Abstract
Aims
The spatial resolution of light microscopy is limited by the wavelength of visible light (the ‘diffraction limit’, approximately 250 nm). Resolution of sub-cellular structures, smaller than this limit, is possible with super resolution methods such as stochastic optical reconstruction microscopy (STORM) and super-resolution optical fluctuation imaging (SOFI). We aimed to resolve subcellular structures (axons, myelin sheaths and astrocytic processes) within intact white matter, using STORM and SOFI.
Methods
Standard cryostat-cut sections of subcortical white matter from donated human brain tissue and from adult rat and mouse brain were labelled, using standard immunohistochemical markers (neurofilament-H, myelin-associated glycoprotein, glial fibrillary acidic protein, GFAP). Image sequences were processed for STORM (effective pixel size 8–32 nm) and for SOFI (effective pixel size 80 nm).
Results
In human, rat and mouse, subcortical white matter high-quality images for axonal neurofilaments, myelin sheaths and filamentous astrocytic processes were obtained. In quantitative measurements, STORM consistently underestimated width of axons and astrocyte processes (compared with electron microscopy measurements). SOFI provided more accurate width measurements, though with somewhat lower spatial resolution than STORM.
Conclusions
Super resolution imaging of intact cryo-cut human brain tissue is feasible. For quantitation, STORM can under-estimate diameters of thin fluorescent objects. SOFI is more robust. The greatest limitation for super-resolution imaging in brain sections is imposed by sample preparation. We anticipate that improved strategies to reduce autofluorescence and to enhance fluorophore performance will enable rapid expansion of this approach.
Keywords: astrocytes, myelin, neurofilaments, super resolution microscopy, white matter
Introduction
The wavelength of visible light imposes a lower size limit on the objects that can be seen with light microscopy. This ‘diffraction limit’ (approximately 250 nm) is close to the diameter of a typical nerve axon. Several modalities of ‘super resolution’ light microscopy have recently been developed which bypass this diffraction limit [1]. One such approach is stochastic optical reconstruction microscopy (STORM) [2,3]. Direct STORM (dSTORM) utilizes fluorophores that can be switched into a dark (non-fluorescent) radical state (see Figure S1). Examples are Alexa568, Alexa647 and Cy5 [2–4]. When the majority of the dye molecules are switched into the dark state, individual fluorescent molecules may be observed, and their positions estimated to a precision of 15 nm or less. Repeating this process many times provides a set of molecular co-ordinates, so as to reconstruct a super-resolution image. A dSTORM image typically results from processing approximately 10 000 individual images, takes a few minutes to acquire, and affords spatial resolution of 20–50 nm (Figure S1) [5]. Achieving such high resolution, artefact-free images requires that the switching process is efficient, so that well-separated fluorophore molecules can be discriminated and precisely localized.
Super-resolution optical fluctuation imaging (SOFI) is an alternative approach which does not require single molecules to be imaged, but exploits the fluctuations in individual fluorophores to retrieve super-resolution information [6]. These fluctuations can be due to a number of behaviours commonly observed in individual fluorophores, such as triplet states, or the dark states utilized in dSTORM. This is done by calculating statistical correlations between adjacent pixels in a sequence of images. These correlations are then used to generate a new image. For example, a first-order SOFI algorithm (such as that used in this paper) will result in pixels half the size of those in the input image sequence (e.g. 80 nm rather than 160 nm). While the spatial resolution obtained is typically lower than dSTORM, SOFI is amenable to samples where molecular switching is less efficient or where fluorophore density is higher. Importantly, a SOFI analysis can be performed on the same raw data acquired in a dSTORM experiment and the results compared. In some material, SOFI can generate a useful image where dSTORM cannot.
These super-resolution fluorescence approaches have several advantages over transmission electron microscopy (TEM; see a recent review [6]). First, they permit molecular labelling, using antibodies or other reagents, as typically used in confocal or epifluorescence microscopy (immuno-TEM is possible but never routine). Second, there is potential to perform multi-channel labelling, so that multiple molecular targets can be visualized simultaneously. The sample preparation work-flow is similar to that for conventional fluorescence microscopy (see Data S1 for protocol) and thus accessible to a greater number of laboratories. Third, thicker tissue sections can be imaged, which in some samples greatly facilitates the interpretation of larger structures – such as neuronal processes – which would pass through multiple TEM sections. Fourth, the costs of super-resolution instrumentation are falling rapidly, which means that they are likely to become accessible to many more researchers. Nevertheless, TEM has an order of magnitude greater spatial resolution relative to dSTORM [7]. Super resolution methods are unlikely to replace TEM and should rather be seen as complementary approaches.
STORM has been previously applied to cell cultures in vitro [2,3,6,8–10]. STORM has also been performed in sections of rodent brain and retinal tissue [11] and has the potential to resolve sub-cellular structures within brain white matter. Here, our aim was to apply super resolution methods to standard histological sections of brain tissue. We report immunofluorescent labelling of human and rodent subcortical white matter, using established markers for axonal neurofilaments, myelin sheaths and astrocyte filamentous processes. We compare imaging with dSTORM and SOFI modalities and discuss caveats.
