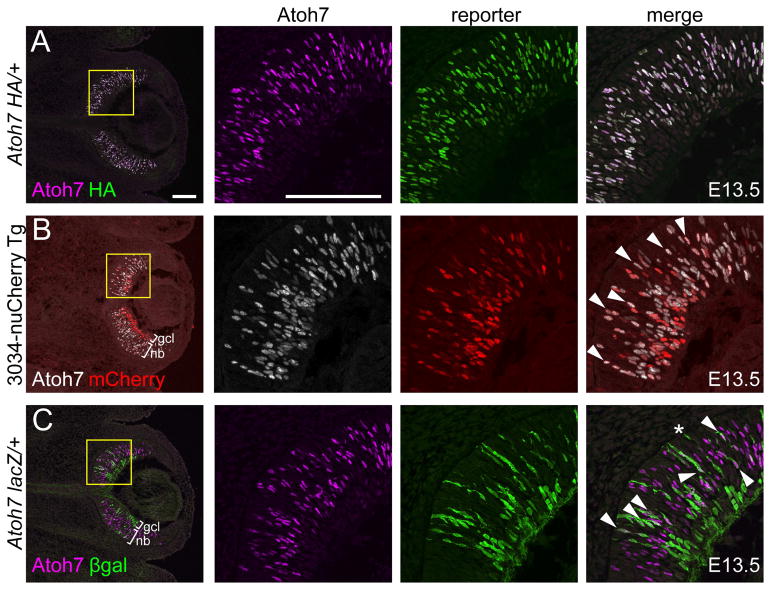
Temporally shifted expression of Atoh7 and transgene reporters
Multiple Atoh7 transgene and knock-in (KI) reporter mouse lines have been generated, including the lacZ KI, which replaces part of the Atoh7 coding sequence; an in-frame C-terminal hemaglutinin (HA) KI; and 3034-nuCherry, a transgene reporter which uses the human ATOH7 remote enhancer to drive expression of red fluorescent protein (Brown et al., 2001; Fu et al., 2009; Ghiasvand et al., 2011). Each line essentially recapitulates the endogenous Atoh7 mRNA pattern. However, the Atoh7lacZ and 3034-nuCherry lines use reporter proteins (βgal and RFP, respectively) that are larger and more stable in vivo (perdurant) than Atoh7. This is expected to cause a delay between transcription and appearance of immunoreactive protein and, because of their relatively slow turnover rates, persistence of reporters long after Atoh7 protein has been degraded. In contrast, the HA knock-in allele adds 27 residues to the Atoh7 coding sequence, which is unlikely to significantly alter translation or stability, and it utilizes endogenous UTRs (Fu et al., 2009). Because of interkinetic nuclear migration (IKNM) within the developing retinal epithelium (Baye and Link, 2007), the temporal features of each reporter create unique laminar effects.
