Abstract
There exits strong correlation between the extended poly-glutamines (polyQ) within exon-1 of Huntingtin protein (Htt) and age onset of Huntington’s disease (HD), however, the underlying molecular mechanism is still poorly understood. Here we apply extensive molecular dynamics simulations to study the folding of Htt-exon-1 across five different polyQ-lengths. We find an increase in secondary structure motifs at longer Q-lengths, including β-sheet content that seems to contribute to the formation of increasingly compact structures. More strikingly, these longer Q-lengths adopt super-compact structures as evidenced by a surprisingly small power-law scaling exponent (0.22) between the radius-of-gyration and Q-length that is substantially below expected values for compact globule structures (~0.33) and unstructured proteins (~0.50). Hydrogen bond analyses further revealed that the super-compact behavior of polyQ is mainly due to the “glue-like” behavior of glutamine’s sidechains with significantly more sidechain-sidechain H-bonds than regular proteins in the Protein Data Bank (PDB). The orientation of the glutamine sidechains also tend to be “buried” inside, explaining why polyQ domains are insoluble on their own.
Introduction
Huntington’s disease (HD) is caused by the expansion of nucleotide CAG repeats in exon-1 of the HD gene, a mutation that encodes an elongated polyglutamine tract (polyQ) within the N-terminal region of the Huntingtin (Htt) exon-1 protein 1. In adult-onset HD, the pathogenic threshold polyQ length is above 36 glutamine (Q) repeats 1,2. Although a negative correlation between polyQ length and the onset age of HD symptoms has been well established 2–4, the mechanisms by which extended polyQ regions cause neuronal dysfunction and cell death remain unclear. Mutant Htt (mtHtt) variants (here we call Q-lengths above 36 as mutants, and Q22 as the “wild-type” wtHtt; more below) are known to form oligomers that aggregate into large, insoluble protein complexes (such as amyloid fibrils) that were previously believed to constitute the principle source of toxicity in HD 5. Recent in situ immunocytochemistry assays, however, have revealed that monomeric and lightweight oligomeric forms of mtHtt are also associated with neuronal death 6. As these experimental results emphasize, elucidating the relationship between polyQ length and protein structure is crucial for understanding the driving forces behind mtHtt oligomer formation and identifying the processes by which these oligomers and larger aggregates lead to neural toxicity.
The Htt exon-1 protein is intrinsically disordered and, as a direct consequence, difficult to characterize structurally. Exon-1 monomers can access a range of structures at physiological temperatures, and it is not known which of these configurations form the basis for toxicity. Previous studies have focused on determining the structures of individual exon-1 domains, including 1) its 17-residue N-terminal region (N17), 2) its polyQ tract, and 3) its 38-residue, polyproline-rich segment near its C-terminus (C38). Experimentally determined structures suggest that N17 folds into an amphipathic α-helix when it is attached to protein tags or placed in a micellar environment 7–9. Nuclear magnetic resonance (NMR) measurements, however, indicate that N17 is intrinsically disordered in solution 10; however, it has been suggested the N17 helices in solution may aggregate to initialize polyQ aggregation11. The polyQ region is also intrinsically disordered in solution, but collapses into a compact globule when both N17 and C38 are absent and the Q-length is 20 or greater 5,12–16. Notably, the polyQ region within an Htt36Q3H variant of Htt (polyQ= Q7HQHQHQ27) was found to adopt β-hairpin configurations, which may play a role in the formation of mtHtt fibrils 15,17.
Molecular dynamics (MD) simulations have well complemented experimental studies by sampling ensembles of possible intermediate states 18–20, a scheme that is particularly crucial for understanding the thermodynamics of intrinsically disordered proteins (IDPs). In this regard, computational studies have provided fruitful insight into the structures of N17 and various polyQ domains 21–24. However, due to the intrinsic complexity of these large IDPs and limitations on computational resources, these early simulation efforts have yielded mixed results (despite numerous attempts with different simulation techniques involving discrete molecular dynamics engines, implicit solvents, low resolution coarse-grained (CG) models, etc). Some of these computational studies have suggested that α-helices dominate the secondary structural profile of monomeric Htt exon-1 21–23, while others have reported significant β-sheet formation in the monomer that increases with Q-length 24. These contradictory observations highlight the need for more extensive simulations conducted with higher resolution models, such as fully atomistic protein representations simulated in explicit solvent.
In this article, we present the results from all-atom, explicit solvent MD simulations of the “wild type” (Q22) and representative “mutant” (Q36, Q40, Q46, and Q56) Htt proteins. To effectively capture the many configurations these IDPs adopt, we carried out extensive temperature replica exchange molecular dynamics (T-REMD) simulations across 128 replicas for at least 500 ns per replica, a technique that substantially improves sampling diversity while maintaining thermodynamic rigor. By leveraging this enhanced sampling to estimate rugged conformational free energy landscapes over a large set of Q-lengths, we are able to codify the Q-length-dependent structural and polymeric properties of Htt exon-1. Intriguingly, we find that the propensity to form β-sheet content increases with increasing Q-length, suggesting that mtHtt oligomerization and fibril formation might be influenced by this trend. This increase in β-sheet content within the polyQ region leads to a decrease in the number of hydrogen bonds between the N17 and C38 domains, an observation consistent with recent FRET experiments 25. Our analysis of the polymeric properties of Htt reveals that the wild type Htt (wtHtt) is more flexible than mtHtts, which favor more condensed structures. The scaling behavior for the polyQ segment’s radius of gyration (Rg) implies that at high Q-lengths, the polyQ region adopts super-compact structures not seen in other globular proteins. This increased compactness at long Q-lengths suggests that extended polyQ domains may lead to increased neurotoxicity both by inducing distinct morphological changes throughout the entire exon-1 protein. One may also suspect that the augmented β-content within and suppressed radial extension of the polyQ domain would impact Htt’s interactions with itself, nearby exon-1 monomers, and its distinct protein binding partners.