Methods
Human tissue
Post mortem human frontal cortical tissue was obtained from two established brain banks. One is the MRC London Neurodegenerative Diseases Brain Bank, Institute of Psychiatry, Kings College, London. The second is the University of California, Irvine Alzheimer’s Disease Research Center, Institute for Memory Impairments and Neurological Disorders, University of California, Irvine, USA. All tissue samples were donated following written informed consent by donors or their next of kin. Use of human tissue within Dr Hainsworth’s laboratory has approval from the Local Research Ethics Committee and from the UK National Research Ethics Service (East Midlands – Derby NHS Research Ethics Committee, Study number 12EM_0028).
Rat and mouse tissue
Adult Wistar rats and C57/BL6 mice were killed by the schedule 1 method (cervical dislocation) and brains rapidly removed into iso-pentane immersed in liquid nitrogen. Animal use complied with the Animals (Scientific Procedures) Act 1986 and with EU Directive 63-2010.
Primary antibodies
GFAP mouse monoclonal (clone GA5; Chemicon/Merck-Millipore, Watford, Herts., UK) was raised against purified GFAP from porcine spinal cord. Neurofilament NF200 rabbit polyclonal (Sigma-Aldrich, Poole, UK) was raised against bovine neurofilament-H (MW 200–220 KDa). Myelin proteolipid protein mouse monoclonal antibody (clone PLPC1; BioRad-AbD Serotec, Oxford, UK) was raised against PLP C-terminal hexapeptide GRGTKF. Myelin-associated glycoprotein (MAG) mouse monoclonal antibody (ab89780; Abcam, Cambridge, UK) was raised against amino acids 119–208 of Human MAG. Alexa647 and Alexa568 conjugated secondary antibodies were from Life Technologies-Molecular Probes, Paisley, UK. YOYO-1 (Molecular Probes, Paisley, UK) was stored as 1 mM in DMSO at −20°C and aliquots were diluted shortly before use.
Immunohistochemistry
Human tissue blocks and rodent brains were stored at −80°C. Tissue samples were mounted in standard cryo-mountant (OCT) at −20°C within the cryostat hood. Frozen sections (8–12 µm thick) were cut by standard cryostat methods and retrieved onto 25 mm diameter #1.5 circular cover slips (Electron Microscopy Sciences, Hatfield, PA, USA) then air-dried for 20 min and fixed with ice-cold 100% ethanol. Heat-induced antigen retrieval was performed, using a Menarini-Biocare decloaker, (120°C, 30 s, in citrate buffer pH 6). Sections were treated with a strong reducing agent, sodium borohydride (5% w/v in PBS; Sigma-Aldrich) for 30 min at room temperature, to reduce tissue autofluorescence. After at least three washes with PBS to remove NaBH4, non-specific protein binding was blocked by incubation with 3% w/v BSA (Jackson Immunochemicals, West Grove, PA, USA) in PBS-T for 1 h at room temperature. Sections were incubated with primary antibodies diluted in 3% w/v BSA in PBS-T overnight in a humidified chamber at 4°C. GFAP antibody was diluted 1:3000, NF200 antibody 1:2000, MAG 1:200. Sections were incubated with appropriate secondary antibodies conjugated to Alexa647, diluted 1:1000 in 3% BSA in PBS-T at room temperature for 1 h. Nuclear chromatin labelling with YOYO-1 (20 min, 3 nM in PBS-T) sections were immersed in ice-cold PBS-T, stored overnight at 4°C, and imaged within 24 h. For further details, see Data S1 for protocol.
Sample preparation
For super-resolution imaging, cover slips were placed in Attofluor Cell Chambers (Thermo Fisher Scientific, Waltham, MA, USA), and dSTORM buffer was added to overfill the bottom chamber. A further cover slip was then applied so as to avoid trapping air bubbles in the sample chamber. The cover slip holder was then placed on the microscope stage for observation. The buffer is made up of three stock solutions: 50 µl of enzyme stock solution (A), 400 µl of glucose stock solution (B), 50–100 µl of MEA stock solution (C) made up to 1000 µl with PBS. Enzyme stock solution A is 2 µg/ml catalase, 0.1% w/v glucose oxidase, 4 mM TCEP, 50% v/v glycerol, 25 mM KCl, 20 mM Tris–HCl (pH 7.5). Glucose stock solution (B) is 0.5 M glucose, 10% v/v glycerol in aqueous solution. MEA-stock solution (C) is 1 M MEA–HCl in aqueous solution. A, B and C are stored as aliquots at −20°C.
Super resolution data acquisition
Direct STORM (dSTORM) measurements were performed, using a custom-built total internal reflection fluorescence (TIRF) objective system based on an Olympus IX71 inverted microscope body with a UAPON 100× OTIRF objective [12]. Image sequences at 128 × 128 pixels were acquired, using an Andor iXon Ultra 897 EMCCD camera (Andor Technology Ltd., Belfast, UK), and were saved in a raw binary format. Exposure time was 10 ms and the frame interval was 19 ms. In this microscope configuration, the effective pixel size is 160 nm (in raw unprocessed data). Typically 10 000 frames were acquired in each sequence. Fluorescence was excited, using objective TIRF or highly inclined illumination, with the angle of incidence adjusted empirically to obtain the best contrast fluorescence. Laser excitation was at 488, 561 or 640 nm as appropriate for the fluorophore. The system was equipped with a quad-band dichroic and emission filter; additional bandpass emission filters were used to reduce cross-talk between fluorescence channels. For multi-colour images, the channels were acquired sequentially. Imaging at lower laser power (1–10% of maximum) was used to select promising fields of view, whereas dye ‘blinking’ was initiated by increasing to maximum power. In addition, a 405 nm laser was used to ‘re-activate’ fluorophores which had entered the dark state when the number of molecules visible in each frame fell due to photo-bleaching.
