Abstract
Herbicidins are adenosine-based nucleoside antibiotics with an unusual tricyclic undecose core decorated with a (5-hydroxy)tiglyl moiety. Feeding studies are herein reported demonstrating that the tricyclic core is derived from d-glucose and d-ribose, whereas the tiglyl moiety is derived from an intermediate of l-isoleucine catabolism. Identification of the gene cluster for herbicidin A biosynthesis in Streptomyces sp. L-9-10 as well as its verification by heterologous expression in a non-producing host are described, and the results of in vitro characterization of a carboxyl methyltransferase encoded in the cluster, Her8, are presented. Based on these observations, a biosynthetic pathway is proposed for herbicidins.
Insert Table of Contents artwork here
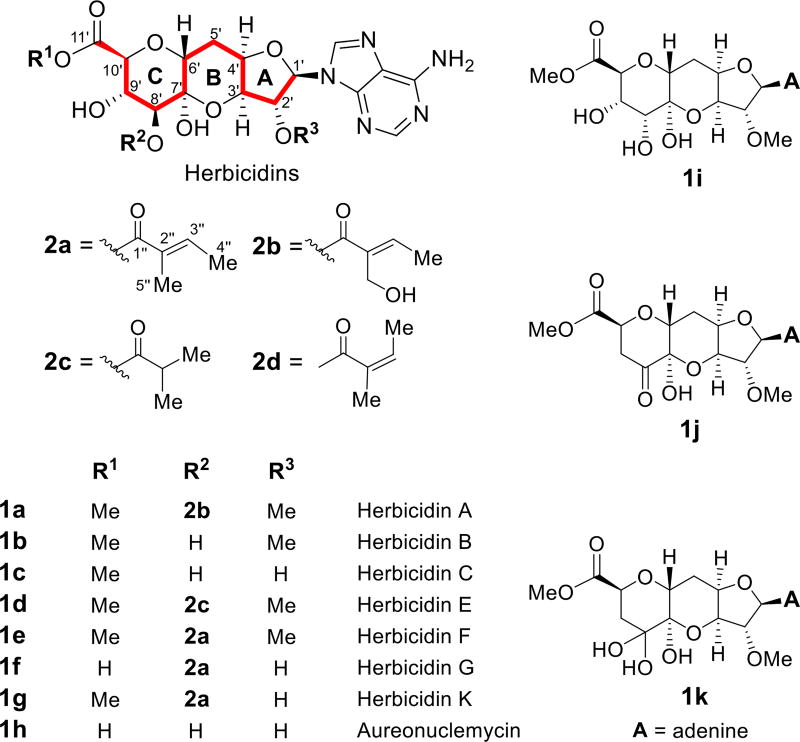
Herbicidins (1a through 1k, Figure 1) are adenosine-derived nucleoside antibiotics that have been isolated from Streptomyces saganonensis,1 Streptomyces sp. L-9-10,2 and Streptomyces scopuliridis RB72.3 Herbicidin variants differ with respect to methylation at the C2′-OH and C11′-COOH functionalities as well as the short, branched-chain fatty acid attached at C8′ via an ester linkage. The most extensively-decorated herbicidin reported to date is herbicidin A (1a), which features a 5-hydroxytiglyl group (2b) at C8′. Herbicidins show selective herbicidal activity toward dicotyledonous plants and inhibit the germination of Chinese cabbage and rice seeds.1b Furthermore, they can protect rice plants from leaf blight1b and also exhibit antialgal1c and antifungal1d activities. However, their biological mode of action4 and biosynthetic origins remain unclear.
Figure 1.
Structures of herbicidins.