To compare differential expression of these reporters and endogenous Atoh7, we analyzed their extent of concordance by immunostaining retinas at E13.5 (Fig 5). As expected, Atoh7 overlapped differently with each reporter. The HA knock-in (Fig 5A) most closely matched endogenous Atoh7 (100% cell concordance), followed by 3034-nuCherry (47% ± 3.8%) and the lacZ knock-in (15% ± 2.7%). In Atoh7 lacZ/+ heterozygotes, the marked shift in onset and decay of βgal expression creates three informative populations – Atoh7+ βgal-, Atoh7+ βgal+ and Atoh7- βgal+ – which are temporally related through a tracer effect. Atoh7+ βgal- cells (youngest) are restricted to the neuroblast layer, whereas Atoh7- βgal+ cells (oldest) represent differentiating neurons, e.g. RGCs in the GCL and cones at the apex (Fig 5C). Atoh7+ βgal+ cells (intermediate stage) were mainly located in the neuroblast layer, but a subset of weakly Atoh7+ cells were observed in the GCL. The Atoh7-βgal concordance (15%) is smaller than the HA-βgal comparison value (65% of HA+ cells are βgal+) reported by Kiyama et al. (2011) in Atoh7lacz/HA heterozygotes (Fu et al., 2009; Wang et al., 2001). The discrepancy may reflect experimental differences in timing (E13.5 vs. E14.5), βgal localization (nuclear vs. cytoplasmic) and/or regional sampling (central vs. temporal retina). Indeed, the number of Atoh7+ cells exceeded the number of βgal+ cells at E13.5, but the ratio was reversed at E16.5, after Atoh7 expression has peaked (Table S1), consistent with the relatively short half-life of Atoh7 protein compared to βgal. Likewise, the overlap between Atoh7 and 3034-nuCherry was highest in the neuroblast layer, and a subset of cells in the GCL were Atoh7- RFP+ (Fig 5B). In Atoh7HA/+ heterozygotes, Atoh7 and HA signals were completely overlapping, with relevant controls excluding cross-species reactivity between the anti-rabbit and -rat IgG secondary antibodies (Fig. S1). Moreover, the Novus antibody detected Atoh7 antigen in Atoh7HA homozygotes (not shown), similar to wild-type embryos, suggesting that internal epitopes are the major determinants of reactivity.
Fig. 5. Atoh7 overlaps reporters with different kinetics.
[A] Atoh7 (magenta) and HA (green) immunoreactivity are completely concordant in Atoh7 HA/+ retina. [B] Atoh7 (white) and nuCherry (red) overlap extensively in the neuroblast layer (nb) but not in the ganglion cell layer (gcl) in 3034-nuCherry transgenic retina. [C] Atoh7 (magenta) and βgal (green) overlap partly in the neuroblast layer but not the ganglion cell layer in Atoh7 lacZ/+ retina. The asterisk (*) indicates a presumptive cone. All tissues are from E13.5 embryos. In each row, the right three panels show single-channel and merged views at higher magnification (yellow boxes). Co-localized signals (arrowheads) appear white in the merged views (panels A and C). Scale bars, 100 μm.
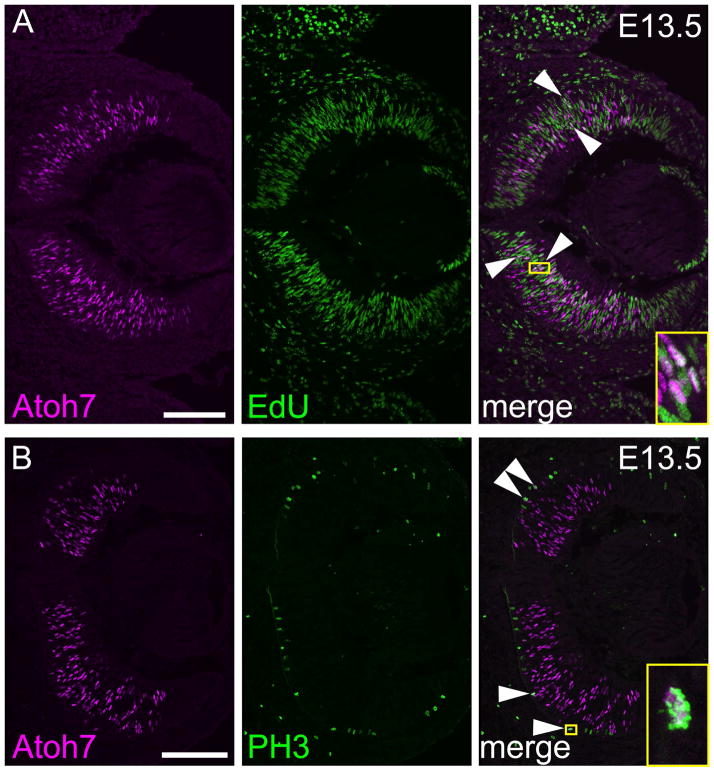
Atoh7 protein in dividing retinal progenitor cells
At the tissue level, Atoh7 is transiently expressed during retinal neurogenesis, reflecting the envelope of RGC birthdates. At the cellular level, Atoh7 is also transiently expressed in individual progenitors, beginning in S/G2 phase of the terminal cell cycle and continuing for a brief time in the post-mitotic period as differentiation proceeds (Brzezinski et al., 2012; Fu et al., 2009; Kiyama et al., 2011; Le et al., 2006). Previous studies have assessed the proliferative status of Atoh7+ cells with variable results, depending on the Atoh7 reporter (denominator) and method used to identify diving cells (numerator), and their unique kinetic properties. For example, during peak Atoh7 expression (E13.5–14.5), 3% to 25% of Atoh7+ progenitors have been reported to be in S-phase, when lacZ (slow) or HA (fast) reporters were used to mark Atoh7+ cells, respectively (Brzezinski