Results
Secondary Structural Analysis
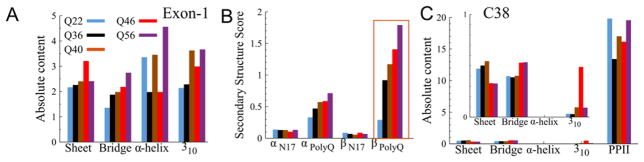
Experimental studies of polyQ chain aggregation have suggested that monomeric proteins with β-sheet content associated with longer Q-lengths may play a role in fibril formation14,26. Artificially constructed β-sheet motifs in polyQ domains have also been shown to increase the rate of polyQ aggregation 27, suggesting that the appearance of β-sheet structures in the polyQ domain represents a key step for the formation of exon-1 fibrils. Here, we have indeed found that the amount of β-sheet content in exon-1 increases with Q-length, as Fig. 1A illustrates. The plot shows that, in general, all of the mtHtts (Q ≥ 36) adopt structures with more residues in β-sheet conformations than are seen in the wtHtt (Q22) structural ensemble. At the extreme, approximately twice as many residues assume β-sheet conformations in the Q46 mutant as compared to the wtHtt. Helical structures are more abundant in the mtHtts, as well.
Figure 1.
Secondary structural analysis shows that the β-sheet content of Huntingtin (Htt) increases with Q-length faster than the α-helical content. (A) Absolute secondary structure content of the full Htt exon-1 for Q22, Q36, Q40, Q46, and Q56 (B) The average secondary structure scores of first 17 residues near N-terminus (N17) and polyglutamine (polyQ) regions of Htt for all Q-lengths. A sharp increase in β-sheet content emerges between Q22 and the pathogenic threshold Q-length, Q36. The estimated errors are less than 3%. (C) Absolute secondary structure content of the proline-rich segment near C-terminus (C38) for all Q-lengths. Because the content of the polyproline II (PPII) helices is so high, the inset shows other secondary structural motifs on a reduced scale.
Because of the transient nature of secondary structure in IDPs, we employed the continuous secondary structure scoring functions developed by Pietrucci and Laio in our analysis 28. These functions assign a continuous β-sheet or α-helix score based on the root mean square deviation between the given structure and an ideal structure determined from the CATH database (http://www.cathdb.info/). A value of 1.0 represents a sliding segment of six residues having very high similarity to the idealized structure. The analog nature of these scoring functions allows us to compare Q-length dependent trends with observed experimental results. The secondary structure scores for α-helices and β-sheets within polyQ domains of all Q-lengths are shown in Fig. 1B. Both α-helix and β-sheet contents increase with Q-length inside the polyQ region. The increase in α-helix content is consistent with the recent Time-Resolved Fluorescence Energy Transfer (TR-FRET) and Electron Paramagnetic Resonance (EPR) data from Langen and co-workers 17. Fig. 1B shows a monotonic increase α-helix content with Q-length that is similar to experimentally observed trends26,29. The observed increase in β-sheet content with increasing Q-length, however, is greater than the increase in α-helix content, especially between Q22 and Q36. This rise in β content has been seen in polyQ studies that found that increasing Q-lengths lead to increases in β-sheet transitions14,26. In particular, the monotonic increase in secondary structure content is manifested only when using a continuous measure of secondary structure (Fig. 1B) and not a binary secondary structure measure (Fig. 1A). This indicates that the emerging α-helix and β-sheet content in longer Q-lengths is transient rather than static.
By contrast, we find that secondary structural motifs within N17 and C38 do not change significantly with increasing Q-length. Fig. 1B illustrates that both α-helix and β-sheet scores for N17 are low in the wild type and remain so in the mtHtts. These results suggest that the N17 domain of the exon-1 monomer is mostly disordered in solution, an observation consistent with previous NMR measurements 10. Compared to N17, C38 is relatively rigid due to the polyproline chains that dominate its sequence. We assigned the left-handed polyproline II helix (PPII) designation to any two sequential proline residues within C38 that adopt backbone dihedral angles (ϕ, ψ) inside the range (−75°±15°, 150°±15°). C38’s polyproline chains assume PPII secondary structures to a large and effectively constant extent among wt and mtHtts (Fig. 1C) 30, a result also consistent with previous experiments 31.