The characteristic tricyclic core of herbicidins is composed of a pyran (ring-C) connected to a furan (ring-A) via a methylene bridge and a hemiketal linkage to form a third six-membered heterocycle (ring-B). This tricyclic furano-pyrano-pyran structure locks all of the ring-C substituents into an axial conformation (similar to 1C4),5 and its reconstruction has drawn the interest of synthetic chemists.6 While the ether and hemiketal linkages in the herbicidin core structure are suggestive of a ladderether polyketide,7 an alternative hypothesis is that the furano-pyrano-pyran heterocycle is constructed from carbohydrate precursors. For example, while a hemiketal linkage between pyran and furan subunits has been reported in several polyketide-derived natural products such as the ionophore monensin A,7 spliceostatin FR9014648 and phorboxazole A,9 a similar linkage has also been reported in the carbohydrate-based metabolite spectinomycin.10 The potential for a carbohydrate-based origin of the herbicidins is of particular interest, because undecosyl skeletons (highlighted in Figure 1) are rare among carbohydrate-based natural products,11,12 with the tunicamycins13 and hikizimycin14 being the only other known examples. Moreover, a carbohydrate-based origin would raise additional questions as to how the methylene-bridged linkage between the A and C rings might be introduced.
The tiglyl and 5-hydroxytiglyl (2a and 2b) esters present in some herbicidin variants are also atypical substituents among bacterial natural products. Tiglyl (2a) along with the isomeric angelyl (2d) groups are more commonly found in plant metabolites such as 3β-tigloyloxytropane, where they are derived from the corresponding CoA esters produced via the catabolism of l-isoleucine.15 However, the only known angelyl ester substituent in bacterial metabolites is found in SF2575 and heliosupine, where it is hypothesized to be of polyketide origin.16 Likewise, there has been little work17 done to determine whether 2a and 2b originate from the catabolism of l-isoleucine or arise though polyketide-medicated biosynthetic pathways.
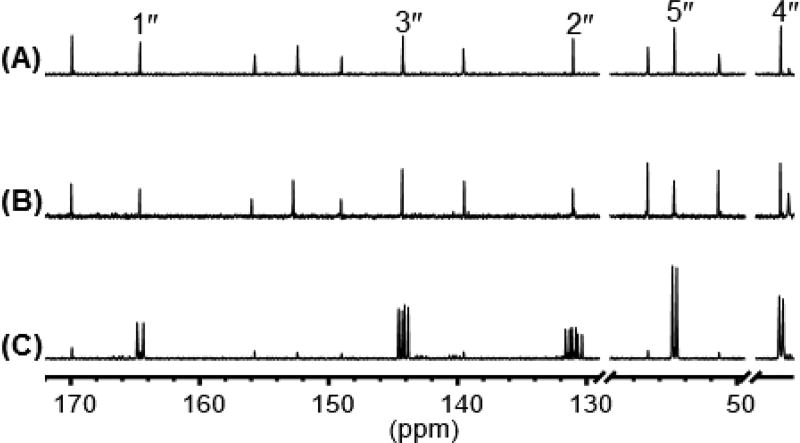
To investigate the biosynthesis of herbicidins, conditions were first optimized for the production and isolation of herbicidin A (1a). Ha and coworkers previously identified growth conditions that optimized the production of 1a from Streptomyces scopuliridis.3b Building on this, it was found that reducing the culture time and simplifying the chromatographic steps further improved the isolable yield of 1a from cultures of Streptomyces sp. L-9-10 to 50–100 mg/L compared to 0.135 mg/L as previously reported2 (see Section S2). Using these conditions, cultures of Streptomyces sp. L-9-10 were fed with [U-13C,15N]isoleucine, and herbicidin A was isolated prior to 13C NMR analysis. As shown in Figure 2, isoleucine is incorporated into 2b as an intact entity. In contrast, no significant 13C enrichment of 2b was observed when the same experiment was performed with sodium [2-13C]acetate. These results indicated that biosynthesis of the 2b moieties in herbicidin A follows a path similar to what is observed in plant metabolites. Likewise, analysis of 13C NMR of herbicidin A isolated from the Streptomyces sp. L-9-10 culture fed with [5-13C]ribose suggested a ribosyl origin of the C5′ carbon of ring-B in herbicidin A. Additional feeding experiments with uniquely-labeled glucose, [U-13C]ribose, and [Me-13C]methionine also revealed that ring-A in herbicidin A is derived from d-ribose, the carbons of ring-C are from d-glucose, and the two methyl groups are from l-methionine (see Section S3).