et al., 2012; Kiyama et al., 2011). We therefore reinvestigated this question using the Novus antibody to detect endogenous Atoh7 (Fig 6). We found that 22 ± 2.3% of Atoh7+ cells in the E13.5 neuroblast layer were positive for the S-phase marker EdU following a 90 min chase (58 of 261 Atoh7+ cells, n = 3 sections). In a parallel experiment, 21 ± 5.2% of HA+ cells in E13.5 Atoh7HA/+ retinas were EdU-positive (56 of 266 HA+ cells, n = 3 sections), in close agreement with previous results (Kiyama et al., 2011). Consistent with these findings, we also detected Atoh7 protein during M-phase, in PH3+ (phosphohistone) cells at the retinal apex (Fig 6B). These mitotic cells must be in their final neurogenic division, however, since lineage-marked cells, identified using an Atoh7-Cre BAC transgene, are invariably PH3- and EdU-negative (Brzezinski et al., 2012). Our results confirm previous data showing that Atoh7 is expressed in dividing progenitors, from S/G2 to M phase of the terminal division.
Fig. 6. Coexpression of Atoh7 and proliferative markers in E13.5 RPCs.
Co-labeling E13.5 retina with Atoh7 (magenta) and [A] EdU (green) or [B] PH3 (green). A subset of Atoh7+ PH3+ cells at the apical surface of the retina are indicated (arrows). Insets (yellow) show magnified views of double-positive cells. Each panel is a composite of two images collected at 200X magnification. Scale bars, 100 μm.
DISCUSSION
In this study, we have verified the specificity of an Atoh7 antibody, which recognizes mouse Atoh7 protein in embryonic tissue and denatured protein extracts. Using this antibody, we documented the dynamic expression pattern of Atoh7 protein during retinal neurogenesis and compared the pattern with three mouse Atoh7 reporter lines. We also assessed colocalization of Atoh7 with established RGC transcription factors, lineage and cell cycle markers. Our results validate the utility of this antibody as a resource to study retinal neurogenesis and disease in mammals.
This tool is an important advance for retinal histogenesis research, which has relied on proxy methods to visualize Atoh7 for two decades. Generating and maintaining mouse lines is time consuming and expensive. The validated antibody will simplify and extend experimental approaches, allowing study of Atoh7 transcriptional regulation and protein dynamics without needing an exogenous tag or reporter line. To be sure, the Atoh7HA knock-in line completely overlaps native Atoh7, whereas lacZ and RFP are temporally shifted. These observations are generally relevant to other reporter systems. Moreover, they suggest that C-terminal addition of HA does not significantly alter the stability of Atoh7 protein in the developing retina. These dual reagents will facilitate investigation of Atoh7 protein modification in vivo. Mounting evidence in fruit flies and vertebrates suggests that some proneural bHLH factors are functionally regulated by post-translational modification (Guillemot and Hassan, 2017; Moore et al., 2002; Philpott, 2015). For example, phosphorylation of serine/threonine residues in Drosophila Atonal and Scute, and vertebrate Neurog2 and Atoh1, can change protein stability, target gene activation, and neurogenesis (Ali et al., 2011; Quan et al., 2016; Xie et al., 2017). These residues are conserved in Atoh7 and may control rates of protein turnover. Moreover, the Notch pathway may modulate the stability of proneural transcription factors via this mechanism, to integrate signaling during neurogenesis. In this way, the stability of Scute, Atonal, Neurog3 and Ascl1 proteins are affected by Notch signaling (Kiparaki et al., 2015; Qu et al., 2013; Sriuranpong et al., 2002; Weinberger et al., 2017). With validation of this Atoh7 antibody, some of these questions can now be investigated to further understand the molecular basis of retinal neurogenesis.