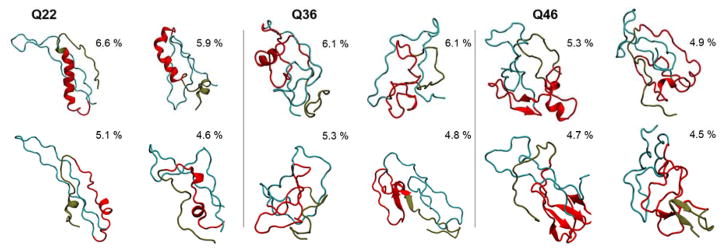
As noted above, β-sheet rich conformations, in particular, become more prevalent as Q-length increases. To further illustrate this trend, Fig. 2 shows high-probability structures derived from clustering analyses of three representative polyQ domains: Q22 (wtHtt), Q36 (mtHtt at the pathogenic threshold), and Q46 (mtHtt far above the pathogenic threshold). Clusters were derived from the GROMACS 5.0.5 cluster analysis tool 32 based on the algorithm developed by Daura et al 33, applying a 3.5 Å RMSD cut-off. The images highlighted in Fig. 2 represent the four most populated clusters within each Q-length’s dataset. The wild type Q22 exhibits more propensity to form helical structures, although these structures are transient, with the majority of sampled structures being disordered. A recent solution NMR study found a propensity to form helical structures in an N17Q17P6 model of Htt exon-1 at low pH and disorder protein behavior at neutral pH34. Additionally, we found that the Q22 variant features more elongated C38 configurations than seen in Q36 and Q46. The structures of Q36 are the most disordered among the three Q-lengths, though some β-sheet motifs do start to appear within its polyQ domain. Q46 configurations, however, feature pronounced β-sheet content, particularly within the third cluster wherein four β-strands coalesce inside the polyQ tract. These types of β-sheet conformations may represent structural motifs capable of seeding the oligomerization processes seen in some polyQ aggregation studies 14,26,27. Although interactions between helical N17 have been shown to be an important nucleation event in Htt exon-1 oligomerization and may be the most kinetically favorable process29,35, competing pathways have been suggested where the Htt exon-1 monomer misfolds into a β-sheet conformation that then aggregates via β-sheet interactions35–37. Although these β-sheet dependent oligomerization pathways may be much less kinetically favorable, they may contribute to the polymorphic nature of the Htt exon-1 aggregates and may play an important role in its toxicity.
Figure 2.
Representative structures determined from clustering analysis of Q22, Q36, and Q46 datasets. The populations of each cluster, as a percentage from sampled conformations, is shown above each structure. In general, as the Q-length increases, the β-sheet content of the polyQ region increases. The N17 domain can also form more β-sheet structures as the Q-length increases. The N17 region is shown in tan, the polyQ in red, and the C38 in cyan. The proteins are rendered using the New Cartoon representation.
Intra- and interdomain tertiary contacts
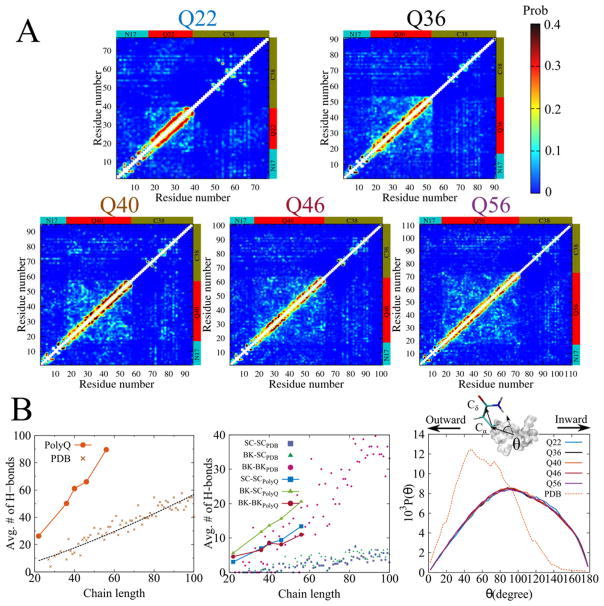
In order to characterize the tertiary contacts between the different domains of the exon-1 monomer, we calculated the average number of hydrogen bonds occurring among all residue pairs (Fig. 3A) and in summation within and among various domains (Fig. 3B). In general, we did not see any persistent contacts among individual residues (Fig. 3A). When examining the average total numbers of hydrogen bonds, however, the polyglutamine region displays an unusually high hydrogen bonding propensity in relation to typical proteins (Fig. 3B, left panel). To conduct this comparison, we calculated the total number of hydrogen bonds within all standard globular α/β proteins in the Protein Data Bank (PDB), limiting our computation to structures that have at least 20% α-helical and 20% β-sheet content. Strikingly, the number of hydrogen bonds within the polyQ region of exon-1 increases much more rapidly than it does in globular proteins, as a function of chain length (Fig. 3B, left panel). These greatly enhanced hydrogen bonding characteristics likely result from a higher propensity for glutamine residues to form hydrogen bonds among themselves rather than with water. The mid panel of Fig. 3B shows that sidechains of the polyQ form more sidechain-sidechain and sidechain-backbone hydrogen bonds than general structured proteins with comparable sizes from PDB. Interestingly, the number of sidechain-backbone interactions within the polyQ follows a trend similar to the number of backbone hydrogen bonds found in structured proteins. This observation demonstrates glutamine sidechain’s “glue”-like property (glutamine acting as “glue”), caused by strong hydrogen-bonding tendency with backbones as well as other GLN sidechains. Furthermore, when only buried glutamines are considered, defined as amino acids having less than 4 water molecules within 3Å, the increase in sidechain-sidechain hydrogen bonds becomes even more prominent (Fig. S1), which implies strong intra-hydrogen bonding propensity of polyQ. Because of this strong intra-hydrogen bonding, the sidechains of glutamine residues are more likely to point inward to the center of polyQ. In contrast, in general structured proteins, the sidechains of glutamine residues mostly point outward (right panel of Fig. 3B). To further illustrate this point, we analyzed the average number of sidechain-water and sidechain-protein hydrogen bonds for surface GLN residues in polyQ and in globular proteins (Table. S1). Surface GLN residues were defined as having at least 6 water molecules within 3Å of the residue. As expected, we did find that polyQ surface GLN residues have more sidechain-protein hydrogen bonds than the globular proteins; while the sidechain-water hydrogen bonds are significantly less, approximately 1.3 less on average, as compared to globular proteins (Table S1). This suggests that surface GLN residues in polyQ have a lower propensity to interact with water and a higher propensity to face inward than GLN residues in globular proteins. This helps to explain the insolubility of polyQ chains without flanking domains. The less “intrusive” surface GLN sidechains to the first solvation shell are also beneficial to the overall folding free energy due to the less disruption of water hydrogen bonding network by the GLN planar sidechain amide groups38,39.