Figure 2.
Selected 13C NMR spectra of herbicidin A isolated from cultures of Streptomyces sp. L-9-10 fed with (A) no additive, (B) sodium [2-13C]acetate, and (C) [U-13C, 15N]isoleucine. Carbon signals from 2b are labeled with their numberings.
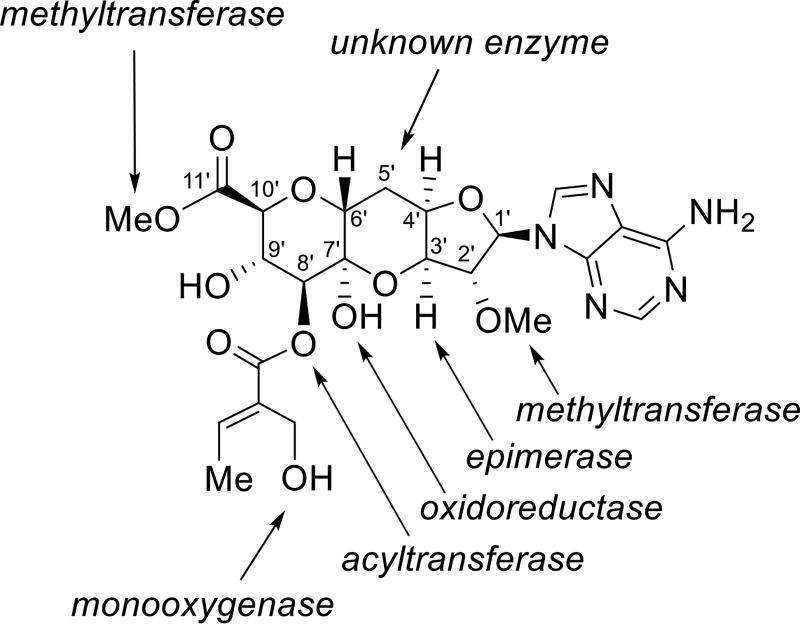
Based on the results from the feeding studies, glucuronic acid and adenosine derivatives were predicted to be building blocks for the herbicidin core. If correct, then the herbicidin gene cluster was expected to encode at least one epimerase for C3 inversion of the adenosine derivative (C3′ in herbicidins), an oxidoreductase for C2 oxidation of the glucuronic acid derivative (C7′ in herbicidins), one or more enzymes for concatenation of the two units, an acyltransferase for installation of the tiglyl moiety, a monooxygenase for the oxidation of the tiglyl moiety, and two methyltransferases as shown in Figure 3. However, when the genome of Streptomyces sp. L-9-10 was sequenced (see Section S1.4), no cluster that closely matched these predictions was identified using the antiSMASH server.18,19 An attempt to locate genes encoding potential O-tiglyl transferases15b in the genome was similarly unsuccessful.
Figure 3.
Enzyme activities initially predicted to be associated with the herbicidin A biosynthetic pathway given d-glucose and d-ribose building blocks for the herbicidin core structure.