Experimental Procedures
Animals
Two mouse Atoh7 alleles – Atoh7lacZ (Atoh7tm1Gla) (Brown et al., 2001) and Atoh7HA (Fu et al., 2009), and two transgenic lines – 3034-BGnCherry (Ghiasvand et al., 2011) and Crx>Atoh7-ires-Cre BAC (Prasov and Glaser, 2012) – were used in this study. The targeted Atoh7lacZ mutation is a null allele, whereas the Atoh7HA allele has wild-type activity, with three HA (hemaglutinin) epitope tags appended to the C-terminus of the Atoh7 polypeptide. The BGnCherry reporter encodes a red fluorescent protein (RFP) with a nuclear localization motif; the lacZ product (βgal) is cytoplasmic. Atoh7lacZ, Atoh7HA, and Crx>Atoh7-ires-Cre congenic mice were maintained on a C57BL/6J background and genotyped as described (Brown et al., 2001; Fu et al., 2009; Prasov and Glaser, 2012). The Atoh7 3034-BGnCherry transgene was maintained on an albino outbred CD1 background and genotyped as described (Ghiasvand et al., 2011). Gestational age was determined by timed matings, with the vaginal plug date as day E0.5. Mice were bred following National Institutes of Health and Association for Research in Vision and Ophthalmology guidelines, with oversight from the UC Davis Institutional Animal Care and Use Committee.
Immunohistochemistry
Embryonic and postnatal heads (E11.5-P0) were fixed by immersion in 4% paraformaldehyde phosphate-buffered saline (PBS) for 1 h at 4°C, processed through graded 10–20% sucrose/PBS solutions, embedded in OCT (optimal cutting temperature compound) and cryosectioned at 10 μm. Immunohistochemistry was performed in Tris-buffered saline 0.1% (w/v) Tween-20 (TBST) and 4% (w/v) nonfat dry milk as described (Brown et al., 1998; Mastick and Andrews, 2001). Primary antibodies were rabbit anti-Atoh7 polyclonal (Novus Biologicals, 1:500, NBP1-88639, RRID:AB_11034390, Lot R12065), rat anti-HA monoclonal (Roche, 1:500, 3F10, RRID:AB_2314622), goat anti-mCherry polyclonal (SICGEN, 1:500, AB0040-200, RRID:AB_2333092), rat anti-βgal polyclonal (1:500, (Saul et al., 2008)), chicken anti-βgal polyclonal (AVES, 1:500, BGL-1040, RRID:AB_2313507), goat anti-PH3 polyclonal (Santa Cruz, 1:200, sc-12927, RRID:AB_2233069), goat anti-Pou4f polyclonal (Santa Cruz, 1:50, sc-6026, RRID:AB_673441), mouse anti-Tubb3 monoclonal IgG2a (Covance, 1:500, TuJ1, MMS-435P, RRID:AB_2314773), mouse anti-Isl1 monoclonal IgG2b (DSHB, 1:50, 39.4D5, RRID:AB_2314683), and mouse anti-Neurog2 monoclonal IgG2a (R&D Systems, 1:500, 117-10C6, MAB3314, RRID:AB_94808). Donkey or goat anti-IgG secondary antibodies were directly conjugated to Alexa Fluor 488 (Invitrogen, A21208), Alexa Fluor 594 (Jackson Immuno-Research, 712-586-153), Alexa Fluor 647 (Invitrogen, A21244) or Dylight 649 (Jackson ImmunoResearch, 711-495-152), immunosorbed to minimize species cross-reactivity (Fig S1), and used at 1:500 dilution. In most experiments, cell nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole, 50 μg/mL). Digital images were obtained using a Leica DM5500 microscope with SPEII confocal optics and solid state laser excitation, and were processed using Leica LASAF and Adobe Photoshop (CS5) software. All images were adjusted equivalently for brightness and contrast, and pseudocolored for optimal display.