Super-resolution Image reconstruction
For dSTORM, reconstructions were performed in rain-STORM software [5], using the ‘jittered histogram’ visualization method. This approach adaptively selects the best pixel size for reconstructions based on the localization precision determined by the analysis. The software also returns information about the quality of the reconstruction including the mean localization precision and the number of molecules localized to generate the image. For SOFI, first order SOFI analyses were performed, using a Localizer implementation [13] and custom MATLAB scripts, and deconvolved by the Lucy–Richardson method, using a Gaussian point spread function (PSF). The pixel size for first-order SOFI images is 80 nm.
Simulated data for dSTORM and SOFI was generated, using a modified form of the testSTORM programme [14]. Parameters were set to emulate the experimental data, except that the time constants τon and τoff for the blinking process were varied, to illustrate the effect of overlapping localizations.
Ridge detection (ImageJ). To measure the thickness of processes, images were further analysed in FIJI/ImageJ [14,15]. Processes were identified and their widths measured using the ImageJ ‘Ridge Detection’ plugin [15] (https://doi.org/10.5281/zenodo.35440). This algorithm is a generic machine vision tool for identifying and measuring linear and curved structure in images, and this is ideal for this application. The results were saved as text files for further analysis.
Statistical analysis
Custom MATLAB scripts were used to import the process width data and to convert the width measurements from pixels to nanometres. The resulting data were then analysed statistically to derive mean widths and 95% confidence limits for each combination of species, process type and method.
Results
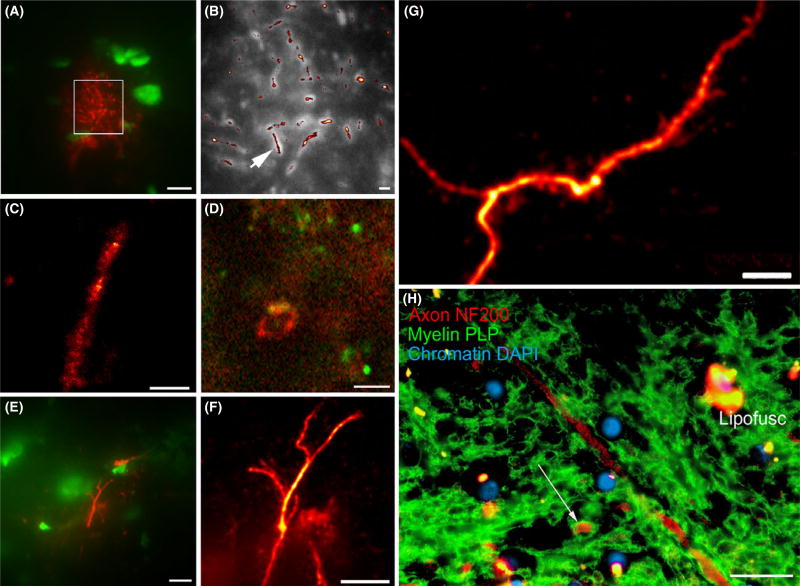
STORM imaging was achieved in frozen sections of subcortical white matter from adult mouse, adult rat and older adult human brain tissue (tissue samples used are listed in Table 1). Immunofluorescent labelling of axonal cytoskeleton (labelled with neurofilament-H), myelin sheaths (labelled with MAG) and astrocytic processes (labelled with GFAP) were clearly seen in super resolution images. Examples are shown in Figure 1A–G). The nuclear chromatin marker YOYO-1 was helpful for tissue orientation (Figure 1A,E). For comparison a multiple-labelled conventional immunofluorescence image is shown, derived from the same human tissue material (Human sample #5, Figure 1H). Tissue autofluorescence, due to lipofuschin and other tissue components, was an issue with human samples. Technical issues with STORM imaging of cryo-cut intact brain tissue are outlined in Supporting Information.
Table 1.
Tissue samples used
| Sample number |
Laboratory identifier |
M/F, age, PMI | Notes |
|---|---|---|---|
| Human #1 | C2 | Male, age 71 year. PMI: 5 h | Normal adult brain |
| Human #2 | C3 | Male, age 61 year. PMI: 53 h | Normal adult brain |
| Human #3 | C8 | Female, age 68 year. PMI: 9 h | Normal adult brain |
| Human #4 | AD10 | Male, age 71 year. PMI: 5 h | Braak stage V–VI. |
| Human #5 | UCI-41-08 | Female, age 91 year. PMI: 5 h | Aged brain |
| Mouse #1 | MS #12 | C57BL6, Male, age 10 weeks | Young wild-type mouse |
| Rat #1 | WKY 13A | Wistar, Male, age 12 months | Aged normotensive rat |
| Rat #2 | S3 | Wistar, Male, age 3 months | Young control rat |
PMI, post mortem interval (hours)
Figure 1.