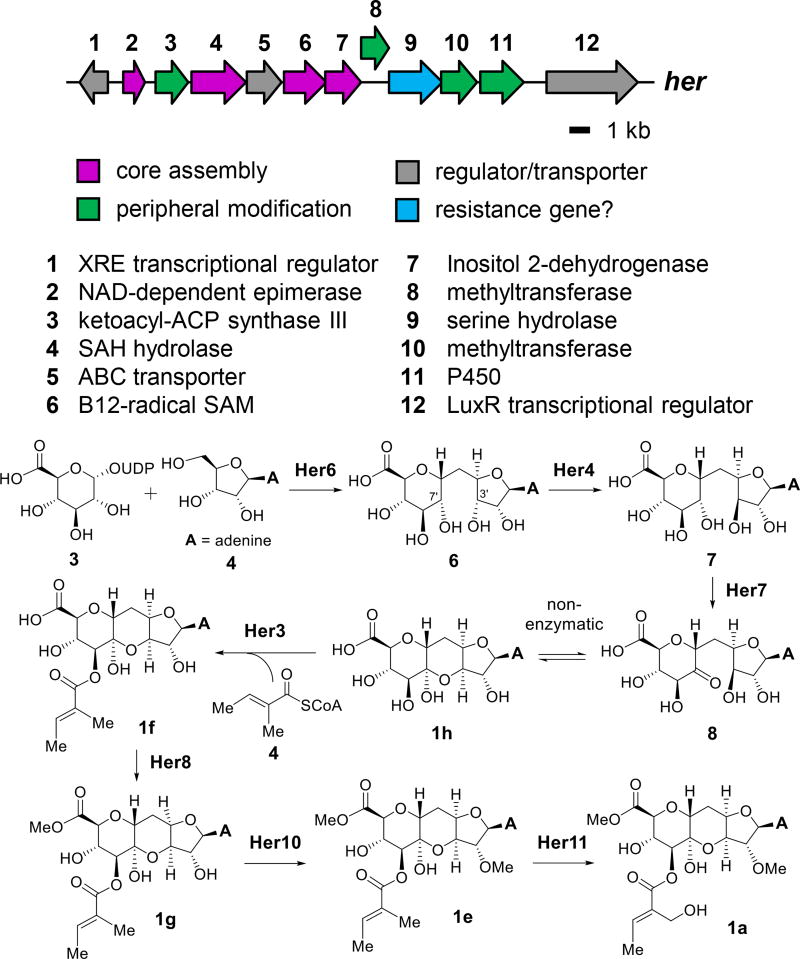
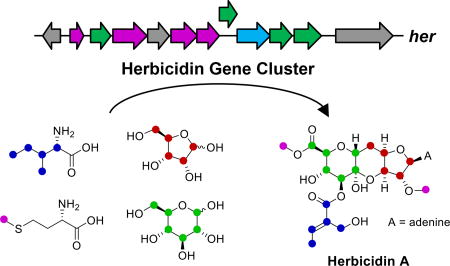
A partial match for the predicted gene cluster was identified, however, when a single locus across the entire genome was found to include two genes annotated as encoding methyltransferases. As these methyltransferases may be responsible for catalyzing methylation of the C2′-OH and C11′-COOH, the surrounding genomic context was analyzed to reveal approximately twelve clustered genes (denoted her) that loosely matched the original predictions (see Figure 5). Homologous clusters were also found in two other species of Streptomyces, including the herbicidin-producing strain S. scopuliridis RB72 (see Section S4), suggesting that the her cluster may indeed be responsible for herbicidin biosynthesis. Interestingly, O-methyltransferase-encoding genes have been used to locate other natural product biosynthetic gene clusters. For example, the identification of three genes in close proximity to each other encoding predicted O-methyltransferases helped to locate the griseofulvin gene cluster.20 In another case, degenerate primers based on O-methyltransferases led to the cloning of the caprazamycin gene cluster.21
Figure 5.
Annotated her gene cluster responsible for the biosynthesis of herbicidin A. A biosynthetic pathway consistent with the gene annotations is proposed beginning with UDP-glucuronic acid and adenosine; however, other immediate precursors are possible.