EdU labeling and cell counts
To identify S-phase cells, pregnant dams (E13.5) were injected with 12 μg/gm EdU (5-ethynyl-2′-deoxyuridine) (Invitrogen, C10339) 90 min prior to embryo harvest. Following dissection, embryonic heads were processed and cryosectioned (10 μm) for immunohistochemistry as described above. EdU was visualized in immunostained sections by click chemistry (Click-iT Alexa Fluor 488, Thermo-Fisher). Cell counts were performed manually using Leica LASX software from single confocal plane mid-retinal sections, imaged at 200X magnification, which included the optic nerve. The percentage of Atoh7 (or HA) positive cells that were EdU-labeled was determined for each section, averaged (n = 3 sections per eye), and is provided as mean ± standard deviation (SD). All Atoh7, HA and EdU signals were coextensive with DAPI+ nuclei.
Concordance between Atoh7, RGC markers and transgene reporters was assessed by counting single- and double-positive cells in temporal retinal sections imaged at 400X, using DAPI to resolve individual βgal+ soma. Percent concordance was calculated using the formula in Table S1 and is presented as the mean ± SD (for n ≥ 2 sections).
Western analysis
Total protein was extracted from mouse tissues – embryonic (E17.5) CD1 wild-type and Atoh7 −/− retinas; adult C57BL/6J and Crx>Atoh7-ires-Cre BAC transgenic retinas; and adult C57BL/6/J cerebral cortex, cerebellum and liver – and transfected HEK293T cell cultures expressing human ATOH7 (± 6xMyc N-terminal epitope tag) or mouse Atoh7 (± 6xMyc tag) following homogenization in ice-cold PBS and low-speed (1000 × g) centrifugation. Cell pellets were lysed in RIPA buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 1% NP40) containing multiple protease inhibitors (Complete™, Roche). Protein concentration was determined by Bradford assay (Biorad, Hercules, CA). Lysates were denatured in 1X NuPAGE LDS (lithium dodecyl sulfate, pH 8.4) sample buffer (Invitrogen, NP0007) and electrophoresed through NuPAGE 4–12% polyacrylamide Bis-Tris gels (Invitrogen, Carlsbad, CA) in 2-(N-morpholino) ethanesulfonate (MES) running buffer pH 7.3 with 0.1% SDS, for 40 min at 25 V/cm with 25 μg protein per lane. Protein gels were transferred to nitrocellulose membranes (GE Life Sciences, Piscataway, NJ) in a XCell SureLock Mini-Cell (Invitrogen) at 30 V for 1 h at 25°C in NuPAGE transfer buffer (25 mM Bicine, 25 mM Bis-Tris, 1 mM EDTA, pH 7.2) with 10% methanol.. Membranes were blocked for 1 h at 25°C in 1:1 TBS:Odyssey Blocking Buffer (Licor, 927-50000) and incubated for 16 h at 4°C with primary antibodies (rabbit anti-Atoh7, Novus Biologicals, 1:1000, NBP1-88639 and mouse anti-Myc, 9E10 monoclonal, Roche, 1:1000). Membranes were then rinsed in TBST, incubated at 25°C for 1 h with secondary antibodies (IRDye 800CW donkey anti-rabbit IgG, 1:20,000, Licor 926-32213 and IRDye 680LT donkey anti-mouse IgG, 1:20,000, Licor 926-68022) in 1:1 TBS:Odyssey Blocking Buffer, and washed in TBS. Signals were detected using the infrared Odyssey imaging system (Licor, Lincoln, NE).