Example direct stochastic optical reconstruction microscopy (dSTORM) images of axonal neurofilaments, myelin and astroglial processes within brain white matter. (A) axons immunolabelled with neurofilament (red) in human subcortical white matter, context image under conventional light microscopy. Nuclei are labelled by the chromatin marker YOYO-1 (green). Region of interest for STORM imaging marked with box. (B) STORM image (red) from A, overlaid on conventional image (greyscale). An arrow indicates the axon shown in panel C. (C) STORM imaged individual axon, from the field highlighted in panel B. (D) myelin sheath in cross-sectional profile in human sub-cortical white matter, immunolabelled with myelin-associated glycoprotein. (E) GFAP-immunolabelled astrocyte filamentous processes in mouse corpus callosum, context images under conventional light microscopy. (F) Super-resolution optical fluctuation imaging (SOFI) image of astroglial filamentous process, from field of view shown in E. (G) SOFI image of GFAP-immunolabelled astrocyte filamentous process in rat corpus callosum. (H) conventional epifluorescence image of human subcortical white matter, double labelled for axonal neurofilaments (red) and the myelin sheath protein PLP (green). Nuclear chromatin is counterstained with DAPI (blue). Note a myelinated axonal profile (arrow). Autofluorescent bundles of lipofuscin (‘Lipofusc’) are relatively sparse in this field of view. Scale bars: 10 µm (A,E), 1 µm (B), 500 nm (C), 2 µm (D,G), 5 µm (F), 20 µm (H).
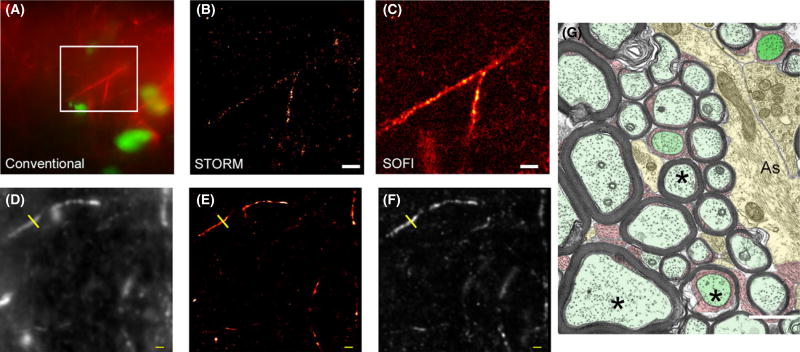
We performed quantification of specific cellular features (Figures 2,3). In particular we attempted to measure the apparent diameters of astrocyte filamentous processes (GFAP immunolabelled, example in Figure 2A–C) and apparent axonal diameter (derived from neurofilament immunolabelling, Figure 2D–F). Diameter measurements derived from dSTORM processing were internally consistent (Table 2), as were those derived from SOFI. Surprisingly, diameter measurements of any given structure differed substantially between the two modalities (Figure 3A–C; Table 2). White matter tissue from primate brain, imaged by TEM, is shown in Figure 2G for comparison.
Figure 2.
Example of quantitative measurements from direct stochastic optical reconstruction microscopy (dSTORM) and super-resolution optical fluctuation imaging (SOFI) experimental data. (A–C) Comparison of conventional epifluorescence (A), dSTORM (B) and SOFI (panel C) images of the same field of view. Rat corpus callosum immunolabelled for astrocyte filaments (GFAP, red). Nuclear chromatin is stained with YOYO-1 (green). A branched astrocytic process is clearly seen in all three images. The branching process visible in the epifluorescence image is not well resolved by dSTORM, probably because fluorophore localizations are too sparse to fully define the structure. When the same raw data are analysed by SOFI the complete structure can be seen, albeit at somewhat lower resolution. Scale bars 2 µm. (D–F) Mouse brain section immunolabelled for axonal neurofilament NF200. (D) Conventional diffraction-limited image; (E) dSTORM image; (F) SOFI image. Yellow line indicates the location of intensity profiles shown in Figure 3. Scale bar 1 µm. (G) For comparison, transverse electron microscopy images are shown for the corpus callosum of an adult monkey (age 6 year). For nerve axons (marked with asterisks, *), a wide range of axonal diameters is clearly apparent. Myelin sheaths are visible, again with a range of thickness. Astrocyte processes (labelled ‘As’) can be seen, containing bundles of filaments. Myelinated axons- pale green; astrocytes- yellow: paranodes and nodes- dark green: oligodendrocyte process- red. Scale bar 1 µm. Reproduced from Boston University website www.bu.edu/agingbrain/.
Figure 3.