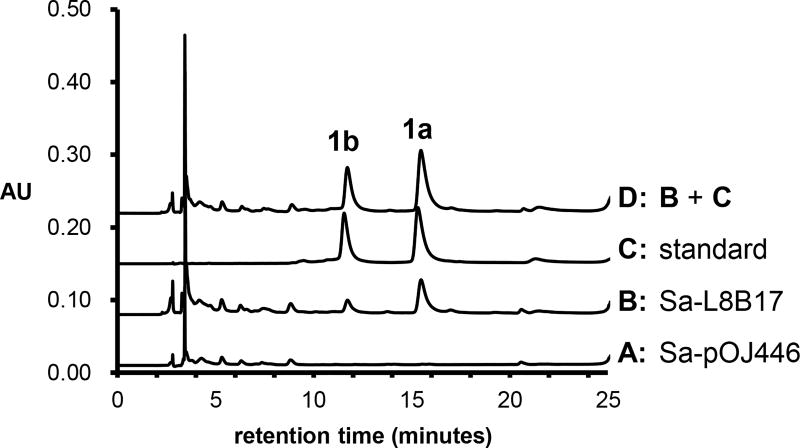
To determine the biological function of the her cluster, the cosmid L8B17, which included the her cluster, was isolated from a pOJ446-based cosmid library22 of Streptomyces sp. L-9-10 (see Section S5). Intergeneric conjugal transfer of cosmid L8B17 from Escherichia coli S17-1 to the non-producing heterologous host Streptomyces albus J1074 was carried out to generate strain Sa-L8B17. The same conjugal transfer was also performed in parallel using the empty pOJ446 vector to yield the strain Sa-pOJ446 as a negative control. The positive exoconjugants were cultured under producing conditions, and the production of herbicidins A (1a) and B (1b) was only observed in the culture of Sa-L8B17 as shown in Figure 4. The identities of the observed products were established by co-elution with herbicidin standards on HPLC (Figure 4, trace D) and electrospray ionization-mass spectrometry (ESI-MS) of the collected HPLC peaks (see Section S5). These results imply that the her cluster encodes the enzymes responsible for the biosynthesis of herbicidins.
Figure 4.
HPLC traces of (A) 10-day extract of Sa-pOJ446, (B) 10-day extract of Sa-L8B17, (C) herbicidin standard, and (D) co-injection of the samples in traces B and C.
A possible biosynthetic pathway for herbicidins based on sequence analysis of the genes in the her cluster is shown in Figure 5. The tricyclic core is proposed to form by the coupling of a d-ribose derivative such as adenosine (4) and a d-glucose derivative such as UDP-glucuronic acid (3) in a reaction catalyzed by Her6, which is annotated as a B12-dependent radical SAM enzyme. This hypothesis is similar to the proposed reaction for the putative radical-SAM enzyme TunB, which may catalyze the addition of a 5′-uridyl radical to an exo-glycal during biosynthesis of tunicamycins.23 Subsequent C3′-epimerization may be mediated by the SAH hydrolase homolog Her4 (see Section S6). Oxidation of the resulting undecose (7 → 8) may then be catalyzed by the putative dehydrogenase Her7 predisposing the resulting intermediate 8 to non-enzyme-catalyzed hemiketal formation and generation of the tricyclic herbicidin core.
Decoration of the core is proposed to be catalyzed by the ketosynthase homolog Her3 (see Section S6), the two putative methyltransferases Her8 and Her10, and the annotated P450 hydroxylase Her11. Herbicidin A (1a) is proposed to be hydrolyzed to herbicidin B (1b) by the serine hydrolase homolog Her9, because 1b accumulates later than 1a in cultures of Streptomyces sp. L-9-10 (see Section S2). Alternatively, substrate promiscuity of the decorating enzymes may explain the production of 1b and other herbicidins. Consequently, the unassigned gene product Her2 may be responsible for generating 1i through 1k that are only produced by Streptomyces sp. L-9-10, since no homolog of Her2 was identified in the corresponding clusters of other producing strains (see Section S4). Finally, the absence of polyketide modules in the her cluster is consistent with the feeding results indicating that the tiglyl moiety is derived from the catabolism of L-isoleucine.