Cell transfection
Human embryonic kidney cells (HEK293T) were cultured in Dulbecco’s modified Eagle media (DMEM) supplemented with 10% fetal bovine serum (FBS) in a humidified 10% CO2. atmosphere at 37°C. Subconfluent cultures were co-transfected with 4 μg expression plasmid DNA, 1 μg pUS2-EGFP (control plasmid), and 25 μL Fugene6 reagent (Promega, Madison, WI) per 100 mm dish. Cell pellets were harvested 48 h after transfection, in PBS with protease inhibitors, and snap-frozen at −80°C. Human and mouse Atoh7 expression plasmids were pCS2-huATOH7, pCS2MT-huATOH7, pCS2-mAtoh7 and pCS2MT-mAtoh7, respectively (Prasov et al. 2010). The pCS2MT (myc tag) vector encodes 6 tandem N-terminal myc epitope motifs (EQKLISEEDL) (Rupp et al., 1994; Turner and Weintraub, 1994)
Supplementary Material
[A–C]To rule-out cross-species reactivity between secondary antibodies used to assess colocalization of Atoh7 and HA antigens, we performed control immunostaining experiments as described in the Methods. E13.5 Atoh7 HA/+ retinal cryosections were treated with a single primary antibody (rabbit anti-Atoh7 [A] or rat anti-HA [B], or no primary [C], in parallel, left labels) prior to treatment with both secondary antibodies (Alexa Fluor 488-conjugated anti-rat IgG or Alexa Fluor 647-conjugated anti-rabbit IgG, in parallel, top labels). The panels in each column were imaged identically. Scale bar, 50 μm.
Cell counts in each row were obtained from single-plane confocal sections imaged at 400X magnification. Each value represents the sum of three fields.
Acknowledgments
We thank Jennifer Aparicio for suggesting we test this reagent in mouse retina, Xiuqian Mu for sharing Atoh7HA mice, and the UC Davis Training Program in Vision Science. This work was supported by NIH grants T32 EY15387 (JBM), R01 EY19497 (TG), R01 EY14259 (TG) and R01 EY13612 (NLB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ali F, Hindley C, McDowell G, Deibler R, Jones A, Kirschner M, Guillemot F, Philpott A. Cell cycle-regulated multi-site phosphorylation of Neurogenin 2 coordinates cell cycling with differentiation during neurogenesis. Development. 2011;138:4267–4277. doi: 10.1242/dev.067900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio JG, Hopp H, Choi A, Mandayam Comar J, Liao VC, Harutyunyan N, Lee TC. Temporal expression of CD184(CXCR4) and CD171(L1CAM) identifies distinct early developmental stages of human retinal ganglion cells in embryonic stem cell derived retina. Exp Eye Res. 2017;154:177–189. doi: 10.1016/j.exer.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baye LM, Link BA. Interkinetic nuclear migration and the selection of neurogenic cell divisions during vertebrate retinogenesis. J Neurosci. 2007;27:10143–10152. doi: 10.1523/JNEUROSCI.2754-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, Glaser T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski JAt, Prasov L, Glaser T. Math5 defines the ganglion cell competence state in a subpopulation of retinal progenitor cells exiting the cell cycle. Dev Biol. 2012;365:395–413. doi: 10.1016/j.ydbio.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Kiyama T, Li R, Russell M, Klein WH, Mu X. Epitope-tagging Math5 and Pou4f2: new tools to study retinal ganglion cell development in the mouse. Dev Dyn. 2009;238:2309–2317. doi: 10.1002/dvdy.21974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiasvand NM, Rudolph DD, Mashayekhi M, Brzezinski JAt, Goldman D, Glaser T. Deletion of a remote enhancer near ATOH7 disrupts retinal neurogenesis, causing NCRNA disease. Nat Neurosci. 2011;14:578–586. doi: 10.1038/nn.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F, Hassan BA. Beyond proneural: emerging functions and regulations of proneural proteins. Curr Opin Neurobiol. 2017;42:93–101. doi: 10.1016/j.conb.2016.11.011. [DOI] [PubMed] [Google Scholar]