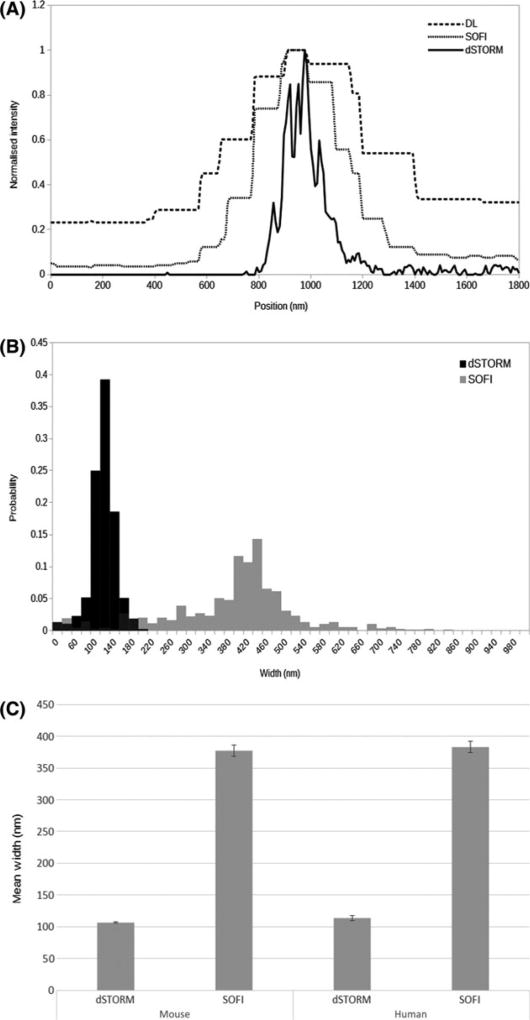
Comparison of quantitative results from direct stochastic optical reconstruction microscopy (dSTORM) and super-resolution optical fluctuation imaging (SOFI) experimental data. (A) Comparison of line profiles through an axonal process as indicated in Figure 2D–F, for diffraction-limited (dashed line) SOFI (dotted line) and STORM (solid line) images. Note the stepped nature of the diffraction-limited and SOFI graphs, due to the larger pixel sizes (160, 80 and 8 nm for diffraction-limited, SOFI and dSTORM, respectively) and the progressively narrower axonal profiles reported. (B) Normalized histogram of mean axonal widths determined for NF200 immunolabelled neurofilaments in mouse and human brain white matter, derived from dSTORM (solid bars) and SOFI analyses (grey bars). (C) Mean axonal widths, derived from neurofilament (NF200) immunolabelled mouse brain tissue by dSTORM and SOFI. Error bars indicate 95% confidence limits.
Table 2.
Diameters of axonal filaments and astrocytic processes, estimated from direct stochastic optical reconstruction microscopy (dSTORM) and super-resolution optical fluctuation imaging (SOFI) images
| dSTORM* | SOFI* | Prior literature | |
|---|---|---|---|
| Axons (neurofilament labelled) | 107 ± 1.1 nm (mouse) | 377 ± 8.5 nm (mouse) | Unmyelinated: 100–500 nm [21,25] |
| 114 ± 4.0 nm (human) | 383 ± 8.8 nm (human) | Myelinated: 300–1100 nm [20,22,25,26] | |
| Astroglial processes (GFAP labelled) | 102 ± 1.5 nm (rat)† | 492 ± 7.8 nm (rat) | 200–500 nm [20,27] |
| 388 ± 2.4 nm (human) |
Each value comes from measurements of at least two individual objects (range 2–277) estimated from multiple automated diameter measurements (range 319–9896, see Table S1). Data values shown are mean values with 95% confidence limits.
Insufficient dSTORM data for GFAP labelling in human tissue was suitable for quantitative analysis.
To estimate diameter, cross-sectional width was estimated at multiple locations along the length of a long filamentous structure (axon or astrocyte filament). For a given structure, the estimates of diameter were narrowly distributed (Figure 3A–B). The mean diameter estimates derived from human tissue were similar to those derived from mouse tissue (Figure 3C). Mean diameters derived from SOFI were substantially greater than equivalent dSTORM-derived values (Figure 3B,C; Table 2). Further details are given in Figure S2 and Table S1.
We carried out analyses of simulated data, using thin lines of different widths, to assess the accuracy of the two modalities (see Figure S3). In conditions where overlap of adjacent fluorophores is likely, dSTORM consistently under-estimated the width of thin objects (Figure S3). The likely rationale for this consistent error is overlap of the probability density functions for adjacent fluorophore molecules, resulting in cancellation of their signals (see Figures S4,S5).
Discussion
dSTORM and SOFI images of axonal neurofilament bundles, myelin sheaths and astrocytic filamentous processes were obtained from mouse, rat and human subcortical white matter. The greatest technical challenge was autofluorescence in human white matter tissue. This was mitigated by ethanol fixation (avoiding aldehydes), intensive quenching of autofluorescence with sodium borohydride, and use of a far-red fluorophore, Alexa 647 (see Data S2 for technical issues).
Other laboratories have reported super resolution images of neuronal cells in culture or in rodent brain tissue. Rat hippocampal neurone cultures were transfected with STORM-compatible fluorophores (YFP, TagBFP and mCherry) [8]. This strategy yielded high fluorescent signal throughout neuronal processes, requiring little histological processing. Axonal structural components (adducin, β-II and β-IV spectrin) have also been immunolabelled in neuronal cultures [16] and in tissue sections of mouse hippocampus, labelled with the actin marker phalloidin or immunolabelled with β-IV spectrin [16]. STORM imaging of presynaptic and postsynaptic membranes, immunolabelled with the markers Bassoon and Homer1, respectively, was performed in cryo-cut (10 µm) sections of mouse olfactory bulb and ventral orbital cortex [17]. STORM imaging of Bassoon and the glutamate receptors GluR1 and GluR2 was also achieved in 4% paraformaldehyde-fixed mouse brain tissue [18]. A combination of immunohistochemistry for the synaptic scaffolding protein gephyrin, with transgenic expression of the fluorophores GFP and YFP, was performed in mouse retinal sections [11]. This permitted detailed structural imaging of pre-and postsynaptic domains in glycinergic and GABAergic synapses within the retina. The tissue preparation protocol included refinements such as post-fixing with glutaraldehyde and epoxy resin embedding followed by ultra-thin sectioning (70 nm) [11]. Similar protocols have been used for mitochondrial labelling in 10–20 µm sections of mouse brain, heart and kidney [19].