Figure 3.
Hydrogen bond analysis shows that the contacts between N17 and C38 are reduced as the Q-length is increased. Panel (A) Pairwise residue hydrogen bonding probability map for all Q-lengths. Panel (B) (Left) Average number of hydrogen bonds in the polyQ region for all Q-lengths (orange circles). For comparison, the average numbers of hydrogen bonds in globular proteins selected from PDB structures are also plotted (brown crosses). The dashed line is a power-law fit with the scaling exponent of 1.28. As seen in the figure, the number of hydrogen bonds within the polyQ region increases at a faster rate than in other globular proteins. (Middle) Average number of sidechain-sidechain (blue square), backbone-sidechain (green triangle) and backbone-backbone (red circle) hydrogen bonds for polyQ and PDB proteins. The sidechains of polyQ show more hydrogen-bonding propensity than the PDB proteins with comparable sizes. (Right) Sidechain orientation with respect to protein surface. θ is an angle between a vector from center of mass of a protein to Cα atom and another vector from Cα to Cδ. Sidechain of GLN in polyQ tends to point inward as compared to general PDB proteins, suggesting that the GLN residues will not be available to form favorable interactions with the solvent.
Figure S2A shows the average total numbers of hydrogen bonds between N17 and polyQ and C38 and polyQ. Both sets of interdomain hydrogen bonds also increase as Q-length increases, indicating augmented interactions between N17 and C38 with the central polyQ domain. Interestingly, hydrogen bonding between the N17 and C38 regions actually decreases to a small extent as Q-length increases (Fig S2B).
Recent FRET experiments by Caron et al. indicated that the N17 and C38 domains of exon-1 are spatially proximate in Htts of Q < 36 but distant in mutants at Q ≥ 36 25. As a result, Caron et al. surmised that the polyQ region in wtHtt favors disordered states whereas the polyQ in mtHtts tends to form β-sheets. Such a secondary structural shift in polyQ should result in a decrease in contacts between N17 and C38, as we indeed observed in our MD data. To further confirm this correlation, we plotted 2D histograms of N17-C38 hydrogen bond counts against α-helix or β-sheet scores for the polyQ region (Fig. S3), and we estimated correlation coefficients using simple linear regression. These calculations clearly demonstrate that the number of hydrogen bonds between N17 and C38 is inversely correlated with β-sheet content in the polyQ region (Fig S3A). The negative correlation coefficients between β-sheet content and N17-C38 hydrogen bonding, though moderate, seem to generally increase with Q-length (Q56 being a notable exception). By contrast, α-helix scores show very little correlation with hydrogen bonding between N17 and C38 (Fig. S3B).
Wild-type Htt exhibits more diverse conformations than mutant Htts
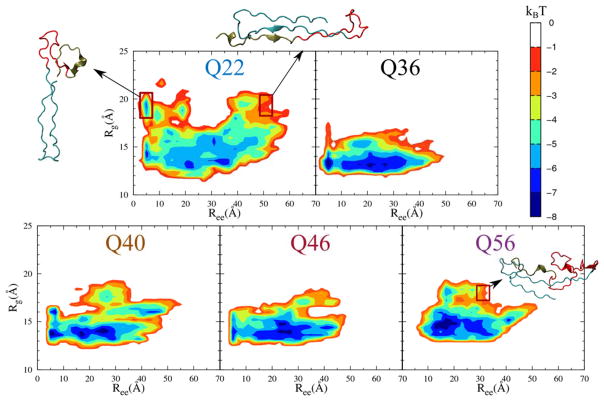
We also computed potentials of mean force (PMFs) for the full exon-1 monomer at all Q-lengths, projected onto radius of gyration (Rg) and end-to-end distance (Ree) order parameters (Fig. 4). Specifically, the PMFs were calculated as F(Rg, Ree) = −kBT [ln P(Rg, Ree) − ln P min (Rg, Ree)], where kB is the Boltzmann constant, T is room temperature, P(Rg, Ree) is the probability of a given (Rg, Ree) bin, and Pmin is the non-zero minimum value in the 2D probability distribution. Put in simple terms, the PMF for Q22 covers a much wider area than any of the other PMFs corresponding to mtHtts. This wide range is partially due to Q22 being able to sample structures that include an extended C38 domain (Fig. 4 Q22), which is not seen in longer Q-lengths (Fig. 4, Q56). This observation also suggests that wtHtt is more flexible and adopts a more diverse set of conformations than the longer polyQ mutants, which favor more compact structures.