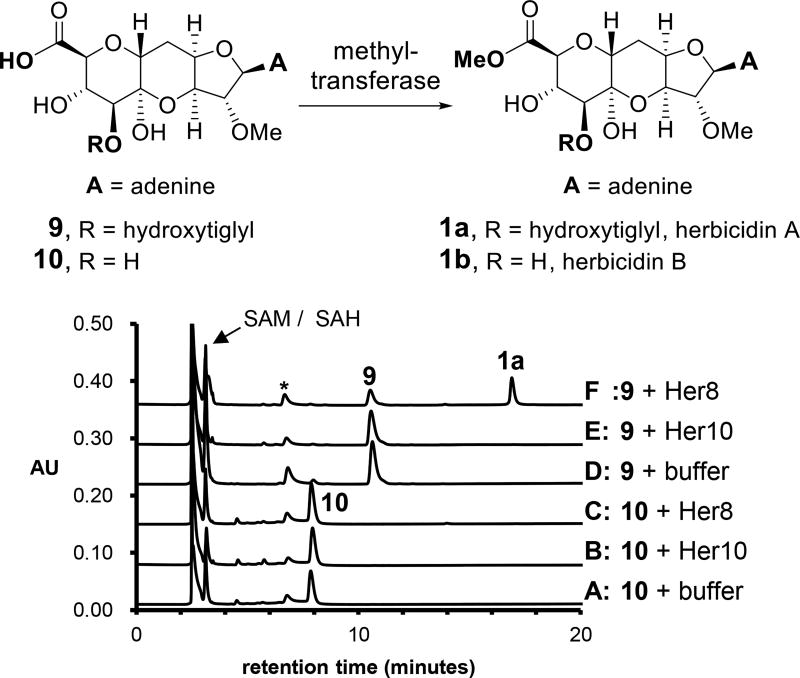
It was hypothesized that one of the two methyltransferases, Her8 or Her10, is responsible for catalyzing the methylation of the C11′-carboxylate of the herbicidins. To test this hypothesis, the putative substrates 9 and 10 were prepared by hydrolyzing herbicidin A using LiOH. Her8 and Her10 were overexpressed in E. coli and purified as N-His6-tagged and maltose-binding protein (MBP)-fusion constructs, respectively. The MBP fused to Her10 was subsequently cleaved using TEV protease (see Section S7.2). A solution of 100 µM of 9 or 10 was incubated with 1 µM Her8 or Her10 in the presence of 0.5 mM S-adenosyl-l-methionine (SAM) in potassium phosphate buffer (pH 6.5, 50 mM) at room temperature for 24 h. Among all the combinations, only Her8 was able to convert 9 to herbicidin A (1a) as shown in Figure 6, which was confirmed by co-injection with a standard and LC-ESI-MS analysis (see Section S7.3). All assays excluding Her8 showed no consumption of 9 or 10. These observations suggested that Her8 is the C11′-O-methyltransferase. Furthermore, 10 was not methylated in the presence of Her8 and SAM suggesting that esterification of C8′ occurs prior to methylation of C11′. The observation of equilibrium between the hemiketal and free carbonyl forms of 9 and 10. (see Section S7.1) is consistent with the proposed nonenzymatic cyclization.
Figure 6.