Our measurements of axonal diameter and astroglial process diameter (Table 2) are comparable with stereology values in the EM literature [20–25]. There is a wide range of axonal diameters within any given brain region (Figure 2) though the distribution is quite similar across species (rodent, cat, primate, human) [21,23,24,26]. Unmyelinated axon diameters were in the range 100–500 nm in rodent, cat, primate and human white matter, though the majority were 100–200 nm [21]. An average width of 250 nm is reported for unmyelinated axons in the mouse corpus callosum [25]. Myelinated axonal diameters were generally larger, most in the range 300–1100 nm [21], though even greater axonal diameters are observed (up to 3 µm), especially at the axonal initial segment [20–22]. For myelinated axons within brain tissue, average widths of 460 nm are reported in murine corpus callosum [25], 620 nm in human corpus callosum [22] and 670 nm in the cingulate bundle of young monkeys [26,27]. The myelin sheath thickness is naturally variable, depending on the number of lamellae (Figure 2). Trans-myelin diameter is typically 800–1100 nm in central myelinated fibres [20,22–24,26]. In non-elderly human corpus callosum an average myelin thickness of 180 nm was measured [22]. While axonal diameter did not differ significantly between young adult and old monkeys, myelin sheaths were thicker and more variable in older monkeys [26]. EM measurements of astroglial processes also show a range of thickness (200–500 nm) in rodents and primates, Table 2 [20,27]. Fibrous astrocytes (the type most likely to be seen in our material) have thicker processes than protoplasmic astrocytes and processes that are more distant from the cell body are generally thinner [28]. The filamentous bundles within astrocytic processes are typically 150–250 nm in diameter [20,27].
There clearly is a difference between detecting a structure and accurately measuring its size. In conventional microscopy, the apparent width of a structure is given by a convolution of the true width with the PSF of the imaging system [29]. For structures much wider than the PSF accurate widths can be measured, whereas for structures narrower than the PSF, the apparent size will be spuriously enlarged. While a similar relationship applies in super-resolution microscopy, the relationship is more complex and is also sample-dependent [5]. In addition, super-resolution can result in apparent narrowing of structures [14]. This arises because the analysis software assumes that each ‘spot’ in an image corresponds to a single fluorophore. However, because the ‘blinking’ of the molecules is stochastic, occasionally adjacent fluorophore molecules will blink at the same time (Figures S4,S5), resulting in an erroneous localization (Figures S3,S5). This effect is more common where fluorophores are present at a high density, or where the switching process is inefficient. Using simulated data, we demonstrated this effect in dSTORM and compared the results with a SOFI analysis of the same data (Figure S3). It is readily apparent that SOFI is not affected by this artefactual narrowing (Figure S3). However, the lower resolution of SOFI (relative to dSTORM) means that thin structures may appear wider than is in fact the case. For example, in our data, the pixel size is 80 nm, therefore the minimum apparent size of any structure will be at least 80 nm. Therefore, the technique giving the most robust estimates of the size of features will vary depending on a) the frequency of overlapping images of fluorophore molecules b) the true size of the structure to be measured.
Our results show that it is important to critically appraise the quality of the raw data obtained from super-resolution microscopy, particularly where challenging samples are being imaged. While super-resolution microscopies have provided new insights into neuronal structure, this has been achieved almost entirely with cultured cells. Obtaining high-quality data from sectioned brain tissue, in particular human post mortem brains from elderly patients, is much more challenging due to the higher levels of background fluorescence [30–32]. The wide-field super-resolution techniques, such as SIM and STORM [6], are in general very vulnerable to background fluorescence, as they require a contrast between the patterned fluorescence (in SIM) or the single fluorophore molecules (in dSTORM) and the background in the raw data. Methods using a pinhole or 2-photon excitation may have an advantage in this scenario, but are also vulnerable to aberrations which may perturb imaging at a depth more than a few microns into a tissue section, due to variations in the refractive index and light scattering.
We conclude that while super-resolution imaging has great potential for imaging in brain sections, the experimenter needs to understand the limitations of each approach. We deduce that the dSTORM modality can underestimate diameters of thin fluorescent objects within the interact tissue samples. SOFI serves as a useful adjunct when high background or poor photo-switching makes dSTORM analysis problematic, since SOFI is more robust to the presence of overlapping single-molecule images. The biggest limitation for super-resolution imaging in brain sections is currently imposed by sample preparation. We anticipate that improved methods to reduce autofluorescence and to enhance the photo-switching performance of fluorophores will enable a rapid expansion of this approach.
Supplementary Material
Figure S1. The direct stochastic optical reconstruction microscopy (dSTORM) imaging process.
Figure S2. Comparison of automated measurement of diameter with manual measurement.
Figure S3. Simulated direct stochastic optical reconstruction microscopy (dSTORM) data is used to demonstrate the effect of overlapping localizations on the apparent widths of structures.
Figure S4. Overlapping ‘blinks’ may cause artefactual narrowing of processes.