Figure 4.
Two-dimensional PMFs projected onto Ree and Rg for the full exon-1 across all Q-lengths. The PMFs are plotted as differences from a global PMF maximum. Q22 has much wider distributions of Rg and Ree values than the other Q-lengths, indicating a more flexible structure. The reduction in the sampled Rg and Ree regions indicates that increases in Q-length lead to more compact structures. For Q22, the overlaid representative structures from the highlighted regions show that the maximum Rg values can be sampled from both the minimum and maximum Ree values, indicating that the extended C38 region is very flexible. On the other hand, the representative structure highlighted in Q56 shows that the protein is relatively compact with the end-end distance less variant even for maximum Rg structures.
Scaling behavior of polyglutamines
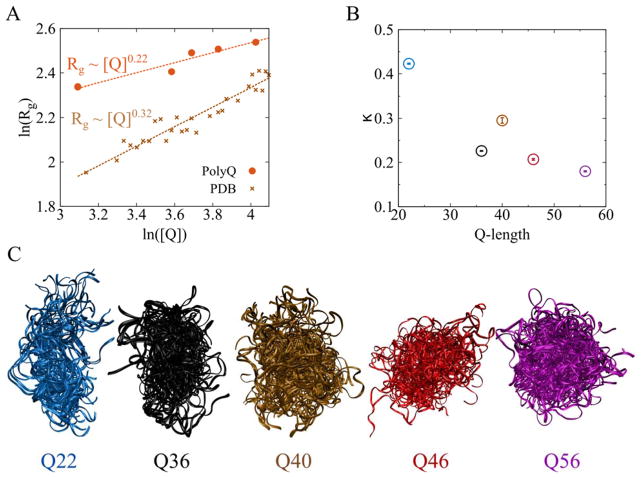
Intrigued by this striking compactness in mtHtts, we next investigated the scaling behavior of polyQ packing density as a function of Q-length.. For purposes of comparison, we calculated the average Rg, as a function of chain length, for both polyQ domains studied here and globular protein structures taken from the PDB. Interestingly, we find that as Q-length increases, the compactness of each polyQ domain (as evaluated by Rg) increases at a much faster rate than expected for standard proteins or polymers (Fig. 5A). The power-law scaling exponent for the Rg of a structured globular protein is expected to be 0.33 40, while the scaling exponent for a relatively unfolded (well-solvated) protein is expected to be ~0.5 (0.60 for an ideal polymer in good solvent) 41,42. Our calculation shows that the proteins from PDB meet these theoretical expectations (Fig. 5A, dashed brown line); however, the scaling exponent for polyQ is estimated at a surprisingly low 0.22, despite that glutamine itself is hydrophilic. These scaling data indicate a Q-length dependent super-compactness in polyQ, a phenomenon that most likely results from the strong internal hydrogen bonding between glutamine residues (Fig. 3B).
Figure 5.
Increasing Q-length leads to super-compact conformations in the polyQ region. (A) Radii of gyration for the polyQ region (orange ⊙) and structured proteins from the PDB (brown ×), as a function of chain length. The dashed lines indicate power-law fitting. The scaling exponents are displayed in the figure, showing that the polyQ will adopt super-compact structures at longer Q-lengths. (B) Relative anisotropy, κ, for all Q-lengths. κ roughly decreases as Q-length increases except for Q40, which indicates that the chain becomes more spherical. (C) Ensemble of polyQ structures. 40 randomly selected polyQ structures are rendered together to illustrate the morphological change in polyQ with increasing Q-length. Q22 has a more aspherical shape than the other mtHtts, and the shape of the protein becomes more spherical as Q-length increases.
Along with this super-compactness, we also find that the shape of the polyQ tract becomes more spherical as the Q-length increases. In order to assess such a change, we estimated the relative anisotropy parameter, κ, which measures the shape anisotropy of a polymer. For perfect spheres, κ=0; for a perfectly straight polymer, κ=1. Fig. 5B shows that κ generally decreases with increasing Q-length, indicating that the polyQ changes from a rod-like morphology to a spherical shape at longer Q-lengths. The polyQ structural ensembles shown in Fig. 5C — each generated from 40 random snapshots — illustrate this trend.