- Hufnagel RB, Le TT, Riesenberg AL, Brown NL. Neurog2 controls the leading edge of neurogenesis in the mammalian retina. Dev Biol. 2010;340:490–503. doi: 10.1016/j.ydbio.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufnagel RB, Riesenberg AN, Saul SM, Brown NL. Conserved regulation of Math5 and Math1 revealed by Math5-GFP transgenes. Mol Cell Neurosci. 2007;36:435–448. doi: 10.1016/j.mcn.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutcheson DA, Vetter ML. The bHLH factors Xath5 and XNeuroD can upregulate the expression of XBrn3d, a POU-homeodomain transcription factor. Dev Biol. 2001;232:327–338. doi: 10.1006/dbio.2001.0178. [DOI] [PubMed] [Google Scholar]
- Jameson BA, Wolf H. The Antigenic Index - a Novel Algorithm for Predicting Antigenic Determinants. Comput Appl Biosci. 1988;4:181–186. doi: 10.1093/bioinformatics/4.1.181. [DOI] [PubMed] [Google Scholar]
- Kiparaki M, Zarifi I, Delidakis C. bHLH proteins involved in Drosophila neurogenesis are mutually regulated at the level of stability. Nucleic Acids Res. 2015;43:2543–2559. doi: 10.1093/nar/gkv083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyama T, Mao CA, Cho JH, Fu XY, Pan P, Mu XQ, Klein WH. Overlapping spatiotemporal patterns of regulatory gene expression are required for neuronal progenitors to specify retinal ganglion cell fate. Vision Res. 2011;51:251–259. doi: 10.1016/j.visres.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TT, Wroblewski E, Patel S, Riesenberg AN, Brown NL. Math5 is required for both early retinal neuron differentiation and cell cycle progression. Dev Biol. 2006;295:764–778. doi: 10.1016/j.ydbio.2006.03.055. [DOI] [PubMed] [Google Scholar]
- Mastick GS, Andrews GL. Pax6 regulates the identity of embryonic diencephalic neurons. Mol Cell Neurosci. 2001;17:190–207. doi: 10.1006/mcne.2000.0924. [DOI] [PubMed] [Google Scholar]
- Moore KB, Schneider ML, Vetter ML. Posttranslational mechanisms control the timing of bHLH function and regulate retinal cell fate. Neuron. 2002;34:183–195. doi: 10.1016/s0896-6273(02)00666-9. [DOI] [PubMed] [Google Scholar]
- Mu X, Fu X, Sun H, Beremand PD, Thomas TL, Klein WH. A gene network downstream of transcription factor Math5 regulates retinal progenitor cell competence and ganglion cell fate. Dev Biol. 2005;280:467–481. doi: 10.1016/j.ydbio.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Philpott A. Multi-site phospho-regulation of proneural transcription factors controls proliferation versus differentiation in development and reprogramming. Neurogenesis (Austin) 2015;2:e1049733. doi: 10.1080/23262133.2015.1049733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasov L, Brown NL, Glaser T. A critical analysis of Atoh7 (Math5) mRNA splicing in the developing mouse retina. PLoS One. 2010;5:e12315. doi: 10.1371/journal.pone.0012315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasov L, Glaser T. Pushing the envelope of retinal ganglion cell genesis: context dependent function of Math5 (Atoh7) Dev Biol. 2012;368:214–230. doi: 10.1016/j.ydbio.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Afelik S, Jensen JN, Bukys MA, Kobberup S, Schmerr M, Xiao F, Nyeng P, Veronica Albertoni M, Grapin-Botton A, Jensen J. Notch-mediated post-translational control of Ngn3 protein stability regulates pancreatic patterning and cell fate commitment. Dev Biol. 2013;376:1–12. doi: 10.1016/j.ydbio.2013.01.021. [DOI] [PubMed] [Google Scholar]