Assays for the methyltransferase activity of Her8 and Her10 showing HPLC chromatograms following incubation of substrate with the gene product and SAM as indicated next to each trace. The peak labeled with an asterisk is 5′-deoxy-5′-methylthioadenosine from the degradation of SAM.24
In summary, feeding results demonstrate that the precursors for herbicidin biosynthesis are d-glucose, d-ribose, l-isoleucine, and l-methionine. This establishes a carbohydrate origin for the herbicidin core and an amino acid origin for the tiglyl and 5-hydroxytiglyl functionalities. The biosynthetic gene cluster was identified in the draft genome and subsequently verified by production of herbicidins in a non-producing host expressing this cluster. This was further substantiated by the successful reconstitution of Her8 activity in vitro. To the best of our knowledge, this is the first example of a bacterial pathway that may borrow tiglyl-CoA (5) from the catabolism of l-isoleucine to modify a secondary metabolite. Only a small number of undecoses have been discovered to date, and our understanding of their biosynthesis is still in its infancy. Nevertheless, our work not only represents a major advancement towards future studies characterizing herbicidin biosynthesis but also lays the foundation for identification of related nucleoside natural product gene clusters in hitherto unexplored microbial genomes.
Supplementary Material
Acknowledgments
We thank Prof. Julian Davies at The University of British Columbia for generously providing Streptomyces sp. L-9-10 used in this study. This work was supported by grants from the National Institutes of Health (GM035906) and the Welch Foundation (F-1511).
Footnotes
ASSOCIATED CONTENT
Details regarding the experimental procedures, molecular cloning, enzyme assays, and spectroscopic characterization of new compounds. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
References
- 1.(a) Haneishi T, Terahara A, Kayamori H, Yabe J, Arai M. J. Antibiot. (Tokyo) 1976;29:870–875. doi: 10.7164/antibiotics.29.870. [DOI] [PubMed] [Google Scholar]; (b) Arai M, Haneishi T, Kitahara N, Dnokita R, Kawakubo K, Kondo Y. J. Antibiot. (Tokyo) 1976;29:863–869. doi: 10.7164/antibiotics.29.863. [DOI] [PubMed] [Google Scholar]; (c) Takiguchi Y, Yoshikawa H, Terahara A, Torikata A, Terao M. J. Antibiot. (Tokyo) 1979;32:857–861. doi: 10.7164/antibiotics.32.857. [DOI] [PubMed] [Google Scholar]; (d) Takiguchi Y, Yoshikawa H, Terahara A, Torikata A, Terao M. J. Antibiot. (Tokyo) 1979;32:862–867. doi: 10.7164/antibiotics.32.862. [DOI] [PubMed] [Google Scholar]
- 2.Chai X, Youn UJ, Sun D, Dai J, Williams P, Kondratyuk TP, Borris RP, Davies J, Villanueva IG, Pezzuto JM, Chang LC. J. Nat. Prod. 2014;77:227–233. doi: 10.1021/np4006635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Choi CW, Choi J-S, Ko YK, Kim C-J, Kim YH, Oh JS, Ryu SY, Yon GH. Bull. Korean Chem. Soc. 2014;35:1215–1217. [Google Scholar]; (b) Ha S, Lee KJ, Lee SI, Gwak HJ, Lee J-H, Kim T-W, Choi H-J, Jang J-Y, Choi J-S, Kim C-J, Kim J-C, Kim HH, Park HW. J. Microbiol. Biotechnol. 2017;27:947–955. doi: 10.4014/jmb.1611.11005. [DOI] [PubMed] [Google Scholar]
- 4.Won OJ, Kim YT, Choi JS, Oh T-K, Shinoki Y, Park KWJ. Fac. Agr. Kyushu Univ. 2016;61:47–51. [Google Scholar]