DISCUSSION
In this work, we performed the first systematic, all-atom MD study of the full Huntingtin exon-1 protein across five different polyglutamine segment lengths. These simulations have provided new insights into the effects of elongated polyQ tracts on Htt’s structure, hinting at the molecular basis for the relationship between CAG repeats and Huntington’s disease pathology. As Q-length is increased, the Htt exon-1 proteins exhibit more β-sheet content in general, which acts to reduce contact between the flanking N17 and C38 domains and results in enhanced compactness in the polyQ region. As such, the toxicity of the elongated polyQ tract can perhaps be partially attributed to two sources: aggregation that is aided by an increase β-sheet propensity and an increase in compactness that likely changes the ways in which Htt interacts with itself, other Htts, and distinct protein binding partners.
Our clustering analysis highlights the emergence of β-sheet rich conformations with increasing Q-length. We find that the Q22 polyQ tract does not contain any β-sheet content in its most populous clusters, instead favoring very helical structures. Contrastingly, the elongated polyQ tracts (especially Q46) feature structures that are rich in β-sheet content. Such β-sheet rich conformations are typical of the motifs sometimes associated with oligomer and fibril formation, suggesting that these configurations might contribute to the increased aggregation of the mtHtt.
Our findings also demonstrate how the elongation of the polyQ segment alters interactions between the N17 and C38 domains. While the number of hydrogen bonds within the polyQ region itself increases with Q-length (in fact, at a much faster rate than expected for globular proteins), the number of hydrogen bonds between N17 and C38 decreases with increasing Q-length. The reduction in N17-C38 hydrogen bonds is correlated with an increase of β-sheet structures in the polyQ region. This observation confirms previously reported experimental FRET results and a proposed model by Truant and co-workers 25.
As the polyQ-length increases, the overall conformational flexibility of the protein is diminished, and the exon-1 mutants adopt increasingly compact structures. Along with the appearance of β-sheet rich conformations, increased compactness and reduced flexibility appear to represent key differences between the wild type and pathogenic Htt variants. It is important to emphasize, however, that even though extending the Q-length leads to reduced conformational flexibility, the exon-1 protein remains disordered and does not adopt a native folded state.
Ultimately, this loss of flexibility stimulates a surprising finding: the radial scaling behavior of polyQ domains is very different from that of standard globular proteins, such that the polyQ region adopts super-compact structures at longer Q-lengths. The Rg scaling exponent for polyQ was estimated to be 0.22, a considerably smaller value than the exponent typical of globular proteins (0.33). Super-compactness is an astonishing characteristic for an IDP to possess: based on conventional wisdom, one expects IDPs to abide by a scaling exponent of approximately ~0.5. This unanticipated scaling behavior likely results from the explosion in internal hydrogen bonding seen with increasing Q-length. The resultant reduction in flexibility and super-compactness should impact the way individual mtHtts interact with other proteins, possibly inducing a loss of function and thereby accessing an orthogonal mechanism of toxicity. One possibility is that the enhanced compactness and resulting mechanical stability may alter the proteolysis mechanism of mtHtts (e.g., exceeding the capability of proteasome to digest them), thus suppressing the degradation of these mutant proteins and leading to their accumulation in vivo43.
Although our findings suggest these possible origins for the toxicity of mtHtts, further work is needed to discover just how these changes in structure affect protein-protein interactions and what roles the N17 and C38 regions play in mediating those downstream effects. Additionally, comparison of Htt with other disease-causing polyglutamine proteins may provide useful insights into the role of the flanking domains play in modulating the pathogenic Q-length. While a long road lies ahead with respect to comprehending the molecular underpinnings of Huntington’s disease, the Q-length-dependent perturbations to Huntingtin exon-1 monomer structure identified here represent encouraging early steps toward that direction.
Materials and Methods
Molecular dynamics simulations were conducted on Htt exon-1 proteins (MATLEKLMKAFESLKFSQnP11QLPQP3QAQPLLPQPQP10) featuring five different Q-lengths: Q22, Q36, Q40, Q46, and Q56. All simulated exon-1 proteins included both the N-terminal (N17) and C-terminal (C38) domains flanking the central polyQ tract. Due to the intrinsic disorder of the Htt exon-1, we used temperature replica exchange molecular dynamics (T-REMD) 44 to enhance the sampling of the conformational free energy surface. Furthermore, to remove biases from any known structural motif, we prepared initial structures for T-REMD using the following procedure45–47: polypeptide chains with fully extended conformations were constructed, collapsed in vacuo through minimization and equilibration at 700K, solvated with a 2.0 nm buffer of TIP3P water, and neutralized with NaCl to a concentration of 100 mM. To obtain fully randomized conformations, the system was heated from 298K to 600K and maintained at that high temperature for 400 ns. Over the final 128 ns of this trajectory, 128 initial conformations for T-REMD seeding were selected at a rate of one per 1 ns.
After a 1 ns equilibration, we carried out standard T-REMD simulations with 128 replicas at constant volume for c.a. 500 ns per replica, using temperature ranges from 298K to 600K. The replicas were allowed to exchange temperatures every 1000 time steps and the resulting acceptance ratio was in the range of 0.25–0.3. A time step of 4 fs with virtual sites was used. All simulations were performed with GROMACS 5.0.5 32 using the OPLS-AA/L force field 48.