- Quan XJ, Yuan L, Tiberi L, Claeys A, De Geest N, Yan J, van der Kant R, Xie WR, Klisch TJ, Shymkowitz J, Rousseau F, Bollen M, Beullens M, Zoghbi HY, Vanderhaeghen P, Hassan BA. Post-translational Control of the Temporal Dynamics of Transcription Factor Activity Regulates Neurogenesis. Cell. 2016;164:460–475. doi: 10.1016/j.cell.2015.12.048. [DOI] [PubMed] [Google Scholar]
- Riesenberg AN, Le TT, Willardsen MI, Blackburn DC, Vetter ML, Brown NL. Pax6 regulation of Math5 during mouse retinal neurogenesis. Genesis. 2009;47:175–187. doi: 10.1002/dvg.20479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp RA, Snider L, Weintraub H. Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 1994;8:1311–1323. doi: 10.1101/gad.8.11.1311. [DOI] [PubMed] [Google Scholar]
- Saul SM, Brzezinski JAt, Altschuler RA, Shore SE, Rudolph DD, Kabara LL, Halsey KE, Hufnagel RB, Zhou J, Dolan DF, Glaser T. Math5 expression and function in the central auditory system. Mol Cell Neurosci. 2008;37:153–169. doi: 10.1016/j.mcn.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronska-Krawczyk D, Chiodini F, Ebeling M, Alliod C, Kundzewicz A, Castro D, Ballivet M, Guillemot F, Matter-Sadzinski L, Matter JM. Conserved regulatory sequences in Atoh7 mediate non-conserved regulatory responses in retina ontogenesis. Development. 2009;136:3767–3777. doi: 10.1242/dev.033449. [DOI] [PubMed] [Google Scholar]
- Sriuranpong V, Borges MW, Strock CL, Nakakura EK, Watkins DN, Blaumueller CM, Nelkin BD, Ball DW. Notch signaling induces rapid degradation of achaete-scute homolog 1. Mol Cell Biol. 2002;22:3129–3139. doi: 10.1128/MCB.22.9.3129-3139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- Wang SW, Kim BS, Ding K, Wang H, Sun D, Johnson RL, Klein WH, Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger S, Topping MP, Yan J, Claeys A, Geest N, Ozbay D, Hassan T, He X, Albert JT, Hassan BA, Ramaekers A. Evolutionary changes in transcription factor coding sequence quantitatively alter sensory organ development and function. Elife. 2017:6. doi: 10.7554/eLife.26402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Kaczynski TJ, Sethuramanujam S, Li R, Jain V, Slaughter M, Mu X. Two transcription factors, Pou4f2 and Isl1, are sufficient to specify the retinal ganglion cell fate. Proc Natl Acad Sci U S A. 2015;112:E1559–1568. doi: 10.1073/pnas.1421535112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie WR, Jen HI, Seymour ML, Yeh SY, Pereira FA, Groves AK, Klisch TJ, Zoghbi HY. An Atoh1-S193A Phospho-Mutant Allele Causes Hearing Deficits and Motor Impairment. J Neurosci. 2017;37:8583–8594. doi: 10.1523/JNEUROSCI.0295-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[A–C]To rule-out cross-species reactivity between secondary antibodies used to assess colocalization of Atoh7 and HA antigens, we performed control immunostaining experiments as described in the Methods. E13.5 Atoh7 HA/+ retinal cryosections were treated with a single primary antibody (rabbit anti-Atoh7 [A] or rat anti-HA [B], or no primary [C], in parallel, left labels) prior to treatment with both secondary antibodies (Alexa Fluor 488-conjugated anti-rat IgG or Alexa Fluor 647-conjugated anti-rabbit IgG, in parallel, top labels). The panels in each column were imaged identically. Scale bar, 50 μm.
Cell counts in each row were obtained from single-plane confocal sections imaged at 400X magnification. Each value represents the sum of three fields.