- 5.Terahara A, Haneishi T, Arai M, Hata T, Kuwano H, Tamura C. J. Antibiot. (Tokyo) 1982;35:1711–1714. doi: 10.7164/antibiotics.35.1711. [DOI] [PubMed] [Google Scholar]
- 6.(a) Ichikawa S, Shuto S, Matsuda A. J. Am. Chem. Soc. 1999;121:10270–10280. [Google Scholar]; (b) Hager D, Mayer P, Paulitz C, Ti´ebes J, Trauner D. Angew. Chem. Int. Ed. 2012;51:6525–6528. doi: 10.1002/anie.201201826. [DOI] [PubMed] [Google Scholar]
- 7.Gallimore AR. Nat. Prod. Rep. 2009;26:266–280. doi: 10.1039/b807902c. [DOI] [PubMed] [Google Scholar]
- 8.Eustáquio AS, Janso JE, Ratnayake AS, O’Donnell CJ, Koehn FE. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E3376–E3385. doi: 10.1073/pnas.1408300111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Searle PA, Molinski TF. J. Am. Chem. Soc. 1995;117:8126–8131. [Google Scholar]
- 10.Wiley PF, Argoudelis AD, Hoeksema H. J. Am. Chem. Soc. 1963;85:2652–2659. [Google Scholar]
- 11.Lin C-I, McCarty RM, Liu H-w. Chem. Soc. Rev. 2013;42:4377–4407. doi: 10.1039/c2cs35438a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elshahawi SI, Shaaban KA, Kharel MK, Thorson JS. Chem. Soc. Rev. 2015;44:7591–7697. doi: 10.1039/c4cs00426d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Takatsuki A, Arima K, Tamura G. J. Antibiot. (Tokyo) 1971;24:215–223. doi: 10.7164/antibiotics.24.215. [DOI] [PubMed] [Google Scholar]; (b) Takatsuki A, Tamura G. J. Antibiot. (Tokyo) 1971;24:224–231. [PubMed] [Google Scholar]
- 14.Uchida K, Ichikawa T, Shimauchi Y, Ishikura T, Ozaki A. J. Antibiot. (Tokyo) 1971;24:259–262. doi: 10.7164/antibiotics.24.259. [DOI] [PubMed] [Google Scholar]
- 15.(a) Leete E. Planta Med. 1979;36:97–112. doi: 10.1055/s-0028-1097249. [DOI] [PubMed] [Google Scholar]; (b) Rabot S, Peerless AC, Robins RJ. Phytochemistry. 1995;39:315–322. [Google Scholar]
- 16.(a) Pickens LB, Kim W, Wang P, Zhou H, Watanabe K, Gomi S, Tang Y. J. Am. Chem. Soc. 2009;131:17677–17689. doi: 10.1021/ja907852c. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Inahashi Y, Shiraishi T, Palm K, Takahashi Y, Ōmura S, Kuzuyama T, Nakashima T. Chembiochem. 2016;17:1442–1447. doi: 10.1002/cbic.201600208. [DOI] [PubMed] [Google Scholar]
- 17.Yoshikawa H, Takiguchi Y, Terao M. J. Antibiot. (Tokyo) 1983;36:30–35. doi: 10.7164/antibiotics.36.30. [DOI] [PubMed] [Google Scholar]
- 18.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Mller R, Wohlleben W, Breitling R, Takano E, Medema MH. Nucleic Acids Res. 2015;43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The cluster was not identified by antiSMASH 3.0. a recent version of antiSMASH 4.0 did find the cluster discussed below as a putative fatty-acid type gene cluster using ClusterFinder algorithm.
- 20.Chooi Y-H, Cacho R, Tang Y. Chem. Biol. 2010;17:483–494. doi: 10.1016/j.chembiol.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaysser L, Lutsch L, Siebenberg S, Wemaker E, Kammerer B, Gust B. J. Biol. Chem. 2009;284:14987–14996. doi: 10.1074/jbc.M901258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bierman M, Logan R, O’Brien K, Seno ET, Rao RN, Schoner BE. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 23.Wyszynski FJ, Lee SS, Yabe T, Wang H, Gomez-Escribano JP, Bibb MJ, Lee SJ, Davies GJ, Davis BG. Nat. Chem. 2012;4:539–546. doi: 10.1038/nchem.1351. [DOI] [PubMed] [Google Scholar]
- 24.Iwig DF, Booker SJ. Biochemistry. 2004;43:13496–13509. doi: 10.1021/bi048693+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.