Figure S5. Simulated images illustrate the effect of overlapping ‘blinks’ in direct stochastic optical reconstruction microscopy (dSTORM), assuming equal intensities.
Table S1. Additional statistics relating to data in Table 1 and in Figure 3 of the main text.
Data S1. Standard protocol for tissue preparation for stochastic optical reconstruction microscopy (STORM) fluorescent imaging.
Data S2. Technical issues with stochastic optical reconstruction microscopy (STORM) imaging of intact brain tissue.
Acknowledgments
This study received financial support from Alzheimer’s Research UK (ARUK PPG2014A-8) to AHH and AEK. AHH gratefully acknowledges financial support from Alzheimer’s Society (UK) (PG146/151), Alzheimer’s Drug Discovery Foundation (Project Ref 20140901) and St George’s Hospital Charity. Funding for the University of California, Irvine-ADRC is provided by NIH/NIA Grant P50 AG016573. The MRC London Neurodegenerative Diseases Brain Bank is supported by the UK Medical Research Council (Centre grant Number: NMRC/CG/NUHS/2010) and Brains for Dementia Research (BDR). Ultrastructural EM data in Figure 2 are reproduced from the following Boston University website with permission: www.bu.edu/agingbrain/ The fine structure of the ageing brain (Supported by the Institute on Aging of the National Institute of Health, grant number P 01-AG 000001).
Footnotes
Author contributions
Atticus H. Hainsworth contributed to study design, experimental work, data analysis, first draft, and the final manuscript. Sarah Lee, Peter Foot and Anita Patel also contributed to experimental work and writing of the final manuscript. Wayne W. Poon performed data analysis and writing of the final manuscript. Alex E. Knight performed study design, experimental work, data analysis and writing of the final manuscript.
Conflict of interest
All the authors have no conflict of interest.
References
- 1.Hell SW, Sahl SJ, Bates M, Zhuang X, Heintzmann R, Booth MJ, Cordes T. The 2015 super resolution microscopy roadmap. J Phys D: Appl Phys. 2015;48:443001. [Google Scholar]
- 2.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–5. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bates M, Huang B, Dempsey GT, Zhuang X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 2007;317:1749–53. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Metcalf DJ, Edwards R, Kumarswami N, Knight AE. Test samples for optimizing STORM super-resolution microscopy. J Vis Exp. 2013 Sep 6;79 doi: 10.3791/50579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rees E, Erdelyi M, Pinotsi D, Knight AE, Metcalf D, Kaminski CF. Blind assessment of localisation microscope image resolution. Opt Nanoscopy. 2012;1:12. [Google Scholar]
- 6.Sydor AM, Czymmek KJ, Puchner EM, Mennella V. Super-resolution microscopy: from single molecules to supramolecular assemblies. Trends Cell Biol. 2015;25:730–48. doi: 10.1016/j.tcb.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Henstridge CM, Jackson RJ, Kim JM, Herrmann AG, Wright AK, Harris SE, Bastin ME, Starr JM, Wardlaw J, Gillingwater TH, Smith C, McKenzie CA, Cox SR, Deary IJ, Spires-Jones TL. Post-mortem brain analyses of the Lothian Birth Cohort 1936: extending lifetime cognitive and brain phenotyping to the level of the synapse. Acta Neuropathol Commun. 2015;3:53. doi: 10.1186/s40478-015-0232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lakadamyali M, Babcock H, Bates M, Zhuang X, Lichtman J. 3D multicolor super-resolution imaging offers improved accuracy in neuron tracing. PLoS ONE. 2012;7:e30826. doi: 10.1371/journal.pone.0030826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cella ZF, Lavagnino Z, Perrone DM, Del BA, Furia L, Faretta M, Diaspro A. Live-cell 3D super-resolution imaging in thick biological samples. Nat Methods. 2011;8:1047–9. doi: 10.1038/nmeth.1744. [DOI] [PubMed] [Google Scholar]
- 10.Vaziri A, Tang J, Shroff H, Shank CV. Multilayer three-dimensional super resolution imaging of thick biological samples. Proc Natl Acad Sci U S A. 2008;105:20221–6. doi: 10.1073/pnas.0810636105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sigal YM, Speer CM, Babcock HP, Zhuang X. Mapping synaptic input fields of neurons with super-resolution imaging. Cell. 2015;163:493–505. doi: 10.1016/j.cell.2015.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erdelyi M, Rees E, Metcalf D, Schierle GS, Dudas L, Sinko J, Knight AE, Kaminski CF. Correcting chromatic offset in multicolor super-resolution localization microscopy. Opt Express. 2013;21:10978–88. doi: 10.1364/OE.21.010978. [DOI] [PubMed] [Google Scholar]
- 13.Dedecker P, Duwe S, Neely RK, Zhang J. Localizer: fast, accurate, open-source, and modular software package for superresolution microscopy. J Biomed Opt. 2012;17:126008. doi: 10.1117/1.JBO.17.12.126008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sinko J, Kakonyi R, Rees E, Metcalf D, Knight AE, Kaminski CF, Szabo G, Erdelyi M. TestSTORM: simulator for optimizing sample labeling and image acquisition in localization based super-resolution microscopy. Biomed Opt Express. 2014;5:778–87. doi: 10.1364/BOE.5.000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steger C. An unbiased detector of curvilinear structures. IEEE Trans Pattern Anal Mach Intell. 1998;20:113–25. [Google Scholar]