The total aggregate simulation time for our dataset reached 320 μs. We recorded atomic coordinates at every 0.1 ns and analyzed the trajectories at temperatures ranging from 298K to 315K, after discarding the first 100 ns of each run. Secondary structure analyses were carried out using STRIDE 49 and the PLUMED software package 50. Convergence checks (Fig. S4-S5) and further analysis can be found in the Supplementary Information. Molecular structures were visualized and rendered using VMD 51 (http://www.ks.uiuc.edu/Research/vmd/)
Supplementary Material
Acknowledgments
We would like to thank Jeffrey K Weber, Tien Huynh, Thomas F. Vogt, Amrita Mohan, Seung-gu Kang, Joseph Morrone and Alicia Preiss for their various help with this work. This work was partially funded by CHDI Foundation Inc., a charitable foundation that funds research into Huntington’s disease. ML acknowledges the support of NIH (GM115749). RZ acknowledges the support of the IBM Blue Gene Science Program (W1258591, W1464125, W1464164).
References
- 1.Group, T. H. s. D. C. R. Cell. 1993;72:971. [Google Scholar]
- 2.Wexler NS. Proceedings of the National Academy of Sciences. 2004;101:3498. [Google Scholar]
- 3.Gray M, Shirasaki DI, Cepeda C, Andre VM, Wilburn B, Lu XH, Tao J, Yamazaki I, Li SH, Sun YE, Li XJ, Levine MS, Yang XW. The Journal of Neuroscience. 2008;28:6182. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saudou F, Finkbeiner S, Devys D, Greenberg ME. Cell. 1998;95:55. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 5.Perutz MF, Johnson T, Suzuki M, Finch JT. Proceedings of the National Academy of Sciences. 1994;91:5355. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller J, Arrasate M, Brooks E, Libeu CP, Legleiter J, Hatters D, Curtis J, Cheung K, Krishnan P, Mitra S, Widjaja K, Shaby BA, Lotz GP, Newhouse Y, Mitchell EJ, Osmand A, Gray M, Thulasiramin V, Frederic, Segal M, Yang XW, Masliah E, Thompson LM, Muchowski PJ, Weisgraber KH, Finkbeiner S. Nature Chemical Biology. 2011;7:925. doi: 10.1038/nchembio.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim MW, Chelliah Y, Kim SW, Otwinowski Z, Bezprozvanny I. Structure. 2009;17:1205. doi: 10.1016/j.str.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim M. Prion. 2013;7:221. doi: 10.4161/pri.23807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michalek M, Salnikov ES, Bechinger B. Biophysical Journal. 2013;105:699. doi: 10.1016/j.bpj.2013.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thakur AK, Jayaraman M, Mishra R, Thakur M, Chellgren VM, Byeon IJL, Anjum DH, Kodali R, Creamer TP, Conway JF, Gronenborn AM, Wetzel R. Nature Structural & Molecular Biology. 2009;16:380. doi: 10.1038/nsmb.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahoo B, Singer D, Kodali R, Zuchner T, Wetzel R. Biochemistry. 2014;53:3897. doi: 10.1021/bi500300c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crick SL, Jayaraman M, Frieden C, Wetzel R, Pappu RV. Proceedings of the National Academy of Sciences. 2006;103:16764. doi: 10.1073/pnas.0608175103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miettinen MS, Knecht V, Monticelli L, Ignatova Z. The Journal of Physical Chemistry B. 2012;116:10259. doi: 10.1021/jp305065c. [DOI] [PubMed] [Google Scholar]
- 14.Heck BS, Doll F, Hauser K. Biophysical Chemistry. 2014;185:47. doi: 10.1016/j.bpc.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 15.Hoop CL, Lin HK, Kar K, Magyarfalvi G, Lamley JM, Boatz JC, Mandal A, Lewandowski JR, Wetzel R, van der Wel PCA. Proceedings of the National Academy of Sciences. 2016;113:1546. doi: 10.1073/pnas.1521933113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S, Ferrone FA, Wetzel R. Proceedings of the National Academy of Sciences. 2002;99:11884. doi: 10.1073/pnas.182276099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fodale V, Kegulian NC, Verani M, Cariulo C, Azzollini L, Petricca L, Daldin M, Boggio R, Padova A, Kuhn R, Pacifici R, Macdonald D, Schoenfeld RC, Park H, Isas JM, Langen R, Weiss A, Caricasole A. PLOS ONE. 2014;9:e112262. doi: 10.1371/journal.pone.0112262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das P, King JA, Zhou R. Proceedings of the National Academy of Sciences. 2011;108:10514. doi: 10.1073/pnas.1019152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou R. Proceedings of the National Academy of Sciences. 2003;100:13280. doi: 10.1073/pnas.2233312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia Z, Yang Z, Huynh T, King JA, Zhou R. Scientific reports. 2013;3:1560. doi: 10.1038/srep01560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelley NW, Huang X, Tam S, Spiess C, Frydman J, Pande VS. Journal of Molecular Biology. 2009;388:919. doi: 10.1016/j.jmb.2009.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Voth GA. The Journal of Physical Chemistry B. 2010;114:8735. doi: 10.1021/jp1007768. [DOI] [PubMed] [Google Scholar]