- 16.Xu K, Zhong G, Zhuang X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 2013;339:452–6. doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dani A, Huang B, Bergan J, Dulac C, Zhuang X. Superresolution imaging of chemical synapses in the brain. Neuron. 2010;68:843–56. doi: 10.1016/j.neuron.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beaudoin GM, III, Schofield CM, Nuwal T, Zang K, Ullian EM, Huang B, Reichardt LF. Afadin, a Ras/Rap effector that controls cadherin function, promotes spine and excitatory synapse density in the hippocampus. J Neurosci. 2012;32:99–110. doi: 10.1523/JNEUROSCI.4565-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barna L, Dudok B, Miczan V, Horvath A, Laszlo ZI, Katona I. Correlated confocal and super-resolution imaging by VividSTORM. Nat Protoc. 2016;11:163–83. doi: 10.1038/nprot.2016.002. [DOI] [PubMed] [Google Scholar]
- 20.Peters A, Palay SL, Webster HF. The Fine Structure of the Nervous System. W.B. Saunders Company; Philadelphia, PA, USA: 1976. [Google Scholar]
- 21.Wang SS, Shultz JR, Burish MJ, Harrison KH, Hof PR, Towns LC, Wagers MW, Wyatt KD. Functional tradeoffs in white matter axonal scaling. J Neurosci. 2008;28:4047–56. doi: 10.1523/JNEUROSCI.5559-05.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wohlschlaeger J, Wenger E, Mehraein P, Weis S. White matter changes in HIV-1 infected brains: a combined gross anatomical and ultrastructural morphometric investigation of the corpus callosum. Clin Neurol Neurosurg. 2009;111:422–9. doi: 10.1016/j.clineuro.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 23.Tang Y, Nyengaard JR, Pakkenberg B, Gundersen HJ. Age-induced white matter changes in the human brain: a stereological investigation. Neurobiol Aging. 1997;18:609–15. doi: 10.1016/s0197-4580(97)00155-3. [DOI] [PubMed] [Google Scholar]
- 24.Riise J, Pakkenberg B. Stereological estimation of the total number of myelinated callosal fibers in human subjects. J Anat. 2011;218:277–84. doi: 10.1111/j.1469-7580.2010.01333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sturrock RR. Myelination of the mouse corpus callosum. Neuropathol Appl Neurobiol. 1980;6:415–20. doi: 10.1111/j.1365-2990.1980.tb00219.x. [DOI] [PubMed] [Google Scholar]
- 26.Bowley MP, Cabral H, Rosene DL, Peters A. Age changes in myelinated nerve fibers of the cingulate bundle and corpus callosum in the rhesus monkey. J Comp Neurol. 2010;518:3046–64. doi: 10.1002/cne.22379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peters A, Sethares CF. The Fine Structure of the Aging Brain. Boston University; 2016. [last accessed 7 July 2016]. Available at: www.bu.edu/agingbrain/ [Google Scholar]
- 28.Oberheim NA, Takano T, Han X, He W, Lin JH, Wang F, Xu Q, Wyatt JD, Pilcher W, Ojemann JG, Ransom BR, Goldman SA, Nedergaard M. Uniquely hominid features of adult human astrocytes. J Neurosci. 2009;29:3276–87. doi: 10.1523/JNEUROSCI.4707-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schermelleh L, Heintzmann R, Leonhardt H. A guide to super-resolution fluorescence microscopy. J Cell Biol. 2010;190:165–75. doi: 10.1083/jcb.201002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsuchida M, Miura T, Aibara K. Lipofuscin and lipofuscin-like substances. Chem Phys Lipids. 1987;44:297–325. doi: 10.1016/0009-3084(87)90055-7. [DOI] [PubMed] [Google Scholar]
- 31.Jones PB, Rozkalne A, Meyer-Luehmann M, Spires-Jones TL, Makarova A, Kumar AT, Berezovska O, Bacskai BB, Hyman BT. Two postprocessing techniques for the elimination of background autofluorescence for fluorescence lifetime imaging microscopy. J Biomed Opt. 2008;13:014008. doi: 10.1117/1.2837169. [DOI] [PubMed] [Google Scholar]
- 32.Boellaard JW, Schlote W, Hofer W. Species-specific ultrastructure of neuronal lipofuscin in hippocampus and neocortex of subhuman mammals and humans. Ultrastruct Pathol. 2004;28:341–51. doi: 10.1080/019131290882330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The direct stochastic optical reconstruction microscopy (dSTORM) imaging process.
Figure S2. Comparison of automated measurement of diameter with manual measurement.
Figure S3. Simulated direct stochastic optical reconstruction microscopy (dSTORM) data is used to demonstrate the effect of overlapping localizations on the apparent widths of structures.
Figure S4. Overlapping ‘blinks’ may cause artefactual narrowing of processes.
Figure S5. Simulated images illustrate the effect of overlapping ‘blinks’ in direct stochastic optical reconstruction microscopy (dSTORM), assuming equal intensities.
Table S1. Additional statistics relating to data in Table 1 and in Figure 3 of the main text.
Data S1. Standard protocol for tissue preparation for stochastic optical reconstruction microscopy (STORM) fluorescent imaging.
Data S2. Technical issues with stochastic optical reconstruction microscopy (STORM) imaging of intact brain tissue.