- 23.Dzugosz M, Trylska J. Journal of Physical Chemistry B. 2011;115:11597. doi: 10.1021/jp206373g. [DOI] [PubMed] [Google Scholar]
- 24.Lakhani VV, Ding F, Dokholyan NV. PLoS Comput Biol. 2010;6:e1000772. doi: 10.1371/journal.pcbi.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caron NS, Desmond CR, Xia J, Truant R. Proceedings of the National Academy of Sciences. 2013;110:14610. doi: 10.1073/pnas.1301342110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagai Y, Inui T, Popiel HA, Fujikake N, Hasegawa K, Urade Y, Goto Y, Naiki H, Toda T. Nat Struct Mol Biol. 2007;14:332. doi: 10.1038/nsmb1215. [DOI] [PubMed] [Google Scholar]
- 27.Kar K, Hoop CL, Drombosky KW, Baker MA, Kodali R, Arduini I, van der Wel PCA, Horne WS, Wetzel R. Journal of molecular biology. 2013;425:1183. doi: 10.1016/j.jmb.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pietrucci F, Laio A. Journal of Chemical Theory and Computation. 2009;5:2197. doi: 10.1021/ct900202f. [DOI] [PubMed] [Google Scholar]
- 29.Jayaraman M, Kodali R, Sahoo B, Thakur AK, Mayasundari A, Mishra R, Peterson CB, Wetzel R. Journal of Molecular Biology. 2012;415:881. doi: 10.1016/j.jmb.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stapley BJ, Creamer TP. Protein Sci. 1999;8:587. doi: 10.1110/ps.8.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darnell GD, Derryberry J, Kurutz JW, Meredith SC. Biophysical Journal. 2009;97:2295. doi: 10.1016/j.bpj.2009.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berendsen HJC, van der Spoel D, van Drunen R. Computer Physics Communications. 1995;91:43. [Google Scholar]
- 33.Daura X, Gademann K, Jaun B, Seebach D, van Gunsteren WF, Mark AE. Angewandte Chemie International Edition. 1999;38:236. [Google Scholar]
- 34.Baias M, Smith PES, Shen K, Joachimiak LA, Żerko S, Koźmiński W, Frydman J, Frydman L. Journal of the American Chemical Society. 2017 doi: 10.1021/jacs.6b10893. [DOI] [PubMed] [Google Scholar]
- 35.Jayaraman M, Mishra R, Kodali R, Thakur AK, Koharudin LMI, Gronenborn AM, Wetzel R. Biochemistry. 2012;51:2706. doi: 10.1021/bi3000929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kokona B, Rosenthal ZP, Fairman R. Biochemistry. 2014;53:6738. doi: 10.1021/bi500449a. [DOI] [PubMed] [Google Scholar]
- 37.Kokona B, Johnson KA, Fairman R. Biochemistry. 2014;53:6747. doi: 10.1021/bi501066q. [DOI] [PubMed] [Google Scholar]
- 38.Liu P, Huang X, Zhou R, Berne BJ. Nature. 2005;437:159. doi: 10.1038/nature03926. [DOI] [PubMed] [Google Scholar]
- 39.Zhou R, Huang X, Margulis CJ, Berne BJ. Science. 2004;305:1605. doi: 10.1126/science.1101176. [DOI] [PubMed] [Google Scholar]
- 40.Dima RI, Thirumalai D. The Journal of Physical Chemistry B. 2004;108:6564. [Google Scholar]
- 41.Rawat N, Biswas P. J Chem Phys. 2009;131:165104. doi: 10.1063/1.3251769. [DOI] [PubMed] [Google Scholar]
- 42.Hofmann H, Soranno A, Borgia A, Gast K, Nettels D, Schuler B. Proceedings of the National Academy of Sciences. 2012;109:16155. doi: 10.1073/pnas.1207719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dougan L, Li J, Badilla CL, Berne BJ, Fernandez JM. Proceedings of the National Academy of Sciences. 2009;106:12605. doi: 10.1073/pnas.0900678106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugita Y, Okamoto Y. Chemical Physics Letters. 1999;314:141. [Google Scholar]
- 45.An D, Su J, Weber JK, Gao X, Zhou R, Li J. J Am Chem Soc. 2015;137:8412. doi: 10.1021/jacs.5b00888. [DOI] [PubMed] [Google Scholar]
- 46.Kang SG, Huynh T, Xia Z, Zhang Y, Fang H, Wei G, Zhou R. J Am Chem Soc. 2013;135:3150. doi: 10.1021/ja310989u. [DOI] [PubMed] [Google Scholar]
- 47.Xia Z, Das P, Shakhnovich EI, Zhou R. J Am Chem Soc. 2012;134:18266. doi: 10.1021/ja3031505. [DOI] [PubMed] [Google Scholar]
- 48.Kaminski GA, Friesner aRA, Tirado-Rives J, Jorgensen WL. The Journal of Physical Chemistry B. 2001;105:6474. [Google Scholar]
- 49.Frishman D, Argos P. Proteins: Structure, Function, and Bioinformatics. 1995;23:566. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 50.Bonomi M, Branduardi D, Bussi G, Camilloni C, Provasi D, Raiteri P, Donadio D, Marinelli F, Pietrucci F, Broglia RA, Parrinello M. Computer Physics Communications. 2009;180:1961. [Google Scholar]
- 51.Humphrey W, Dalke A, Schulten K. Journal of molecular graphics. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.